Open Access Article
Open Access ArticleIn vitro bioaccessibility of proteins and lipids of pH-shift processed Nannochloropsis oculata microalga
L R.
Cavonius
*,
E.
Albers
and
I.
Undeland
Chalmers University of Technology, Biology and Biological Engineering, Kemigården 4, Gothenburg, Sweden. E-mail: lillie@chalmers.se
First published on 15th March 2016
Abstract
The pH-shift process fractionates biomass into soluble proteins and insoluble fractions, followed by precipitation and recovery of the solubilized proteins. Nannochloropsis oculata in seawater was subjected to the pH-shift process, followed by digestion of various intermediates and product fractions of the process, using the Infogest in vitro digestion model (Minekus et al., 2014) with added gastric lipase. As measures for protein and lipid accessibility, degrees of protein hydrolysis and fatty acid liberation were assessed post-digestion and compared to the amounts of peptide bonds and total fatty acids present in the raw materials. Results showed that neither proteins nor lipids of intact Nannochloropsis cells were accessible to the mammalian digestive enzymes used in the digestion model. Cell disruption, and to a lesser extent, further pH-shift processing with protein solubilisation at pH 7 or pH 10, increased the accessibility of lipids. For proteins, differences amongst the pH-shift processed materials were non-significant, though pre-freezing the product prior to digestion increased the accessibility from 32% to 47%. For fatty acids, pH-shift process-products gave rise to 43% to 52% lipolysis, with higher lipolysis for products solubilised at pH 10 as opposed to pH 7. Our results indicate the importance of processing to produce an algal product that has beneficial nutritional properties when applied as food or feed.
Introduction
Microalgae have been proposed as food for humans: some species contain, amongst other valuable nutrients, n-3 polyunsaturated fatty acids (PUFAs) and have balanced amino acid profiles.1,2 Furthermore, marine microalgae can be cultivated off-shore in seawater, offering a sustainable source of nutrients. As the demand for n-3 PUFAs and proteins is expected to increase as the planet's increasing population grows more affluent, microalgae can contribute to fill the gap between supply and demand. As part of the development of microalgae as food and feed, we have previously applied the pH-shift process to the n-3 PUFA-producing marine microalgae Nannochloropsis oculata and proposed the product as a functional food or feed ingredient.3The pH-shift process solubilises proteins at extreme pH-values and subsequently precipitates them at the proteins’ iso-electric point after non-solubilised, undesirable components have been removed. On an industrial scale, the pH-shift process-principle is currently applied to certain animal by-products and plant material such as fish and soybeans, to recover a refined protein fraction. The refined protein can show an altered technical functionality, e.g. gelling properties, since proteins are partially denatured in the process and may take on non-native conformations when re-folding.4,5 In a first application of the pH-shift process to N. oculata in seawater, the nutritional composition was studied before and after processing: a modest reduction in the ash content was observed, while the concentration of proteins, lipids and carbohydrates remained stable or increased slightly.3 The study also included a comparison of protein solubilisation at either the algae suspension's native pH 7 or pH 10. Although the compositions of the products were similar, it cannot be excluded that the solubilisation pH impacted the degree of protein unfolding or refolding and thereby altered the technical functional properties of the product, as well as the digestibility. To the best of our knowledge, the impact of pH-shift processing on algal digestibility has not been studied earlier.
The digestibility or accessibility of a food matrix is a measure of how much of the food component is available for uptake. Generally, microagal in vitro digestion studies have focused on proteins,6–11 though lipid-soluble compounds have also been assessed.12 Using sundry different methods to assess the digestibility of proteins and lipid-soluble compounds, these studies’ conclusions can be summarised as follows: the protein digestibility of Scenedesmus obliquus was increased by cell disruption (bead-milling);6 the protein digestibility of Spirulina platensis was greater when the algae are fresh as opposed to dried (either freeze-dried or sun-dried);7Chlorella vulgaris algae subjected to mechanical disruption, drying, and ethanol-extraction were more susceptible to digestion by pancreatin than disrupted, dried, non-extracted algae;9 treatment of dried Galdieria sulphuraria with a carbohydrase cocktail increased the protein digestibility;10 accessibility of β-carotene, lycopene and α-tocopherol from Nannochloropsis oculata and Chaetoceros calcitrans increased when the lipophilic compounds were extracted into specific solvents.12
Nannochloropsis has been subjected to both in vitro digestion and a feeding trial.11,13 In the in vitro digestion, both whole Nannochloropsis granulata and lipid-extracted N. granulata were compared, with low protein digestibility (15–28%) reported for the whole algae, while digestibility improved somewhat for lipid-extracted algae.11 A feeding trial on mink with Nannochloropsis oceanica demonstrated that the apparent crude-protein and lipid digestibility decreased as increasing amounts of Nannochloropsis were added to the mink chow.13 The authors reached the conclusion that cell disruption would likely increase the protein and lipid digestibility of Nannochloropsis.13 A recent publication offers an explanation for the poor digestibility of whole Nannochloropsis cells: the cell wall of Nannochloropsis is composed primarily of cellulose, surrounded by an outer layer of algaenan, which could be expected to block enzymes from acting on the cell.14
In the various aforementioned digestibility studies, not only the analysed end-points are different, but the applied in vitro digestion protocols also differ. This makes results from different in vitro digestion studies difficult to compare. The Infogest consortium (encompassing over 250 scientists from 32 different countries, coordinated by the French National Institute for Agricultural Research) has been working towards harmonising digestion models, which has resulted in published guidelines (Minekus et al., 2014).15 The published guide, however, omits gastric lipases, as these were not commercially available at the time of publication, even though it is established that gastric lipases have a significant impact on lipolysis.15,16 Recently, rabbit gastric lipase has become purchasable. We believe that the protocol suggested by Minekus et al., 2014,15 but with a minor change to include gastric lipase according to Capolino et al., 2011,17 would provide data closer to an in vivo scenario, yet comparable to other in vitro studies.
Heat-treatment of food is common, either as part of the pre-consumer processing or immediately prior to consumption. Heat-treatment can change the nutritive value of a food, e.g. by protein denaturation or the Maillard reaction.18Nostoc commune has been subjected to mild cell disruption and heat-treatment with dry and wet heat at 100 and 120 °C, resulting in no statistically significant difference in protein digestibility compared to non-heated Nostoc commune.8 Since Nostoc commune is a cyanobacterium while Nannochloropsis oculata is a eukaryote, it is conceivable that the protein and lipid digestibility of Nannochloropsis is affected differently by cell disruption and heating.
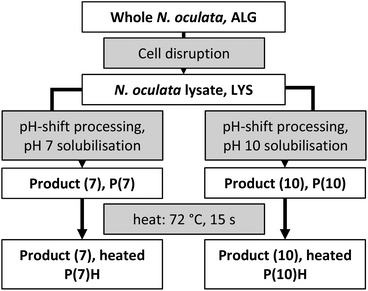
The aim of this paper was to investigate the accessibility of proteins and lipids of pH-shift processed N. oculata using enzyme levels of the standardised Infogest in vitro digestion model with added gastric lipase.15,17 To answer how the process per se affects accessibility and also what difference a heat-treatment makes, the following materials were included in the comparison: whole-algae-in-seawater (ALG), disrupted algae (LYS), and products of the pH-shift process with solubilisation at pH 7 or 10 with and without heat-treatment [P(7)H, P(10)H, P(7), P(10)]. Responses analysed were (i) the relative amount of broken peptide bonds (degree of protein hydrolysis, DH) and (ii) the relative amount of liberated free fatty acids (FFA).
Materials and methods
pH-Shift processing of Nannochloropsis
The product of Necton, microalgae Nannochloropsis oculata “Phytobloom ice” for aquaculture, was purchased in March 2012, delivered as a frozen wet paste (ca. 30% dry weight) and stored at −80 °C. Seawater was obtained from the marine research station of the Sven Lovén Centre for Marine Sciences at Tjärnö, Sweden, and was filtered through a 0.22 μm membrane and then autoclaved. The materials ALG, LYS, P(7), P(10), P(7)H and P(10)H were snap-frozen immediately after production, with one protein of P(7) remaining non-frozen: P(7)NF. Fig. 1 shows an overview of the algae-containing materials prepared for later in vitro digestion. The pH-shift process was carried out on ice, according to the process reported previously:3 one part of wet microalgal paste was thawed and mixed with four parts seawater. Algae in seawater were shaken until dissociation to mimic a microalgal culture which had been harvested by dewatering to ca. 10% dry weight. An aliquot of ALG was snap-frozen for later digestion and analysis by plunging the containing tube into a slurry of 95% ethanol and dry ice at ca. −78 °C; after freezing, tubes were stored at −80 °C until use. The remaining algae suspension (20 ml) was transferred to a 50 ml Falcon tube containing 10 ml of glass beads (212–300 μm, Sigma, acid-washed before use). Algae were disrupted by shaking the tube at 30 s−1 for 3 × 200 s in a Retsch MM400 (with adapters for 50 ml Falcon tubes); the tubes were cooled in ice between cycles. Near-complete disruption was confirmed by phase-contrast microscopy (Axiostar plus, Carl Zeiss, Germany at 400× magnification with an A-plan 40×/0.65 objective, ∞/0.17, and appropriate phase plate). Beads were removed by passing the suspension over a 100 μm mesh and LYS was collected on ice. An aliquot of LYS was snap-frozen for later digestion and analysis. The pH was measured with a Radiometer analytical PHM210 pH meter with a Hamilton double pore electrode (Christian Berner, Sweden). The remaining LYS was split into two streams, one in which the solubilisation pH was the native pH of the lysate (pH 7); in the other stream, the pH was adjusted to 10 with 1.0 M NaOH (prepared from NaOH pellets, Scharlau, purity ≥ 98.5%, and ultrapure water). Suspensions were centrifuged (4 °C, 4000g, 10 min; Heraeus multifuge 1 S-R with a swinging-bucket rotor) and the supernatants containing the majority of the proteins and lipids,3 were recovered. The supernatants were precipitated by adjusting the pH to 3 with 1.0 M HCl (prepared from 37% HCl, Scharlau, ACS). The suspensions were centrifuged as above, and the pellets were neutralized to pH 7 with NaOH, yielding a product of both the pH 7-process [P(7)] and pH 10-process [P(10)]. Aliquots of P(7) and P(10) were snap-frozen for later digestion and analysis. Some P(7) was kept liquid [P(7)NF] and was digested on the same day of the pH-shift process to elucidate the effect of snap-freezing. The remaining product was filled into 15 ml test tubes (round-bottom, polypropylene, TPP, Switzerland) and placed into the snug-fitting bore of a heating block (Grant QBH2, Cambridge, United Kingdom) at 95 °C for 360 s, by which time the core temperature was at least 72 °C for not less than 15 s, conditions typical of a high-temperature short-time pasteurization.19 Aliquots of both heat-treated products [P(7)H and P(10)H] were snap-frozen for later digestion and analysis.In vitro digestion
The digestion followed the protocol recommended by Minekus et al., 201415 with a modification to include gastric lipase as recommended by Capolino et al., 2011.17 The modification involved that the gastric step (120 min at pH 3 with pepsin, in the original protocol) was altered to a first step at pH 5.5 with rabbit gastric lipase for 30 min, followed by pH 3 with pepsin for 90 min; a more detailed description of raw materials, digestion enzymes, digestion fluids and bile acid follows below.Enzyme activities were measured according to Minekus et al., 2014:15 α-amylase from porcine pancreas (Sigma, type VI-B) was 13 U mg−1, pepsin from porcine stomach mucosa (Sigma) was 900 U mg−1, and pancreatin from porcine pancreas (Sigma, 4 × USP) was 3.6 U mg−1 (based on lipase activity). Rabbit gastric lipase was purchased from Germe, Marseille, France and according to its specification was 70 U mg−1. The total bile acids of porcine bile extract (Sigma) were determined according to Minekus et al., 201415 to be 1.4 μmol mg−1. The composition of simulated salivary fluid, simulated gastric fluid and simulated intestinal fluid is described by Minekus et al. (2014).15 The Ca2+-content of ALG was determined to be 9 mmol l−1 by high-pressure ion chromatography as reported in our previous work on pH-shift processing, using the same method as that for Na+-quantification.3 Since the Ca2+concentration was well in excess of the concentration recommended by Minekus et al., 2014,15 no Ca2+ was added in the digestion.
An overview of the digestion is given in Fig. 2. For samples containing Nannochloropsis, 2.5 ± 0.5 g of the material was weighed in 50 ml Falcon tubes (tapered bottom, polypropylene, TPP, Switzerland); for seawater blank (SW), 2.5 ml seawater was used and for the DH-control casein (CAS), 75 mg casein [from bovine milk, purchased from Sigma, corresponding to the amount of protein present in 2.5 g P(7)], was mixed with 2.43 ml seawater. For the oral step, 1.75 ml simulated salivary fluid at 37 °C was added and the tube was vortexed briefly to homogenise. Next, amylase solution (1500 U ml−1, in simulated salivary fluid, 0.25 ml at 37 °C) and ultrapure water (0.5 ml at 37 °C) were added and the tube was vortexed to homogenise the content. Thereafter, the tube was shaken in an upright position, at 200 rpm at 37 °C for 2 min (water bath SW22, from Julabo, Seelbach, Germany). For the first gastric step, rabbit gastric lipase solution (1.5 ml, 102 U ml−1 in simulated gastric fluid) and simulated gastric fluid (1.7 ml) were added and the pH was adjusted to 5.5 (using 1.0 M HCl). The tube was shaken as above for 30 min, and the pH was kept between 5.3 and 5.7 with HCl. For the second gastric step, pepsin solution (25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 U ml−1 in simulated gastric fluid, 0.8 ml), was added and the pH was adjusted to 3.0 (with HCl). The volume of the mixture was adjusted to 10 ml with ultrapure water and the tube was shaken as above for 90 min, with addition of HCl as necessary to keep the pH between 2.9 and 3.1. For the intestinal step, simulated intestinal fluid (5.5 ml) was added. Pancreatin solution (800 U ml−1, in simulated intestinal fluid, 2.5 ml) was added, followed by bile solution (160 mmol bile acids per l, in ultrapure water, 1.25 ml). NaOH (1.0 M) was added to adjust the pH to 7.0. Ultrapure water was added to bring the total volume to 20 ml. The tube was shaken as above for 120 min, with addition of HCl as necessary to keep the pH between 7.0 and 7.2. Aliquots for measuring the degree of protein hydrolysis and fatty acid extraction were withdrawn and snap-frozen by plunging the tubes into a slurry of 95% ethanol and dry ice at ca. −78 °C; after freezing, the tubes were stored at −80 °C until analysis.
000 U ml−1 in simulated gastric fluid, 0.8 ml), was added and the pH was adjusted to 3.0 (with HCl). The volume of the mixture was adjusted to 10 ml with ultrapure water and the tube was shaken as above for 90 min, with addition of HCl as necessary to keep the pH between 2.9 and 3.1. For the intestinal step, simulated intestinal fluid (5.5 ml) was added. Pancreatin solution (800 U ml−1, in simulated intestinal fluid, 2.5 ml) was added, followed by bile solution (160 mmol bile acids per l, in ultrapure water, 1.25 ml). NaOH (1.0 M) was added to adjust the pH to 7.0. Ultrapure water was added to bring the total volume to 20 ml. The tube was shaken as above for 120 min, with addition of HCl as necessary to keep the pH between 7.0 and 7.2. Aliquots for measuring the degree of protein hydrolysis and fatty acid extraction were withdrawn and snap-frozen by plunging the tubes into a slurry of 95% ethanol and dry ice at ca. −78 °C; after freezing, the tubes were stored at −80 °C until analysis.
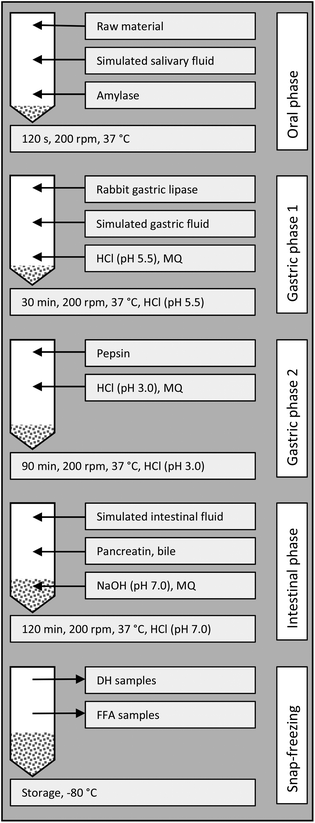 | ||
| Fig. 2 Scheme for the in vitro digestion. MQ = ultrapure water, DH = degree of protein hydrolysis, and FFA = liberated (free) fatty acids. | ||
Compositional analysis of pH-shift process products
The pH-shift process reported above is identical to the one we previously reported, carried out on the same microalgal paste.3 In the mentioned study, the composition of LYS, P(7) and P(10) was reported, but as information to the reader, we include it in Table 1. In brief, the water content was measured by drying at 105 °C, overnight; the protein content was measured according to Slocombe et al. (2013);20 total fatty acids and proportions of individual fatty acids were measured according to Cavonius et al. (2014);21 carbohydrates were measured according to Herbert et al. (1971),22 using a scaled-down method adapted for 96-well plates; and the ash content was determined by heating to 550 °C for 3 h.| Material (pH-shifted Nannochloropsis) | Dry/wet mass (%) | Protein/dry mass (%) | Total fatty acids/dry massa (%) | Carbohydrate/dry mass (%) | Ash/dry mass (%) |
|---|---|---|---|---|---|
| a Fatty acid profile of all algae-containing samples was similar in all cases, with a mean composition of 34% saturated fatty acids, 38% monounsaturated fatty acids and 27% PUFAs. | |||||
| LYS | 10 (0.5) | 19 (0.7) | 11 (0.2) | 37 (3.3) | 34 (4.8) |
| P(7) | 13 (3.3) | 23 (1.4) | 12 (1.5) | 42 (4.5) | 25 (2.4) |
| P(10) | 13 (2.2) | 24 (3.0) | 13 (0.5) | 58 (4.6) | 28 (14.5) |
Analysis of the degree of protein hydrolysis (DH)
For the present study, we define protein accessibility as the amount of peptide bonds cleaved in the in vitro digestion model. The DH was based on the method reported by Adler-Nissen, 1979,23 in which primary amines are detected, with the following adjustments: (i) after inactivating enzymes at 75 °C in 1% w/v sodium dodecyl sulphate (≥98.5% pure, dissolved in ultrapure water), samples were stored at −80 °C awaiting further analysis; (ii) after thawing, samples were centrifuged (2000g, 5 min) to remove light-scattering particles; (iii) spectrophotometric analysis was adapted to a 48-well plate (Costar 3548, Corning) by scaling down all volumes by a factor of 10; and (iv) the amount of broken peptide bonds was measured at 420 nm where there is little absorption by the polystyrene 48-well plate (instead of at 340 nm). Plate-readers used were a Tecan Safire 2 plate reader with Magellan software version 5.03 and a FluoSTAR Omega (BMG Labtech) with Omega software version 1.30. Samples were analysed in triplicate. Leucine (Sigma, purity ≥ 98%) was used as a standard (0–7.5 mM). The DH was expressed as h/htot × 100, where h is the sample's amount of broken peptide bonds after subtracting the digested SW-blank, and htot is the maximum amount of peptide bonds in any given sample. Values for htot, see Table 2, were calculated from the amino acid profiles of Nannochloropsis as reported previously from work on the same material,3 while the htot-value for casein was calculated as suggested by Babault et al., 2014.24| Sample | h tot-Value (mmol l−1) |
|---|---|
| CAS | 220 |
| ALG | Assumed to be as LYS |
| LYS | 189 |
| P(7) | 257 |
| P(10) | 317 |
| P(7)H | Assumed to be as P(7) |
| P(10)H | Assumed to be as P(10) |
| P(7)NF | Assumed to be as P(7) |
Analysis of total and liberated fatty acids
For the present study, we define lipid accessibility as the amount of FFAs liberated from their parent molecule in the in vitro digestion model. The total fatty acids (TFAs, i.e. all fatty acids, regardless if esterified or not) were quantified in all materials prior to digestion using direct transesterification according to Cavonius et al. (methanolic-HCl method);21 we previously have shown this method to recover fatty acids more completely than the Bligh and Dyer extraction, however, it does not distinguish between esterified fatty acids and FFAs.21 The determination of FFAs liberated during digestion was based on the Bligh and Dyer lipid extraction method25 followed by separation of lipid classes by solid phase extraction (SPE) according to Balasubramanian et al., 2013.26 Some minor modifications were applied to the Bligh and Dyer extraction as follows: the extraction was carried out on ice, with all solvents ice-cold. To each digested sample of 1.5 ml, 1.0 mg internal standard (nonadecanoic acid, C19:0, from Larodan in Solna, Sweden, purity ≥ 99%, in 200 μl chloroform) was added. Chloroform–methanol (chloroform: Sigma-Aldrich, purity ≥ 99.8%; methanol: Fluka, purity ≥ 99.9%) with 0.05% w/v butylated hydroxytoluene (BHT from Fluka, purity ≥ 99.0%), 4.5 ml, was added and the tubes were vortexed for 60 s. Chloroform, 1.5 ml, was then added and the tubes were vortexed for 15 s, where after 1.5 ml of aqueous NaCl (0.5% w/v) was added and the tubes were vortexed for 30 s. Phase-separation was aided by centrifugation (2000g, 4 °C, 6 min). The organic phase was transferred to a clean tube and the remaining aqueous phase was re-extracted with 3.0 ml chloroform, followed by 30 s vortexing and centrifugation as above, before pooling the organic phases. The solvent was evaporated under N2(g) at 40 °C and re-dissolved in 0.5 ml chloroform.Lipid classes (neutral lipids, FFAs and polar lipids) were separated by SPE according to the report of Balasubramanian et al., 2013![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 26 which was in turn based on the report of Kaluzny et al., 1985;27 we confirmed that fatty acids eluted in the appropriate fraction by studying the recovery of C21:0 eluted according to the method and found the yield to be close to 100%. Solvents used were hexane (Sigma, purity ≥ 97.0%), chloroform (as above), 2-propanol (Fisher, purity = 99.98%), diethyl ether (Sigma, purity ≥ 99.8%), glacial acetic acid (Scharlau, purity ≥ 99.8%), and methanol as above. The fraction containing FFAs was collected and the solvent was evaporated under N2(g) at 40 °C and re-dissolved in toluene (Scharlau, purity ≥ 99.8%) while awaiting fatty acid quantification. FFAs were converted into fatty acid methyl esters and quantified by the method according to Cavonius et al., 2014, as above.21 FFAs liberated from the parent molecule during digestion are expressed as (FFA − FFASW)/TFA × 100, where FFASW are the fatty acids liberated in the digested SW control; the same calculation was applied to individual fatty acids.
26 which was in turn based on the report of Kaluzny et al., 1985;27 we confirmed that fatty acids eluted in the appropriate fraction by studying the recovery of C21:0 eluted according to the method and found the yield to be close to 100%. Solvents used were hexane (Sigma, purity ≥ 97.0%), chloroform (as above), 2-propanol (Fisher, purity = 99.98%), diethyl ether (Sigma, purity ≥ 99.8%), glacial acetic acid (Scharlau, purity ≥ 99.8%), and methanol as above. The fraction containing FFAs was collected and the solvent was evaporated under N2(g) at 40 °C and re-dissolved in toluene (Scharlau, purity ≥ 99.8%) while awaiting fatty acid quantification. FFAs were converted into fatty acid methyl esters and quantified by the method according to Cavonius et al., 2014, as above.21 FFAs liberated from the parent molecule during digestion are expressed as (FFA − FFASW)/TFA × 100, where FFASW are the fatty acids liberated in the digested SW control; the same calculation was applied to individual fatty acids.
Statistical analysis
To produce all samples, the pH-shift process was performed on two separate days with P(7) produced on either days (n = 2) used to assess the reproducibility of the pH-shift process. Analysis of DH, FFAs and TFAs on non-digested samples was carried out in triplicate with the means being reported without variance. Digestions were performed in triplicate on each type of sample. Analysis of DH and FFAs on digested samples was carried out once for each digestate (n = 3).Results from the two separate days of P(7) were compared by the T-test (independent samples, two-tailed, 95% confidence interval) and analytical data were pooled once it was confirmed that there was no significant difference between production days for either DH or FFAs. The DH and FFAs after digestion in different materials were compared by one-way ANOVA, with both Welch and Brown–Forsythe tests since the data are heteroscedastic, followed by a Games–Howell post-hoc test; p < 0.05 was considered statistically significant.
Results and discussion
Degree of protein hydrolysis (DH) in in vitro digestion
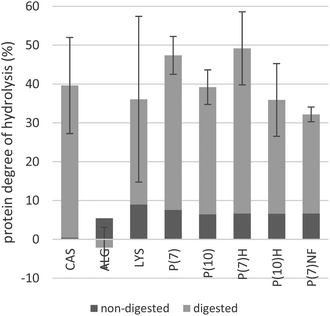
After digestion, 32–50% of the peptide bonds had been hydrolysed in the samples containing algae, which was comparable with the CAS control in which 40% of the bonds had been hydrolysed, see Fig. 4. There was no significant difference in DH between digested P(7)-samples from pH-shift processes performed on two separate days, indicating that the pH-shift process is reproducible. The data of the two separate P(7)-samples are therefore pooled in Fig. 3. However, in the ALG sample (i.e. whole algae), only 3% of the peptide bonds were hydrolysed, a significant difference from most other types of materials [i.e. p ≤ 0.033 for P(7), P(10), P(7)H, P(7)NF]. The only other significant difference regarding DH was between P(7) and P(7)NF (p = 0.004), with the lowest DH for any of the pH-shift products detected in P(7)NF at 32%. A large variance was noted for LYS samples, where two of the replicates were within the range of the pH-shift process-products, while the third was just slightly higher than the ALG sample; analyses were repeated several times to confirm this result. Due to this large variance, there is no statistical significance between the key materials LYS (coefficient of variation = 59%) on the one hand and products of the pH-shift process (coefficient of variation = 10–26%) on the other hand, even if mean values differ. Differences between heat-treated and non-heated materials were small for both P(7) and P(10), suggesting that the heat-treatment had neither a positive nor a negative impact on DH. Thus, the results strongly indicate that disruption of the Nannochloropsis cells is necessary to make the proteins accessible to the digestive enzymes.Prior to digestion, primary amines corresponding to approximately 7% of all peptide bonds were detected in the algae-containing samples. If the variance of analysis is taken into account, there was no significant difference between any of the algae-containing starting materials. Neither was there any indication that DH increased with an increased pH-shift processing time (e.g. by endogenous proteolytic enzymes of the algae), though it cannot be ruled out that the ca. 7% of initial DH is caused by algal proteases, acting during the harvest, handling and storage of the algal raw material. However, microalgae are reported to contain non-protein nitrogen in the form of nucleic acids, amines, glucosamides, cell wall material, pigments and even free amino acids and inorganic nitrogen;2,28 some of these species could contribute towards the primary amines detected prior to digestion.23
CAS was included as a reference protein, since it is known to have high digestibility, i.e. 93%.29 In the present study, digested CAS had a DH of 40%. For comparison, a different study applying the Infogest in vitro digestion model15 reported a DH of 80–84% for goat milk and kefir.30 A similar study on the digestibility of fish protein isolate used a different in vitro digestion method, but the same batch of casein, and reported a DH of 15% for the casein control.31 In the present study, we observed that the casein powder remained in small particles and did not dissolve properly, thus, the large difference in DH between pure casein and milk/kefir may be attributed to a smaller surface area available to the digestive enzymes in pure casein, compared to casein present in its native state in milk.
For the heat treatment, 72 °C for 15 s was chosen, a temperature and time sufficient to kill spoilage micro-organisms in milk.19 The heat-treated pH-shift process products did not differ significantly from the non-heated products, suggesting that the heat-treatment has neither a detrimental nor a beneficial effect on the accessibility of N. oculata protein. Our finding agrees with that of Hori et al. (1990), in which the cyanobacterium Nostoc commune was investigated and it likewise was reported that heat treatment had little effect on the in vitro protein digestibility.8
Comparison of frozen and non-frozen pH-shift process products, i.e. P(7) and P(7)NF was included to assess the impact of freezing on the digestibility of lipids and proteins. Although it may be possible to process microalgae into fresh food or feed for immediate consumption, such an approach was impractical for the study design: storing pH-shift process products, at e.g. 4 °C, over the two weeks it took to perform the in vitro digestions may have altered the composition of the product and further increased the variance. Alternatively, in an industrial food-processing setting, microalgae could be processed and stored frozen prior to consumption. When comparing P(7) and P(7)NF, these two were found to be significantly different, suggesting that freezing improves the accessibility of N. oculata proteins slightly. Cold-induced denaturation of proteins can occur, particularly when proteins are stabilised primarily by non-polar interactions.18 Indeed, our previous experience of N. oculata proteins3 has given us reason to believe that much of the protein is embedded in membranes. To perform SDS-PAGE, Laemmli buffer needed to be augmented with SDS and urea to denature the proteins, indicative of non-polar interactions.3 Thus, while results indicate that freezing increases the accessibility of N. oculata proteins in P(7), future investigations could see if the same holds true for P(7)H, P(10) and P(10)H.
Liberated FFAs in in vitro digestion
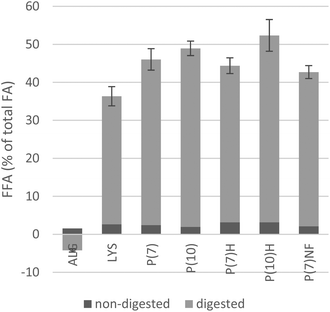
After digestion, 34–55% of the total fatty acids had been liberated in most of the algae-containing materials, with the exception of the ALG sample, where less FFAs were detected than in the SW blank, resulting in a negative value. The difference between ALG and other samples was highly significant (p ≤ 0.007). There was no significant difference in the amounts of FFA present after digestion between P(7)-samples from two separate pH-shift processes; the results are therefore pooled in Fig. 4. Thus, the results strongly indicate that disruption of the Nannochloropsis cells is necessary also to make the lipids accessible to the digestive enzymes.Among the pH-shift processed algae, significantly less FFAs were liberated during the in vitro digestion of the LYS sample (34%) than in the P(7), P(10) and P(10)H samples (44–49%, p ≤ 0.049). When a T-test was applied to the pooled data of pH-shift process-products carried out at pH 7 and compared to products of the pH 10-process, differences were also highly significant (p = 0.001) with the latter showing a higher degree of lipolysis. Indeed, the highest fraction of FFAs liberated by the in vitro digestion model was 52% in P(10)H, closely followed by P(10) and P(7) at 49% and 46%, respectively. These values are comparable to those reported by Lin et al. (2014), in a study that subjected emulsified and non-emulsified algal oil to an in vitro digestion model including gastric lipase.32 Thus, the results further demonstrate that subsequent pH-shift processing can improve lipolysis, which may be due to a conformational change of protein–lipid complexes, resulting in better access by lipases.
Prior to digestion, small amounts of FFAs were detected in all algae-containing materials, corresponding to 2–3% of the TFAs. Possibly, the FFAs present before digestion were a result of endogenous algal lipases, acting during the harvest, handling and storage of the algal raw material. It is well-established that in both plant and animal food raw materials, there is considerable post-harvest hydrolysis of fatty acids attached to phospholipids and for vegetable material also to glycolipids.33,34 While the study of post-harvest lipolysis of microalgal lipids is worth dedicating future investigations to, for this study it is enough to note that there was no statistically significant difference in initial FFAs between analysis replicates of the various algae-containing raw materials. However, the presence of FFAs during digestion is known to inhibit lipases in in vitro digestion.16In vivo, FFAs are removed when they are absorbed by enterocytes, whereas in the static model employed here, some product inhibition may occur. Product inhibition may in part explain why not all fatty acids were released, a maximum release of 52% was determined.
In the current study, only liberation of FFAs was studied as an index for lipid accessibility. Since monoacylglycerides are known to be absorbed in the small intestine, our results on FFA-liberation are not fully equivalent to lipid accessibility, but nonetheless provide an indication. In the limited number of studies available on in vitro algal lipid digestibility, FFAs have been used in the past as a measure of lipid accessibility.32
The separation of FFAs from other lipid classes relied on SPE. However, after performing SPE on the non-digested SW blank, a material which initially contained no other fatty acid apart from the internal standard, palmitic and stearic acids were detected. Therefore, the contamination must be expected to also be present in the algae-containing materials. The amount of palmitic and stearic acids was small relative to the total amount of liberated FFAs of the digested algae-containing materials (<3%) and therefore considered negligible in the context of total liberated FFAs. However, when analysing individual fatty acids liberated during digestion relative to the corresponding individual fatty acids present in the non-digested initial material (in the esterified form), the contamination had a major impact: especially for stearic acid, which is present in very low amounts in Nannochloropsis, the relative increase following digestion was unreasonably high (up to 290% increase). According to Notter et al. (2008), who reported the same contamination, palmitic and stearic acids originate from the polypropylene in the commercial SPE-columns used.35
By considering the profile of individual fatty acids, it was seen that a small contamination of palmitic acid and stearic acid was present in the non-digested SW blank, therefore, differences in the liberation of palmitic and stearic acids were not considered further. More than 70% of the total palmitoleic (C16:1 n7) and oleic acid (C18:1 n9) was released in P(7), P(10), P(7)H, and P(10)H, with less of these fatty acids released from P(7)NF, LYS and ALG, following the same pattern as for total liberated fatty acids (above). 50% or more lauric acid (C12:0) and myristic acid (C14:0) was released in P(7), P(10), P(7)H, and P(10)H. Only 2–3% of total eicosapentaenoic acid (C20:5 n3) was detected as liberated FFA in products of the pH-shift process after digestion, with none detected in LYS and ALG-samples. More liberated arachidonic acid (C20:4 n6) was detected in the digested SW and CAS-blanks than in any of the algae-containing materials. Since PUFAs are sensitive to oxidation (also during in vitro digestion36) it is possible that C20:5 n3 and C20:4 n6 were degraded and therefore could not be detected as such. An alternative explanation is that the PUFAs were not released to the same degree as the more saturated fatty acids: Ryckebosch et al. (2014) have shown that in a study of Nannochloropsis oculata containing ca. 30% lipid per dry weight, only 68% of the C20:5 n3 was located in the glycolipids.37 Further studies could investigate the position of the eicosapentaenoic acid in Nannochloropsis glycolipids and to which degree the eicosapentaenoic acid is accessible to human digestive enzymes.
Although we demonstrated that lipolysis can be slightly increased by applying the pH-shift process, it is less clear whether the pH-shift processing changes the protein accessibility. It could thus be asked if pH-shift-processing beyond the disruption step is justified from a nutritional point of view: the addition of an acid and base, as well as equipment and time needed for the pH-shift process represent significant investments, but resulted in only a modest increase in fatty acid accessibility. Nonetheless, it is possible that the full pH-shift process imparts other advantages on the final product. For example, improved protein functionality has been reported after pH-shift processing of fish38 and is worth exploring for algae-containing materials.
In summary, the most striking feature of the results presented here is the low accessibility of the ALG material, both in terms of DH and liberated FFAs. The low accessibility of whole cells is not surprising in light of previous studies on the cell wall structure of Nannochloropsis: it has been reported that the Nannochloropsis cell wall is composed of cellulose, protected by an outer layer of algaenan.14 Therefore, whole algal cells can be expected to pass through the human digestive tract largely unaffected, since humans lack the enzymes needed to hydrolyse the cellulosic cell wall. Given that the detected DH and liberated FFAs in digested ALG were lower than those detected in the digested SW-control, it can even be speculated that the whole cells adsorb and bind digestive enzymes, acting as anti-nutrients.11,13,39,40 In a rat feeding trial, some animals fed 50 mg whole Nannochloropsis daily for eight weeks developed intestinal atrophy and transmural necrosis not observed in the control group, suggesting that whole algae challenged the rats’ gastrointestinal tract to the point of injury.41 It has thus been confirmed that it is necessary to disrupt the cell wall of Nannochloropsis to make the proteins and lipids accessible, in agreement with earlier studies.2,6
Conclusion
The present study demonstrates that cell disruption is necessary to make Nannochloropsis proteins and lipids accessible to mammalian digestive enzymes used in the applied in vitro digestion model. By processing the algal material with the pH-shift method beyond the initial disruption step, it was further demonstrated that fatty acid liberation could be modestly increased. With good accessibility (>50%) of amino acids and TFAs, pH-shift processed Nannochloropsis has potential as a functional food or feed ingredient. Future studies should investigate the fate of the PUFAs during digestion.Abbreviations
| ALG | Whole algae in seawater |
| CAS | Casein |
| DH | Degree of protein hydrolysis |
| FFA | Free fatty acid |
| LYS | Lysate |
| h | The amount of broken peptide bonds of a sample after subtracting the seawater-blank |
| h tot | The maximum theoretical amount of peptide bonds in a sample |
| P(7) | Product of the pH-shift process with solubilisation at pH 7 |
| P(7)H | Product of the pH-shift process with solubilisation at pH 7, heat-treated |
| P(7)NF | Product of the pH-shift process with solubilisation at pH 7, non-frozen |
| P(10) | Product of the pH-shift process with solubilisation at pH 10 |
| P(10)H | Product of the pH-shift process with solubilisation at pH 10, heat-treated |
| PUFA | Polyunsaturated fatty acid |
| SDS | Sodium dodecyl sulphate |
| SPE | Solid phase extraction |
| SW | Seawater |
| TFA | Total fatty acid |
Acknowledgements
This study was funded by Chalmers University of Technology. We would like to thank the following people for contributing to the study: Cecilia Tullberg, Marie Alminger, Annette Almgren, Charlotte Egger, Cecilia Svelander, Hanna Harrysson, Nikul Soni, Otto Savolainen, Niklas Engström, Cyrielle Bonzom, Nils-Gunnar Carlsson, and Rita Cavonius.References
- M. Vanthoor-Koopmans, R. H. Wijffels, M. J. Barbosa and M. H. M. Eppink, Bioresour. Technol., 2013, 135, 142–149 CrossRef CAS PubMed.
- W. E. Becker, in Handbook of Microalgal Culture: Applied Phycology and Biotechnology, ed. Q. Hu and A. Richmond, John Wiley & Sons, Somerset, NJ, USA, 2nd edn, 2013 Search PubMed.
- L. R. Cavonius, E. Albers and I. Undeland, Algal Res., 2015, 11, 95–102 CrossRef.
- S. K. Marmon, A. Krona, M. Langton and I. Undeland, J. Agric. Food Chem., 2012, 60, 7965–7972 CrossRef CAS PubMed.
- Q. Liu, R. Geng, J. Y. Zhao, Q. Chen and B. H. Kong, J. Agric. Food Chem., 2015, 63, 4853–4861 CrossRef CAS PubMed.
- G. Hedenskog, L. Enebo, J. Vendlova and B. Prokes, Biotechnol. Bioeng., 1969, 11, 37–51 CrossRef CAS PubMed.
- M. A. Devi, G. Subbulakshmi, K. M. Devi and L. V. Venkataraman, J. Agric. Food Chem., 1981, 29, 522–525 CrossRef CAS PubMed.
- K. Hori, T. Ueno-Mohri, T. Okita and G. Ishibashi, Plant Foods Hum. Nutr., 1990, 40, 223–229 CrossRef CAS PubMed.
- H. J. Morris, A. Almarales, O. Carrillo and R. C. Bermudez, Bioresour. Technol., 2008, 99, 7723–7729 CrossRef CAS PubMed.
- G. Graziani, S. Schiavo, M. A. Nicolai, S. Buono, V. Fogliano, G. Pinto and A. Pollio, Food Funct., 2013, 4, 144–152 CAS.
- S. M. Tibbetts, J. E. Milley and S. P. Lall, J. Appl. Phycol., 2015, 27, 1109–1119 CrossRef CAS.
- L. P. Goh, S. P. Loh, M. Y. Fatimah and K. Perumal, Malaysian J. Nutr., 2009, 15, 77–86 Search PubMed.
- A. Skrede, L. T. Mydland, O. Ahlstrom, K. I. Reitan, H. R. Gislerod and M. Overland, J. Anim. Feed Sci., 2011, 20, 131–142 Search PubMed.
- M. J. Scholz, T. L. Weiss, R. E. Jinkerson, J. Jing, R. Roth, U. Goodenough, M. C. Posewitz and H. G. Gerken, Eukaryotic Cell, 2014, 13, 1450–1464 CrossRef PubMed.
- M. Minekus, M. Alminger, P. Alvito, S. Ballance, T. Bohn, C. Bourlieu, F. Carriere, R. Boutrou, M. Corredig, D. Dupont, C. Dufour, L. Egger, M. Golding, S. Karakaya, B. Kirkhus, S. Le Feunteun, U. Lesmes, A. Macierzanka, A. Mackie, S. Marze, D. J. McClements, O. Menard, I. Recio, C. N. Santos, R. P. Singh, G. E. Vegarud, M. S. J. Wickham, W. Weitschies and A. Brodkorb, Food Funct., 2014, 5, 1113–1124 CAS.
- J. C. B. N'Goma, S. Amara, K. Dridi, V. Jannin and F. Carrière, Ther. Delivery, 2012, 3, 105–124 CrossRef.
- P. Capolino, C. Guérin, J. Paume, J. Giallo, J. M. Ballester, J. F. Cavalier and F. Carrière, Food Dig., 2011, 2, 43–51 CrossRef CAS.
- Food Chemistry, ed. O. R. Fennema, CRC Press, Boca Raton, 1996 Search PubMed.
- P. J. Fellows, Food Processing Technology, Woodhead Publishing Limited, Cambridge, England, 2nd edn, 2000 Search PubMed.
- S. P. Slocombe, M. Ross, N. Thomas, S. McNeill and M. S. Stanley, Bioresour. Technol., 2013, 129, 51–57 CrossRef CAS PubMed.
- L. R. Cavonius, N. G. Carlsson and I. Undeland, Anal. Bioanal. Chem., 2014, 406, 7313–7322 CrossRef CAS PubMed.
- D. Herbert, P. J. Phipps and R. E. Strange, in Methods in Microbiology, ed. J. R. Norris and D. W. Ribbons, Academic Press Inc., London, 1971, vol. 5B, ch. 3, pp. 210–383 Search PubMed.
- J. Adler-Nissen, J. Agric. Food Chem., 1979, 27, 1256–1262 CrossRef CAS PubMed.
- N. Babault, G. Deley, P. Le Ruyet, F. Morgan and F. A. Allaert, J. Int. Soc. Sports Nutr., 2014, 11, 36–45 CrossRef PubMed.
- E. G. Bligh and W. J. Dyer, Can. J. Biochem. Physiol., 1959, 37, 911–917 CrossRef CAS PubMed.
- R. K. Balasubramanian, T. T. Y. Doan and J. P. Obbard, Chem. Eng. J., 2013, 215, 929–936 CrossRef.
- M. A. Kaluzny, L. A. Duncan, M. V. Merritt and D. E. Epps, J. Lipid Res., 1985, 26, 135–140 CAS.
- E. Barbarino and S. Lourenço, J. Appl. Phycol., 2005, 17, 447–460 CrossRef CAS.
- J. Boye, R. Wijesinha-Bettoni and B. Burlingame, Br.J. Nutr., 2012, 108(Suppl 2), S183–S211 CrossRef CAS PubMed.
- S. Nehir El, S. Karakaya, S. Simsek, D. Dupont, E. Menfaatli and A. T. Eker, Food Funct., 2015, 6, 2322–2330 CAS.
- S. K. Marmon and I. Undeland, Food Chem., 2013, 138, 214–219 CrossRef CAS PubMed.
- X. Lin, Q. Wang, W. Li and A. J. Wright, Food Funct., 2014, 5, 2913–2921 CAS.
- M. J. Kim, J. M. Oh, S. H. Cheon, T. K. Cheong, S. H. Lee, E. O. Choi, H. G. Lee, C. S. Park and K. H. Park, J. Agric. Food Chem., 2001, 49, 2241–2248 CrossRef CAS PubMed.
- H. A. Roldán, S. I. Roura, C. L. Montecchia, O. Pérez Borla and M. Crupkin, J. Food Biochem., 2005, 29, 187–204 CrossRef.
- S. J. Notter, B. H. Stuart, B. B. Dent and J. Keegan, Eur. J. Lipid Sci. Technol., 2008, 110, 73–80 CrossRef CAS.
- K. Larsson, L. Cavonius, M. Alminger and I. Undeland, J. Agric. Food Chem., 2012, 60, 7556–7564 CrossRef CAS PubMed.
- E. Ryckebosch, C. Bruneel, R. Termote-Verhalle, K. Goiris, K. Muylaert and I. Foubert, Food Chem., 2014, 160, 393–400 CrossRef CAS PubMed.
- S. K. Marmon and I. Undeland, J. Agric. Food Chem., 2010, 58, 10480–10486 CrossRef CAS PubMed.
- O. Marrion, J. Fleurence, A. Schwertz, J. L. Gueant, L. Mamelouk, J. Ksouri and C. Villaume, J. Appl. Phycol., 2005, 17, 99–102 CrossRef CAS.
- E. Rodríguez De Marco, M. E. Steffolani, C. S. Martínez and A. E. León, LWT—Food Sci. Technol., 2014, 58, 102–108 CrossRef.
- K. Nuño, A. Villarruel-López, A. M. Puebla-Pérez, E. Romero-Velarde, A. G. Puebla-Mora and F. Ascencio, J. Funct. Foods, 2013, 5, 106–115 CrossRef.
| This journal is © The Royal Society of Chemistry 2016 |