Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
The role of plant cell wall encapsulation and porosity in regulating lipolysis during the digestion of almond seeds†
Myriam M. L.
Grundy
a,
Frédéric
Carrière
b,
Alan R.
Mackie
c,
David A.
Gray
d,
Peter J.
Butterworth
a and
Peter R.
Ellis
*a
aKing's College London, Diabetes and Nutritional Sciences Division, Biopolymers Group, Franklin-Wilkins Building, London SE1 9NH, UK. E-mail: peter.r.ellis@kcl.ac.uk; myriam.grundy@kcl.ac.uk; peter.butterworth@kcl.ac.uk; Fax: +44 (0)207 8484171; Tel: +44 (0)207 8484238
bCNRS, Aix-Marseille Université, UMR 7282, Enzymologie Interfaciale et Physiologie de la Lipolyse, 31 Chemin Joseph Aiguier, 13402 Marseille Cedex 20, France. E-mail: carriere@imm.cnrs.fr
cInstitute of Food Research, Norwich Research Park, Colney, Norwich NR4 7UA, UK. E-mail: alan.mackie@ifr.ac.uk
dUniversity of Nottingham, Division of Food Sciences, School of Biosciences, Sutton Bonington Campus, Leicestershire, England LE12 5RD, UK. E-mail: david.gray@nottingham.ac.uk
First published on 21st October 2015
Abstract
Previous studies have provided evidence that the physical encapsulation of intracellular nutrients by cell walls of plant foods (i.e. dietary fibre) plays a predominant role in influencing macronutrient bioaccessibility (release) from plant foods during human digestion. One unexplored aspect of this is the extent to which digestive enzymes can pass through the cell-wall barrier and hydrolyse the intracellular lipid in almond seeds. The purpose of the present study was to assess the role played by cell walls in influencing the bioaccessibility and digestibility of almond lipid using a range of techniques. Digestibility experiments were performed on raw and roasted almond cells as well as isolated almond oil bodies using in vitro gastric and duodenal digestion models. Residual triacylglycerols and lipolysis products were extracted after 1 h of incubation and analysed by thin layer chromatography. The lipolysis kinetics of almond cells and oil bodies were also investigated using the pH-stat technique. Finally, the potential penetration of pancreatic lipase through the cell wall matrix was investigated using confocal microscopy. Differences in the rates and extent of lipolysis were clearly seen between almond cells and oil bodies, and these differences were observed regardless of the lipase(s) used. These results also showed that almond cell walls that are completely intact limit lipid digestibility, due to an encapsulation mechanism that hinders the diffusion of lipase into the intracellular environment and lipolysis products out of the cells.
1. Introduction
Recent studies have highlighted the importance of the cell walls of edible plants, which are the main source of dietary fibre, in reducing the rate and extent of bioaccessibility and digestion of lipid and other macronutrients.1–3 The effects of the almond seed cell walls on the kinetics of lipid digestion has received particular attention, mainly because of the putative benefits of almond consumption on human health and risk factors associated with disease (e.g. type 2 diabetes, cardiovascular disease and obesity).4–6 Thus, an important physiological factor seems to be that most of the lipid in almonds remains encapsulated by the cell walls post-mastication and is less available for digestion.7,8 One metabolic consequence of this is a decrease in energy absorption and attenuation in postprandial lipaemia.9,10Almond seeds, like many other oilseeds, store lipids as triacylglycerols (TAG) in oil bodies until they are eventually mobilised upon seed germination. Oil bodies are small, spherical organelles enclosed in a monolayer of phospholipids into which unique proteins, mainly oleosins, are embedded.11,12 The diameter of oil bodies in almond cotyledon cells ranges between 1–5 μm. The TAG constitute about 50% of the total dry weight of the oil bodies, with the predominant fatty acids being, in decreasing order of abundance, oleic (18:1Δ9), linoleic (18:2Δ9,12), palmitic (16:0), stearic (18:0) and palmitoleic (16:1Δ9). The almond cells have an average diameter of about 35 μm (ranging between 20 and 50 μm) and are surrounded by a cell wall of about 0.1–0.3 μm thickness. Plant cell walls, which are largely resistant to digestion in the upper gastrointestinal (GI) tract, consist of complex heterogeneous networks of mainly polysaccharides, namely cellulose, hemicelluloses and pectic components.13 Almond cotyledon cell walls are considered to be predominantly composed of arabinose-rich pectic material, with smaller amounts of xylan, xyloglucan and cellulose.7,14
Lipases (triacylglycerol acylhydrolases EC 3.1.1.3) are a group of enzymes that catalyse the hydrolysis of TAG in a stepwise fashion producing diacylglycerols (DAG) and monoacylglycerols (MAG) accompanied at each step by the release of one free fatty acid (FFA). The two main lipases involved in lipid digestion in humans are gastric and colipase-dependent pancreatic lipases.15–17
Previous studies have shown that the physical encapsulation of intracellular nutrients (i.e. lipid or starch) by intact plant cell walls restricts the access of digestive enzymes and the release of nutrients.1,2,18 In almonds for example, only the lipid in peripheral cells ruptured by mechanical damage or mastication8,19 are easily accessible to lipase action during the early stages of digestion (0–3 h). Some loss of lipid may still occur however from intact cells below the fractured surface, but only at longer digestion times of 3–12 h.3 One possible explanation of this finding is that the cell walls become more permeable as a result of swelling after a prolonged retention time in the GI tract. Nonetheless, there is currently no evidence to indicate whether or not lipases, colipase and the other digestive agents such as bile salts are able to penetrate almonds cells via the cell wall at any stage of the digestion process. The specific mode of action of lipases, especially the difference in water solubility between the lipases and their substrate, and the change in the lipase conformation occurring during lipolysis, makes lipase action particularly difficult to investigate. However, to answer a key question of whether lipase can penetrate the cell walls of almond cells and digest intra-cellular lipid, we have used a novel experimental approach by combining confocal microscopy for locating pancreatic lipase, labelled with a fluorescent probe, with kinetic studies of lipolysis.
The main aim of this work therefore was to measure the rate and extent of lipolysis of cells prepared from raw and roasted almonds and isolated almond oil bodies using in vitro gastric and duodenal digestion models. Mechanistic studies of almond cell wall porosity and lipase diffusion into almond cells were also performed to assess the permeability of the cell walls to lipase and the efflux of products of lipolysis (i.e. FFA release). These in vitro studies have allowed us to obtain a deeper insight of how the cell wall barrier hinders the lipolysis process during the digestion of almond seeds.
2. Materials and methods
2.1. Preparation of almond materials
Raw and roasted almond (Amygdalus communis L.; variety Nonpareil) kernels were produced by Hughson Nut and supplied by the Almond Board of California. Separated cells of raw and roasted almonds were prepared as previously described.2 Oil bodies (crude) were physically isolated from raw and roasted almond seeds by homogenising (Moulinex, Masterchef 650 duotronic, Windsor, UK) the seeds in water (ratio 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4) with 2–3 drops of sodium azide (0.2%, w/v) at full power for 2 min. The slurry was filtered through three layers of cheesecloth to remove almond particles and cell fragments. The filtrate was then centrifuged (Beckman J2-21 centrifuge; fixed rotor JA-10) at 9936g, 4 °C for 20 min. The upper layer (creamy white pad) of each sample was removed and transferred into a 10 mL glass bijou bottle.20 This gentle extraction method aimed at obtaining oil bodies with a composition (including endogenous proteins) similar to the ones present in the separated almond cells. The proteins were however likely to be digested by proteases in the digestion assay and so did not offer much impedance to lipase access.21
4) with 2–3 drops of sodium azide (0.2%, w/v) at full power for 2 min. The slurry was filtered through three layers of cheesecloth to remove almond particles and cell fragments. The filtrate was then centrifuged (Beckman J2-21 centrifuge; fixed rotor JA-10) at 9936g, 4 °C for 20 min. The upper layer (creamy white pad) of each sample was removed and transferred into a 10 mL glass bijou bottle.20 This gentle extraction method aimed at obtaining oil bodies with a composition (including endogenous proteins) similar to the ones present in the separated almond cells. The proteins were however likely to be digested by proteases in the digestion assay and so did not offer much impedance to lipase access.21
2.2. Lipase sources
Different lipases and enzyme mixtures were used to investigate their effect on lipolysis, alone or in combination, on complex almond substrates (i.e. oil bodies and separated cells). Crude lipase preparation used in the digestibility experiments with the pH-stat device and lipase diffusion observations (see sections 2.6. and 2.8.) was from porcine pancreas type II (no. L3126, lipase activity 53 units per mg powder, where 1 unit corresponds to 1 μmol of butyric acid released from tributyrin per minute at 37 °C, pH 8.0) purchased from Sigma. Rabbit gastric extract (RGE) was prepared as previously described.22 RGE contains 77 lipase units and 660 pepsin units per mg powder, as measured with tributyrin as substrate for lipase activity23 and haemoglobin as substrate for pepsin activity.24 Purified rabbit gastric lipase23 and porcine pepsin (Sigma, no. P6887) were used as reference standards for enzyme assays and SDS page analysis of proteins. Porcine pancreatic extract (PPE) was also purchased from Sigma Aldrich (no. P7545; 464 units per mg powder). Porcine pancreatic lipase (PPL) and colipase were purified to homogeneity according to Verger et al.25 and Chapus et al.,26 respectively. Their purity was assessed by SDS-PAGE and enzyme activity measurements based on PPL specific activity.25 RGE, PPE and PPL were used in the thin layer chromatography analysis (see section 2.7.).2.3. Particle size distribution and ζ-potential measurements
Oil body suspensions were formulated by dispersing the crude oil body preparation into water (10% oil, w/w). The average size (d4,3) of the oil bodies was measured using a Beckman Coulter LS13320® and their zeta potential (ζ-potential) determined with a Beckman Coulter Delsa™Nano C (Beckman Coulter Ltd, High Wycombe, UK). The ζ-potential measurements were performed at 25 °C, pH 7.0, with a dispersant (water) refractive index of 1.330, almond oil refractive index of 1.471, viscosity of 0.891 mPa s, and relative dielectric constant of 79.0. The electrode spacing was 50.0 mm. The ζ-potential was calculated by the instrument software fitting data to the Smoluchowski equation.27 Each measurement was obtained as the mean of triplicates.2.4. Crude lipid analysis of oil bodies
The lipids from the oil bodies were extracted by adding 500 μL of isooctane into 0.2 g of oil bodies that were previously dried in a vacuum oven at 40 °C for 48 h. The tubes were homogenised into a FastPrep®-24 Instrument (MP Biomedicals, Cambridge, UK) for 30 s and microcentrifuged (MSE Micro Centaur, Sanyo, London, UK) at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 5 min. A known volume of the supernatant was pipetted into labelled Eppendorf tubes. This extraction step was repeated 3 times. One mL of the pooled supernatant was poured into a 10 mL glass bijou bottle and dried in a vacuum oven for a few hours; this fraction contained the lipids. The isooctane remaining in the tubes was evaporated in vacuo and the pellet stored in a freezer at −20 °C for protein analysis. The lipid content was determined gravimetrically by calculating the difference in the sample weight measured before and after the extraction process.
000g for 5 min. A known volume of the supernatant was pipetted into labelled Eppendorf tubes. This extraction step was repeated 3 times. One mL of the pooled supernatant was poured into a 10 mL glass bijou bottle and dried in a vacuum oven for a few hours; this fraction contained the lipids. The isooctane remaining in the tubes was evaporated in vacuo and the pellet stored in a freezer at −20 °C for protein analysis. The lipid content was determined gravimetrically by calculating the difference in the sample weight measured before and after the extraction process.
2.5. Protein analysis
The protein layer from the defatted oil body samples (section 2.4.) was sonicated for ∼1 min and 1 mL of 2% (w/v) sodium dodecyl sulphate (SDS) added to it. The samples were heated at 60 °C for 30 min, vortexed for 1 min and then centrifuged at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 3 min. The supernatant was collected and diluted 100 fold with 2% (w/v) SDS. Concentrations of protein in the samples were then determined using the bicinchoninic acid (BCA) assay (Sigma, Poole, UK).
000g for 3 min. The supernatant was collected and diluted 100 fold with 2% (w/v) SDS. Concentrations of protein in the samples were then determined using the bicinchoninic acid (BCA) assay (Sigma, Poole, UK).
2.6. In vitro intestinal digestions using the pH-stat method
The in vitro duodenal digestion experiments were performed on both raw and roasted almond cells and oil bodies as detailed elsewhere.2 Briefly, the simulated digestion mixture contained 12.5 mM bile salt solution, 150 mM NaCl solution, 10 mM CaCl2 solution and lipid material dissolved in 1% (w/w) β-lactoglobulin solution (the amount of material used was sufficient to obtain 300 mg of lipid). Test runs were then performed by incubating the mixture for 1 h at 37 °C, pH 7.0, in a mechanically stirred reaction vessel of a pH-stat instrument (Titrino 848 plus, Metrohm UK Ltd) with 1.5 mL of freshly prepared pancreatic lipase type II solution (40 mg mL−1 in 10 mM phosphate buffer). A blank assay in the absence of lipase was performed and the volume of NaOH added to keep the pH constant was then deducted from the NaOH volume added in the course of the lipolysis assay with lipase. Each sample analysis was performed in triplicate.2.7. In vitro lipolysis of almond materials, lipid extraction and analysis by thin layer chromatography (TLC)
Raw almond materials (i.e.; cells and oil bodies) were added into Eppendorf tubes at a weight equivalent to 50 mg of lipids, which corresponded to about 120 mg of cell preparation or 50 mg of oil bodies. Each reaction system had a total volume of 1 mL and was left incubating for 1 h (gastric or duodenal) or 2 h (gastric plus duodenal) at 37 °C. The buffer consisted of either 10 mM MES, pH 5.5 (i.e. representing the gastric phase) or 10 mM Tris, pH 8.0 (duodenal and gastric plus duodenal phases) containing 150 mM NaCl. The reactions were performed using RGE (1 mg mL−1) and PPE (20 mg mL−1), alone and in combination, as well as PPL (1 mg mL−1) with colipase (added at a 2 to 1 excess molar ratio). Total lipids from native and digested samples were then extracted by the Folch method.28 Reference standards (triolein, diolein, monoolein and oleic acid purchased from Sigma-Aldrich, St Quentin-Fallavier, France) and extracted lipids from the native and digested samples (15 μL) were spotted onto thin layer silica gel 60 plates (10 × 20 cm from Merck, Massachusetts, USA) using a Linomat IV apparatus (Camag, Muttenz, Switzerland) equipped with a 100 μL dosage syringe (Camag, Muttenz, Switzerland). The plate was then placed into a tank containing a mixture of heptane/diethyl ether/formic acid (55![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 45
45![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v/v) and left to migrate for about 10 min. Following this stage, the plate was dried at room temperature for 10 min and then sprayed with a mixture of saturated aqueous solution of cupric acetate and 85% phosphoric acid (1
1, v/v/v) and left to migrate for about 10 min. Following this stage, the plate was dried at room temperature for 10 min and then sprayed with a mixture of saturated aqueous solution of cupric acetate and 85% phosphoric acid (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v). The liquid was left to evaporate for 10 min and the plate placed in the oven at 180 °C for 10 min.
1, v/v). The liquid was left to evaporate for 10 min and the plate placed in the oven at 180 °C for 10 min.
2.8. Diffusion experiments
The reaction environment contained 25 μL of diluted (1/10 in 12.5 mM of bile salt solution) cell or oil bodies preparation, 1 μL of Nile red solution (1 mg mL−1 in dimethyl sulphoxide), 1 μL of calcofluor white (2% w/v in deionised water), 25 μL of labelled lipase (0.76 mg mL−1) and 4 μL of colipase (1 mg mL−1). Aliquots were taken at different time points (0, 30, 60 and 120 min, and ∼20 h of digestion) and visualised using confocal laser scanning microscopy (SP1 or SP5 CLSM, Leica Microsystems, Mannheim, Germany). Nile red and calcofluor white were used to detect the lipids and cell walls, respectively.32 Images were captured using both 40× (N.A. 1.25) and 63× (N.A. 1.32) oil immersion objective lenses. The samples were excited using an argon laser at 488 nm for Nile red and Alexa Fluor® 488, and at 405 nm for calcofluor white. The fluorescence emitted by the samples was detected at 630 to 680 nm (Nile red), 505 to 550 nm (Alexa Fluor 488) and 406 to 460 nm (calcofluor white).
2.9. Statistical analysis
The data were analysed using SPSS version 17.0. For all tests, the significance level was set at P < 0.05 (2 tailed). All data are expressed as means ± SEM. Particle size, ζ-potential, and FFA release data were analysed by repeated-measures ANOVA with time and materials as ‘within-sample’ factors. Differences between almond oil body and cell samples were analysed by Student's paired t-test.3. Results
3.1. Characterisation of oil bodies
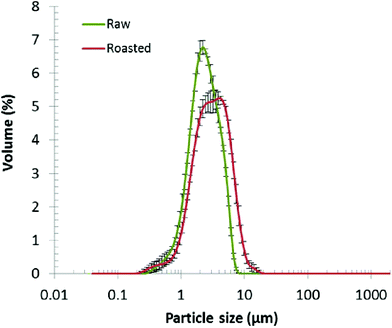
The crude oil bodies contained 89.3 ± 2.0 and 85.1 ± 1.5% of lipid, and 5.8 ± 0.7 and 5.3 ± 1.4% of protein, for raw and roasted almond, respectively. A non-negligible fraction (4.9 and 9.7% for raw and roasted almond oil bodies, respectively) of the total weight could not be identified. The likely cause of the lack of identification was contamination by cell debris such as polysaccharides from the cell wall as has been previously reported.33 This unidentified mass could also have contained some of the degradation products formed from the oil bodies by the seed during the period between harvest and pasteurisation, to generate energy as sugars and carbon chains of amino acids (mainly asparagine, aspartate, glutamine and glutamate) that are required for embryonic growth.34The particle size distributions of raw and roasted almond oil bodies are shown in Fig. 1. The difference in average diameter of the oil bodies was statistically significantly (P < 0.005) between raw and roasted almonds: 2.6 ± 0.09 and 3.8 ± 0.11 μm, respectively. The size of raw almond oil bodies is in agreement with the data from other groups.12,35 The average size increase observed with roasted almonds probably resulted from the partial coalescence of oil bodies upon roasting, probably due to changes in the oil body monolayer (e.g. denaturation of the oleosins).
The ζ-potentials of raw and roasted almond oil bodies were −33.7 ± 1.5 and −27.7 ± 1.3 mV, respectively, values that are consistent with previous results.35,36 The structure of the oil bodies interface (anionic phospholipids and protein molecules) is responsible for the negative surface charges which prevent coalescence of the oil bodies.37 The ζ-potential values confirmed that raw almond oil bodies, similar to oil bodies found in other seeds, are stable even in isolated preparations. On the other hand, roasted almond oil bodies tend to aggregate and coalesce as demonstrated notably by the variability in their particle size. The loss of negative charge in roasted oil bodies may be due to some denaturation of oleosins occurring during the roasting process.
3.2. Analysis of in vitro duodenal digestion with a pH-stat device
The rate and extent of lipolysis in almond oil bodies and cells were measured with the pH-stat technique and porcine pancreatic lipase type II as a source of lipase (Table 1). The initial reaction rate, as well as the amount of FFA released following 1 h of digestion, were somewhat similar for both oil body types isolated from raw and roasted almonds, albeit slightly lower for the latter type (statistically significant at P < 0.05). As indicated in Table 1, the FFA release and reaction rate values for the raw and roasted oil bodies were significantly higher than the corresponding values for the cells (P < 0.05). No significant differences were found between raw and roasted almond cells. Thus, the isolated oil bodies appeared to be a good substrate for the porcine pancreatic lipase (68.8 ± 2.6% hydrolysis after 1 h incubation), whereas the lipolysis of crude oil bodies by purified human pancreatic lipase (HPL) has been reported to be low, particularly when compared to the lipolysis of almond oil emulsion.12 The crude pancreatic lipase solution used here however contains additional enzymes like protease and phospholipase A2, which can act in synergy with pancreatic lipase and trigger lipolysis as indicated by the comparative TLC analysis of digestion experiments performed with PPE and purified PPL (see section 3.3).| Almond material | Form | FFA (%) at 1 h | Initial reaction rate (μmol FFA per min) |
|---|---|---|---|
| a Statistically significant differences compared with raw almond cells (P < 0.05). b Statistically significant differences compared with roasted almond cells (P < 0.05). c Statistically significant differences between raw and roasted oil bodies (P < 0.05). | |||
| Oil bodies | Raw | 68.8 ± 2.64a,c | 71.3 ± 2.04a,c |
| Roasted | 57.5 ± 6.15b,c | 66.0 ± 1.19b,c | |
| Cells | Raw | 21.2 ± 1.59 | 36.5 ± 5.21 |
| Roasted | 22.1 ± 2.04 | 42.5 ± 3.35 | |
A striking finding was the activity of the crude lipase preparation on the whole almond cells. Thus, the 1 h FFA release value for the raw almond cells was only about a third of the value observed for isolated oil bodies (Table 1), although FFA release for the cells was still much higher than anticipated. As suggested from our previous work that showed release of lipid only from ruptured cells,38 we would have expected the hydrolysis from separated cells (assuming that they all had intact cell walls) to be close to 0%. One explanation for this result is that some of the almond cells might be physically disrupted during preparation, thus allowing easier access of the lipase to the intracellular lipid.2
3.3. In vitro duodenal digestion of almond material and lipolysis products analysis by TLC
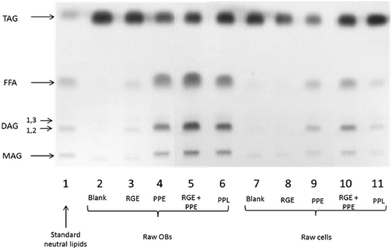
Fig. 2 illustrates the extent of digestion of almond oil bodies and cells by gastric and pancreatic lipases. The TLC-densitometry method permitted the identification of both residual TAG and lipolytic products. The lipid digestion appeared more limited for separated cells than for isolated oil bodies and this was observed regardless of the enzyme preparation used. The direct action of RGE appeared restricted compared with that of PPE, but the samples incubated with RGE and PPE were digested to a greater extent than with PPE alone, thus showing some synergy between gastric and pancreatic lipases. Lipolysis was more effective in the presence of PPE than PPL. Given that the PPE contains a mixture of different enzymes (i.e. pancreatic lipase, carboxylester hydrolase, proteases and phospholipase A2), it is probable that degradation of oleosins and phospholipids on the surface of oil bodies occurred with PPE, which permitted better access of the PPL to its TAG substrate. A synergistic action of lipolytic enzymes may also have increased the overall lipolysis rate.3.4. Diffusion of molecules through the almond cell wall
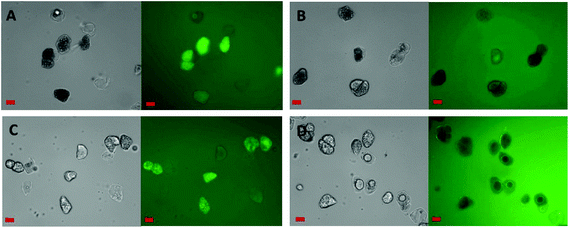
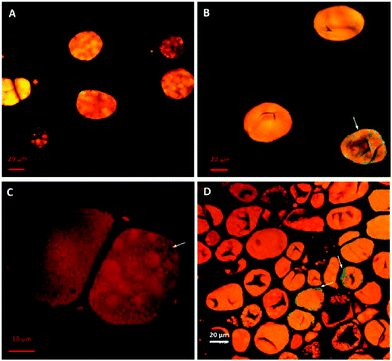
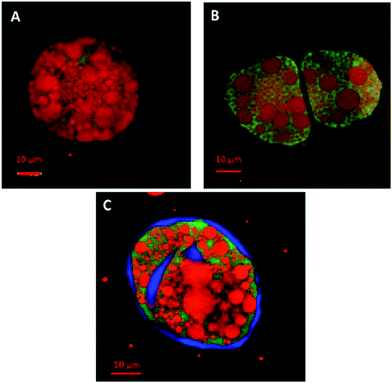
Fig. 3 shows the penetration of 20 kDa dextran (Rg ∼ 3.4 nm) into the cells for both raw and roasted almonds but this did not occur when using 40 kDa dextran (Rg ∼ 5.0 nm). It is presumed therefore that lipases with a molecular weight of ∼50 kDa would not be able to penetrate the cell wall of the almond cells; however, molecular weight alone is not sufficient to characterise the size of a biopolymer. Indeed, other information such as shape, charge (pH environment) and behaviour in solution are necessary. Therefore further experiments were carried out using fluorescently-labelled pancreatic lipase to confirm whether the enzyme was able to penetrate the cell wall.Localisation of fluorescently-labelled PPL was first performed with oil bodies isolated from raw almond cells. In Fig. 4, clusters of labelled PPL (green colour) are visible in the vicinity of the oil droplets. The apparent absence of lipase at the surface of lipid droplets could be due to the fact that only a small fraction of the lipase adsorbed to the interface as has been reported for various systems (e.g. monolayers).39 Since these experiments were performed in the presence of bile salts, it is also known that these strong surfactants have an impact on the partitioning of the lipase between the aqueous phase and the water–lipid interface (i.e. competition for the interface).40 Lipase can thus move to and from the bulk phase and the interface by rapid adsorption–desorption events by a process referred to as the hopping mechanism.41 An apparent reduction in the size of some oil bodies confirmed that lipolysis had taken place (Fig. 4B and C).
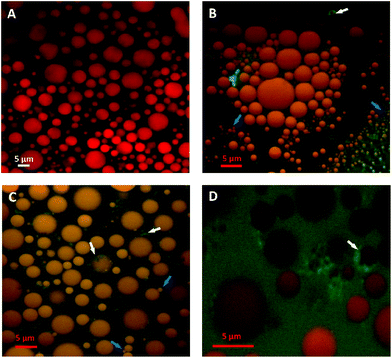
Since pancreatic lipase also displayed some activity towards almond lipids encapsulated in cells, as discussed above, its diffusion into almond cells was studied. However, the experiments with whole almond cells revealed that their lipid content was still mostly intact after extended incubation times, even after 20 h (Fig. 5). One interesting observation was the uneven distribution of the labelled lipases between the intra- and extracellular environments, so that the bulk of green fluorescent areas appeared in some, probably damaged, cells (Fig. 6). The oil bodies inside these cells have lost their integrity (i.e. there has been coalescence), which probably occurred during the preparation of the separated cells. Unfortunately, it was virtually impossible in our laboratory to obtain a preparation devoid of any broken or fragmented cells; nevertheless, the majority of the cells shown in Fig. 5D seemed to be intact.
4. Discussion
The lipids from many foods (e.g. milk, dairy products, egg and meat) are likely to be released (bioaccessible) during the early stages of digestion to form lipid droplets in the proximal GI tract, which are then available for hydrolysis by lipases.42 In some plant foods however, such as legumes, tree nuts and cereals, a variable but significant proportion of the intracellular nutrients, including lipid, can remain entrapped within the cells of the plant tissue at later stages of digestion and reach more distal parts of the gut.7,38,43,44 The results of our present study highlight how important the physical integrity of the cell walls (i.e. structurally-intact dietary fibre) is in regulating lipid bioaccessibility, as seen by the marked reductions in FFA release and lipolysis rates of lipid encapsulated by a cell wall.2Previous studies have already shown that the water-soluble polysaccharides of cell walls (i.e. ‘soluble dietary fibre’) have the capacity to inhibit lipid digestion in different ways including binding to bile salts, interfering with the emulsification process, increasing the viscosity of intestinal content, and by interacting with lipase or lipase substrates.45,46 However, the role of the cell wall barrier in plant foods in restricting lipid digestion has received much less attention. Nevertheless, structurally-intact cell walls also appear to limit lipid digestibility by encapsulating lipid and preventing lipid release and/or lipase from having direct access to intracellular lipid.2,7 In our previous study using 2 mm almond cubes,3 we reported that although most of the lipid remained encapsulated after ≤3 h of digestion in vivo, at later stages of digestion (≥12 h) some of the intracellular lipid was lost from seemingly intact cells located beneath the fractured surface layer. Two hypotheses, which are not mutually exclusive, arise from these observations: (1) the lipids may have diffused out of the intact cells underlying the fractured layer to reach the extracellular environment where they were then hydrolysed by lipase, and/or (2) the lipase may have diffused through the different cell layers and cell walls to degrade the TAG originally inside the ostensibly intact cells. The lipolytic products could then potentially diffuse into the extracellular environment. Both these mechanisms may operate and explain the disappearance of lipid from intact almond cells, but a critical factor in this process could be the permeability of the cell walls. Thus the rate and extent of lipid loss from these cells are likely to be highly dependent on the natural porosity of the cell walls and/or, as previously reported, the introduction of small cracks/fissures during oral and mechanical processing.8,19 The results of the current study, showing hydrolysis of lipid in laboratory-separated cells, suggest that during cell preparation the cell walls became more permeable, perhaps as a result of changes to the pectic material in the middle lamella,7,19,30 or even physical damage, hence exposing the intracellular lipid.
Before reaching the encapsulated lipids inside the almond cell, the enzyme has to cross different barriers, including the cell wall and the oil body monolayer, and perhaps interact with components of a different nature (e.g. polysaccharides, phospholipids and proteins), thus slowing down the lipolytic process. The FFA release and lipolysis rates, reported in the current study are likely to reflect these physico-chemical processes and also the efflux of lipolysis products. However, careful interpretation of the data is required when using separated almond cells in the digestion experiments. Such preparations also contain some damaged cells, in which the lipid substrate is immediately available to the lipase, as well as intact cells that are protected from lipolysis by the cell wall barriers. The high initial reaction rates, but low amount of FFA released from separated cells relative to the oil bodies, provided further evidence that some of the lipid in the preparation was freely available, and thus rapidly hydrolysed, whereas the encapsulated substrate remained undigested.
The almond cell wall is a complex polysaccharide matrix that reduces the accessibility of the lipase to the intracellular TAG and thus impairs hydrolysis as shown by the decrease in lipid digestibility in cells compared with free oil bodies (i.e. more than a 3-fold difference in FFA release). If the TAG hydrolysis takes place in the intracellular compartment, the enzyme has to be able to penetrate the almond cell via ‘pores’ in the cell walls of the polymer matrix, including plasmodesmata. The size range of cell wall pores of different plants has been estimated to be between 3.5 to 5.2 nm.31 Differences in composition and structure of the cell wall matrix can affect the size of these pores.47 Gastric and pancreatic lipases (50 kDa) have a radius of gyration (Rg) of about 1.7 and 1.9 nm, respectively.48,49 This is below the cell wall pore size and so free diffusion of the lipase through the cell wall may be possible theoretically. Diffusion experiments in the current study (Fig. 3) using FITC-labelled dextran revealed that dextran with a Rg of 3.4 nm (20 kDa) penetrated the almond cell wall whereas dextran with a Rg of 5.0 nm (40 kDa) did not. However, despite the relatively lower Rg of pancreatic lipase compared with the dextran, the labelled enzyme did not appear to diffuse into intact almond cells. The pancreatic lipase seemed to penetrate only separated cells with damaged cells walls (i.e. cells with increased porosity).
Pancreatic lipase is active towards emulsions, monolayers and oil bodies.12,50 Consequently, once inside the lipid-rich almond cell, the enzyme should theoretically be able to hydrolyse efficiently the TAG contained in the oil bodies. Lipolysis of oil bodies is facilitated by their small size that provides a large surface area per volume unit (0.27 m2 mL−1 for crude oil bodies, expressed as a fraction of the total volume of oil bodies in one mL). The phospholipids present in oil body membranes are likely to slow down the lipolysis by lipase. Beisson and colleagues showed previously however that the addition of phospholipase did not enhance the hydrolysis of TAG in oil bodies by pancreatic lipase.12 The absence of proteases in that particular investigation may provide an explanation for these results since proteases are also involved in the breakdown of proteins found at the surface of oil bodies. Indeed, phospholipid hydrolysis seems to occur only when the oleosins are removed.51 Beisson et al. also showed that oleosins were partially protected from protease digestion because of the central hydrophobic domain they contain.52 A more recent study performed on almond milk demonstrated that the digestion of the proteins (amandin and oleosin) by pepsin and subsequently trypsin and chymotrypsin affected the microstructure of the oil bodies and permitted their lipolysis.53 Furthermore the bile salts are likely to have displaced any amphiphilic molecules present at the interface including oleosins and phospholipids, the interface thus covered by the bile salts would have promoted colipase and lipase adsorption, and subsequently lipolysis.54
Our results indicate that the roasting process had a relatively minor impact on the extent and rate of lipolysis of almond cells, although lipolysis values were lower for the oil bodies from roasted almonds compared with the raw sample. It appears that the roasting procedure compromised the integrity of the oil bodies, which has encouraged coalescence to occur, as shown by the increase in their particle size with average values of ∼2.6 and 3.8 μm for oil bodies from raw and roasted almonds, respectively. This decrease in the relative surface area to volume ratio of oil bodies from roasted almonds may have reduced the availability of TAG on the oil body surface for lipase action.
Localisation of pancreatic lipase within the almond cells and oil bodies provided further information about the mechanisms governing lipolysis in almonds. The loss of structural integrity of the intracellular oil bodies, caused by the preparation of the separated cells, led to coalescence of these lipids, which could not easily pass through the cell wall and thus remained inside the cell (Fig. 5). Lipase on the other hand appeared to be capable of reaching the intracellular compartment but only as a result of disruption of the cell wall structure and/or increased porosity of the cell wall. It seems reasonable to conclude that the permeability of the cell wall increased because of the treatment used to separate the cells. A video recording (ESI†) of a 3 h digestion of intact and ‘damaged’ almond cells by pancreatic lipase displayed no visual modification of the overall cell structure apart from the diffusion of fluorescently-labelled lipase into the damaged cells and alteration in the size of the oil bodies. Intact cells were identified in these digested samples by the lack of any evidence showing lipase penetration into the cell or damage to the oil bodies. Nevertheless, in all the digestibility experiments most of the lipid was still found to be enclosed inside these separated cells. If this behaviour occurs in humans following the ingestion of almonds, then the lipid content of almond tissue would remain unavailable and undigested on reaching the colon. Previous human studies from our group have already provided evidence of the low digestibility of almond lipid, with some of the intracellular lipid fermented by microflora in the large intestine and the remaining undigested lipid being excreted.3,7,9
5. Conclusions
The results from this work provide clear evidence that the cell walls of almond cells act as a physical barrier to lipid digestibility. Although pancreatic lipase was observed to diffuse through the damaged cell walls to some extent, so that some intracellular lipolysis may have occurred, the majority of the lipids remained enclosed within the intact cells even after 20 h of incubation. Based on the results of the current study and other recent observations,2,3,8,19 we can suggest a possible classification of different populations of almond particles according to the structural integrity and behaviour of their lipid-rich cells during digestion. Thus, depending on lipid release (bioaccessibility) and digestion patterns, we can classify the cells in the following way: (1) cells that are completely ruptured (e.g. by mastication or mechanical processing) on the fractured tissue surfaces have high lipid bioaccessibility and availability for digestion; (2) cells that are less intact and contain microfissures, which are more likely to be located in cell layers immediately below the fractured surfaces, have lower levels of lipid release and digestion than ruptured cells; and (3) cells that are completely intact, with no apparent loss of cell wall integrity, and located in the inner regions of almond particles display negligible lipid release and digestion. Encapsulated lipid contained in intact (undamaged) almond cells can only be digested by lipases that slowly diffuse through the cell wall matrix and even if lipolysis takes place, hydrolysed products have to leak out of the cells before they are available for absorption. Such a mechanism of lipid release and digestion is likely to be very slow.In conclusion, our results provide convincing evidence that, although lipase seems to penetrate the cell wall of the damaged cells, intact almond cells retain intracellular lipid even after long periods of digestion and that the cell wall is an effective physical barrier to lipolysis. These observations explain why in human studies the majority of lipid in almonds is undigested in the upper GI tract.3,7,9 This study also provides further explanation on the discrepancy between the amount of calories present in almond seeds as calculated by the Atwater factor and the actual metabolizable energy.10 We believe these results improve our understanding of the complex physical and biochemical degradation of lipid and other macronutrients in heterogeneous plant foods.
Abbreviations
| BCA | Bicinchoninic acid |
| CLSM | Confocal laser scanning microscopy |
| DAG | Diacylglycerols |
| FFA | Free fatty acid |
| FITC | Fluorescein isothiocyanate |
| GI | Gastrointestinal |
| MAG | Monoacylglycerols |
| PPE | Porcine pancreatic extract |
| PPL | Porcine pancreatic lipase |
| R g | Radius of gyration |
| RGE | Rabbit gastric extract |
| SDS | Sodium dodecyl sulphate |
| TAG | Triacylglycerols |
| TLC | Thin layer chromatography |
Acknowledgements
We thank Dr Sawsan Amara for sharing her expertise on lipid analysis, Dr Balazs Bajka for his help with the microscopy, Neil Rigby for his assistance with the labelling of lipase, Prof. Peter Wilde for valuable discussions and Dr Karen Lapsley (Almond Board of California) for providing the almond seeds. This work was funded by the BBSRC DRINC project BB/H004866/1 and COST action Infogest (FA1005). Dr Myriam Grundy was in receipt of BBSRC studentship award (reference no. BB/H531994/1).References
- C. H. Edwards, F. J. Warren, P. J. Milligan, P. J. Butterworth and P. R. Ellis, Food Funct., 2014, 5, 2751–2758 CAS.
- M. M.-L. Grundy, P. J. Wilde, P. J. Butterworth, R. Gray and P. R. Ellis, Food Chem., 2015, 185, 405–412 CrossRef CAS PubMed.
- G. Mandalari, R. M. Faulks, G. T. Rich, V. Lo Turco, D. R. Picout, R. B. Lo Curto, G. Bisignano, P. Dugo, G. Dugo, K. W. Waldron, P. R. Ellis and M. S. J. Wickham, J. Agric. Food Chem., 2008, 56, 3406–3416 CrossRef PubMed.
- D. J. A. Jenkins, C. W. C. Kendall, A. R. Josse, S. Salvatore, F. Brighenti, L. S. A. Augustin, P. R. Ellis, E. Vidgen and A. V. Rao, J. Nutr., 2006, 136, 2987–2992 CAS.
- S.-C. Li, Y.-H. Liu, J.-F. Liu, W.-H. Chang, C.-M. Chen and O. C. Y. Chen, Metabolism, 2011, 60, 474–479 CrossRef CAS PubMed.
- S. Y. Tan and R. D. Mattes, Eur. J. Clin. Nutr., 2013, 67, 1205–1214 CrossRef CAS PubMed.
- P. R. Ellis, C. W. Kendall, Y. Ren, C. Parker, J. F. Pacy, K. W. Waldron and D. J. Jenkins, Am. J. Clin. Nutr., 2004, 80, 604–613 CAS.
- M. M.-L. Grundy, T. Grassby, G. Mandalari, K. W. Waldron, P. J. Butterworth, S. E. Berry and P. R. Ellis, Am. J. Clin. Nutr., 2015, 101, 25–33 CrossRef CAS PubMed.
- S. E. Berry, E. A. Tydeman, H. B. Lewis, R. Phalora, J. Rosborough, D. R. Picout and P. R. Ellis, Am. J. Clin. Nutr., 2008, 88, 922–929 CAS.
- J. A. Novotny, S. K. Gebauer and D. J. Baer, Am. J. Clin. Nutr., 2012, 96, 296–301 CrossRef CAS PubMed.
- A. H. C. Huang, Curr. Opin. Struct. Biol., 1994, 4, 493–498 CrossRef CAS.
- F. Beisson, N. Ferte, S. Bruley, R. Voultoury, R. Verger and V. Arondel, Biochim. Biophys. Acta, Mol. Cell Biol. Lipids, 2001, 1531, 47–58 CrossRef CAS.
- N. C. Carpita and D. M. Gibeaut, Plant J., 1993, 3, 1–30 CrossRef CAS PubMed.
- F. Dourado, A. Barros, M. Mota, M. A. Coimbra and F. M. Gama, J. Agric. Food Chem., 2004, 52, 1364–1370 CrossRef CAS PubMed.
- E. Bauer, S. Jakob and R. Mosenthin, Asian–Australas J. Anim., 2005, 18, 282–295 CrossRef CAS.
- F. Carriere, J. A. Barrowman, R. Verger and R. Laugier, Gastroenterology, 1993, 105, 876–888 CAS.
- J.-C. Bakala N'Goma, S. Amara, K. Dridi, V. Jannin and F. Carriere, Ther. Delivery, 2012, 3, 105–124 CrossRef.
- E. E. Groopman, R. N. Carmody and R. W. Wrangham, Am. J. Phys. Anthropol., 2015, 156, 11–18 CrossRef PubMed.
- T. Grassby, D. R. Picout, G. Mandalari, R. M. Faulks, C. W. C. Kendall, G. T. Rich, M. S. J. Wickham, K. Lapsley and P. R. Ellis, Food Funct., 2014, 5, 3096–3106 CAS.
- D. A. White, I. D. Fisk, S. Makkhun and D. A. Gray, J. Agric. Food Chem., 2009, 57, 5720–5726 CrossRef CAS PubMed.
- S. Makkhun, A. Khosla, T. Foster, D. J. McClements, M. M.-L. Grundy and D. A. Gray, Food Funct., 2015, 6, 125–134 Search PubMed.
- H. Moreau, R. Verger, D. Lecat and J. L. Junien, France Pat, EP19870401984, 1988 Search PubMed.
- H. Moreau, Y. Gargouri, D. Lecat, J. L. Junien and R. Verger, Biochim. Biophys. Acta, 1988, 959, 247–252 CrossRef CAS.
- M. L. Anson, J. Gen. Physiol., 1938, 22, 79–89 CrossRef CAS.
- R. Verger, G. H. de Haas, L. Sarda and P. Desnuelle, Biochim. Biophys. Acta, 1969, 188, 272–282 CrossRef CAS.
- C. Chapus, P. Desnuelle and E. Foglizzo, Eur. J. Biochem./FEBS, 1981, 115, 99–105 CrossRef CAS PubMed.
- A. Sze, D. Erickson, L. Q. Ren and D. Q. Li, J. Colloid Interface Sci., 2003, 261, 402–410 CrossRef CAS.
- J. Folch, M. Lees and G. H. Sloane Stanley, J. Biol. Chem., 1957, 226, 497–509 CAS.
- K. Andrieux, P. Lesieur, S. Lesieur, M. Ollivon and C. Grabielle-Madelmont, Anal. Chem., 2002, 74, 5217–5226 CrossRef CAS.
- O. Baron-Epel, P. K. Gharyal and M. Schindler, Planta, 1988, 175, 389–395 CrossRef CAS PubMed.
- N. C. Carpita, D. Sabularse, D. Montezinos and D. P. Delmer, Science, 1979, 205, 1144–1147 CAS.
- A. Altan, K. L. McCarthy, R. Tikekar, M. J. McCarthy and N. Nitin, J. Food Sci., 2011, 76, E212–E221 CrossRef CAS PubMed.
- S. Makkhun, PhD, University of Nottingham, 2012.
- M. Barros, L. F. Fleuri and G. A. Macedo, Braz. J. Chem. Eng., 2010, 27, 15–29 CrossRef CAS.
- S. Gallier, K. C. Gordon and H. Singh, Food Chem., 2012, 132, 1996–2006 CrossRef CAS PubMed.
- S. Bonsegna, S. Bettini, R. Pagano, A. Zacheo, V. Vergaro, G. Giovinazzo, G. Caminati, S. Leporatti, L. Valli and A. Santino, Appl. Biochem. Biotechnol., 2011, 163, 792–802 CrossRef CAS PubMed.
- R. J. Hunter, Zeta Potential in Colloid Science: Principles and Applications, Academic Press, London, 1981 Search PubMed.
- G. Mandalari, M. M.-L. Grundy, T. Grassby, M. L. Parker, K. L. Cross, S. Chessa, C. Bisignano, D. Barreca, E. Bellocco, G. Lagana, P. J. Butterworth, R. M. Faulks, P. J. Wilde, P. R. Ellis and K. W. Waldron, Br. J. Nutr., 2014, 112, 1521–1529 CrossRef CAS PubMed.
- A. Benarouche, V. Point, G. Parsiegla, F. Carriere and J. F. Cavalier, Colloids Surf., B, 2013, 111C, 306–312 CrossRef PubMed.
- V. Delorme, R. Dhouib, S. Canaan, F. Fotiadu, F. Carriere and J.-F. Cavalier, Pharm. Res., 2011, 28, 1831–1842 CrossRef CAS PubMed.
- H. Haiker, H. Lengsfeld, P. Hadvary and F. Carriere, Biochim. Biophys. Acta, Mol. Cell Biol. Lipids, 2004, 1682, 72–79 CrossRef CAS PubMed.
- M. C. Michalski, C. Genot, C. Gayet, C. Lopez, F. Fine, F. Joffre, J. L. Vendeuvre, J. Bouvier, J. M. Chardigny and K. Raynal-Ljutovac, Prog. Lipid Res., 2013, 52, 354–373 CrossRef CAS PubMed.
- C. H. Edwards, M. M.-L. Grundy, T. Grassby, D. Vasilopoulou, G. S. Frost, P. J. Butterworth, S. E. Berry, J. Sanderson and P. R. Ellis, Am. J. Clin. Nutr., 2015, 102, 791–800 CrossRef PubMed.
- A. S. Levine and S. E. Silvis, N. Engl. J. Med., 1980, 303, 917–918 CrossRef CAS PubMed.
- D. Lairon, B. Play and D. Jourdheuil-Rahmani, J. Nutr., 2007, 18, 217–227 CAS.
- P. Gunness and M. J. Gidley, Food Funct., 2010, 1, 149–155 CAS.
- A. Fleischer, M. A. O'Neill and R. Ehwald, Plant Physiol., 1999, 121, 829–838 CrossRef CAS PubMed.
- G. H. Peters, D. M. van Aalten, O. Edholm, S. Toxvaerd and R. Bywater, Biophys. J., 1996, 71, 2245–2255 CrossRef CAS.
- A. Selvan, C. Seniya, S. N. Chandrasekaran, N. Siddharth, S. Anishetty and G. Pennathur, FEBS Lett., 2010, 584, 4599–4605 CrossRef CAS PubMed.
- R. Verger, in Lipases, ed. B. Borgström and H. L. Brockman, Elsevier Science, Amsterdam, 1984, pp. 83–150 Search PubMed.
- J. T. C. Tzen and A. H. Huang, J. Cell Biol., 1992, 117, 327–335 CrossRef CAS.
- F. Beisson, N. Ferte, R. Voultoury and V. Arondel, Plant Physiol. Biochem., 2001, 39, 623–630 CrossRef CAS.
- S. Gallier and H. Singh, Food Funct., 2012, 3, 547–555 CAS.
- J. Maldonado-Valderrama, P. Wilde, A. Macierzanka and A. Mackie, Adv. Colloid Interface Sci., 2011, 165, 36–46 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5fo00758e |
| This journal is © The Royal Society of Chemistry 2016 |