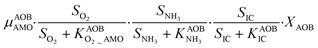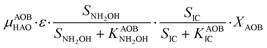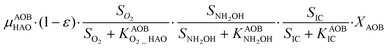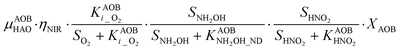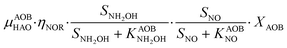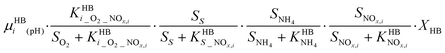A consilience model to describe N2O production during biological N removal†
C.
Domingo-Félez
 and
B. F.
Smets
*
and
B. F.
Smets
*
Department of Environmental Engineering, Technical University of Denmark, Miljøvej Building 113, 2800 Kongens Lyngby, Denmark. E-mail: bfsm@env.dtu.dk; Fax: +45 4593 2850; Tel: +45 4525 1600
First published on 11th October 2016
Abstract
Nitrous oxide (N2O), a potent greenhouse gas, is produced during biological nitrogen conversion in wastewater treatment operations. Complex mechanisms underlie N2O production by autotrophic and heterotrophic organisms, which continue to be unravelled. Mathematical models that describe nitric oxide (NO) and N2O dynamics have been proposed. Here, a first comprehensive model that considers all relevant NO and N2O production and consumption mechanisms is proposed. The model describes autotrophic NO production by ammonia oxidizing bacteria associated with ammonia oxidation and with nitrite reduction, followed by NO reduction to N2O. It also considers NO and N2O as intermediates in heterotrophic denitrification in a 4-step model. Three biological NO and N2O production pathways are accounted for, improving the capabilities of existing models while not increasing their complexity. Abiotic contributions from NH2OH and HNO2 reactions are also included. The consilient model structure can theoretically predict NO and N2O emissions under a wide range of operating conditions and will help develop mitigation strategies.
Water impactWastewater treatment operations are anthropogenic sources of nitrous oxide (N2O), a potent greenhouse gas and ozone depleting compound. While energy efficiency has been the recent focus of technology development in wastewater management, the carbon footprint of a wastewater treatment plant is utmost sensitive to its N2O emissions. Informed by a review of known biological and chemical N2O producing mechanisms, an improved mathematical model structure that may help the development of N2O mitigation strategies for full-scale treatment operations is proposed. |
1. Introduction
Nitrous oxide (N2O) is a potent greenhouse gas emitted from wastewater treatment processes during biological nitrogen conversions. Due to its high radiative forcing, the carbon footprint of wastewater treatment plants (WWTPs) is highly sensitive to N2O emissions,1 which vary largely between WWTPs.2 Biologically mediated, N2O can be produced during nitrification and exists as an obligate intermediate during denitrification.3 The mechanisms and regulations of N2O production in these processes are still under investigation, and identification and better understanding of the key variables driving N2O production are necessary.With the final goal of mitigating N2O emissions, mathematical models are useful tools to translate our understanding of biological phenomena into equations and predictions. Models must be developed by identifying, combining and translating into mathematical equations the key processes and influencing variables that govern N2O dynamics.
The first models that described autotrophic N2O production considered only one of two pathways, either the nitrifier nitrification (NN) or the nitrifier denitrification (ND) pathway. Each pathway was modelled with different levels of complexity affecting the number of considered variables and substrate or inhibition dependencies.4 However, the range of applicability of single pathway models is narrow.5 Newly developed models consider both nitrifier pathways, better capturing the state of knowledge on mechanisms. However, the simplification proposed to one of the N2O pathways might not always be true, thus limiting their applicability.6,7
In combination with N2O production, physicochemical processes transfer N2O from the liquid to the gas phase resulting in actual N2O emissions. Mass-transfer processes are relatively well studied, and our emphasis here is on the production processes.8 A comprehensive model structure should be capable of describing N2O production under a wide range of operating conditions. By increasing the model complexity with additional components and parameters, model predictions can be more accurate. However, model over-parameterization challenges the calibration process and increases parameter identifiability problems. The large variability of reported model parameters in N2O models is likely an indicator of the limited structural and practical identifiability of the models. For example, reported substrate affinity constants for nitrite (NO2−) and nitric oxide (NO) reduction in current N2O models range across almost two orders of magnitude (Table S1†). Assessing calibration results helps one to discriminate between models by comparing parameter identifiability or prediction uncertainty.9 It is therefore necessary to obtain simple, yet sufficiently complete, model structures that capture the fundamental mechanisms of N2O during wastewater treatment operations.
The aims of this communication are (i) to identify key processes and variables driving N2O production during N removal and (ii) to propose a simple yet comprehensive model structure capable of describing reported N2O observations. The model should increase the applicability of existing N2O models and be consistent with current knowledge on N2O production mechanisms.
2. N2O production during wastewater treatment operations
Biological nitrogen removal typically is a two-step process where nitrifying bacteria oxidize ammonia (NH3) to nitrogen oxides (NOx−), followed by anoxic NOx− reduction to dinitrogen gas (N2) with organic matter (COD) as an electron source usually by heterotrophic denitrifying bacteria. N2O can be produced by ammonia oxidizing bacteria (AOB) and archaea during oxidation of ammonia to nitrite (NO2− or, more correctly, nitrous acid (HNO2)) and by heterotrophic bacteria (HB) as an obligate intermediate of denitrification. We do not discuss the scenario of completely autotrophic N removal which would involve a combination of aerobic and anaerobic ammonium oxidation (anammox) as anammox bacteria have no known N2O production mechanisms.Autotrophic N2O production
The oxidation of NH3 with molecular oxygen to hydroxylamine (NH2OH) by ammonia monooxygenase requires two electrons. These electrons are supplied by the subsequent oxidation of NH2OH to HNO2 consuming molecular water, which releases four electrons, while oxygen is reduced in the terminal oxidase. Aerobic NH2OH oxidation is therefore the electron-yielding process for AOB growth10,11 and essential for energy production.AOB can produce N2O from the incomplete oxidation of NH2OH to HNO2via NO or to its reduced form HNO.12 This process is referred to as nitrifier nitrification (NN),13 recently shown to be uncoupled from HNO2 production.14 In addition, AOB have a denitrifying functionality, where a set of NO2−- and NO-reducing enzymes (NIR, NOR) can result in N2O production termed nitrifier denitrification (ND) (this has been confirmed by genomic analysis of Nitrosomonas europaea15). Under low dissolved oxygen (DO) conditions, HNO2 is reduced to N2O via NO in the presence of an electron donor such as NH2OH.11,16,17 DO differently affects the expression of NIR and NOR enzymes. NO production, regulated by NIRK, is favoured under anoxic conditions,18–21 while NORB activity is upregulated under oxic conditions.22 Moreover, the enzymology of AOB suggests the presence of additional NO reducing catalytic units similar to the NOR cluster such as the CYT554.23,24
Varying DO levels are common during wastewater treatment operations which, together with dynamic HNO2 concentrations, can lead to imbalances in NO and N2O emissions.21,25 Thus, process conditions can switch the dominant AOB-associated N2O production pathway between NN and ND.
pH levels have two distinct effects on autotrophic N2O production. First, on the enzymatic level, maximum activities have been described as pH-dependent.26 Second, the true substrates available for AOB enzymes AMO and NIR are NH3 and HNO2. The actual concentrations of these species are in a pH-dependent equilibrium with their ionized counterparts NH4+ and NO2−![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 27 (pKa,HNO2 = 3.25, pKa,NH4+ = 9.25, 25 °C).28
27 (pKa,HNO2 = 3.25, pKa,NH4+ = 9.25, 25 °C).28
Inorganic carbon (IC) is the carbon source subject to C fixation during AOB growth. At limited IC availability, NH3 is oxidized at a lower rate due to increased cellular maintenance energy demand, with a simultaneous decrease in N2O production.29 However, at the same NH3 oxidation rates, low IC levels increase the fraction of N2O produced.30 Depending on the nitrogen removal system, wastewaters can have varying IC levels. Due to the heterotrophic oxidization of the organic content of conventional urban wastewater, IC is typically in excess for autotrophic growth, but high N-strength wastewaters with a lower C/N ratio may result in IC limited AOB growth.31
Heterotrophic N2O production
Under DO limited conditions, canonical denitrifiers respire NO3−, NO2−, NO and N2O anaerobically, catalysed by enzymes encoded by nar, nir, nor, and nosZ genes. Heterotrophic denitrifiers constitute a highly modular microbiome with very different distributions of denitrifying genes.32 Cellular co-occurrence of nar, nir and nor genes without nosZ would yield a net N2O producer, while non-denitrifier N2O reducers carrying an atypical nosZ have been identified and may act as N2O sinks.33 The potential of a heterotrophic community to serve as a N2O source or sink may be governed by the diversity and relative abundance of the nosZ gene with respect to nar, nir and nor genes.33,34The rate of NOx− reduction has been suggested as inhibited by products in the respiratory chain, such as NO3− reduction would be influenced by the concentration of further terminal electron acceptors and the number of other reductases.35 In the presence of both N2O and NO2−, the N2O reductase competes with NO2− reductase for electrons from the reduced cytochrome c.36 In addition, the four enzymes responsible for denitrification may compete for electrons with cytochrome oxidases, where O2 is reduced. The reversible inhibitory effect of DO on NOx− reduction is similar for each step.36,37 N2O reduction is the most sensitive step towards DO, and its inhibition will promote N2O accumulation compared to the other N species.38
A limited flow of electron donors (as provided by the external chemical oxygen demand, COD) can also slow down NOx− reduction rates. Therefore, N2O accumulation may result due to a reduced N2O reduction rate due to a lower electron affinity compared to previous reduction steps. Consequently, side stream processes, characterized by high N content and low COD content, are potential hotspots for heterotrophic N2O production.3
Moreover, the activities of enzymes encoded by the nir, nor and nosZ genes, located in the periplasm, are pH-dependent, with different optima for each denitrification step.39 Thus, pH will have a direct effect on the concentration of intermediates. Specifically for N2O, high and low pH values promote its consumption and accumulation, respectively.40
Abiotic N2O production
Two chemical reactions driven by NH2OH41 can occur at relevant rates under wastewater treatment conditions.42,43| 4NH2OH → N2O + 2NH3 + 3H2O | (1) |
| NH2OH + HNO2 → N2O + 2H2O | (2) |
NH2OH can decompose to N2O at high pH (eqn (1); the acidic form NH3OH+ is more stable,44 pKa = 5.9 at 25 °C). In the second reaction, an N–N linkage is formed by N-nitrosation of NH2OH, a nucleophile, with a nitrosating agent, HNO2, at low pH45 (eqn (2)). Thus, independently from the main driving process (e.g. nitrification or denitrification) and the environmental conditions (e.g. aerobic or anaerobic), biotically-driven (as NH2OH is biotically produced) abiotic N2O production is possible in WWTPs.
While previously considered to be insignificant, NH2OH concentrations from highly N-loaded wastewaters can be substantial (0.03–0.11 mg N L−1),46 and abiotic N2O production may have been underestimated.47 For example, a nitritation reactor treating reject water (high AOB activity and NO2− accumulation) had a 1.1% abiotic N2O emission factor.46
3. Modelling N2O dynamics
With the final purpose of mitigating N2O emissions, it is critical to accurately quantify the contribution of individual N2O production and consumption pathways to the total N2O pool. Process models are useful tools for this purpose, and several models have been proposed for each of the aforementioned biological N2O production pathways.48 Models vary based on the number of processes and variables considered and on the mathematical description of the process rates.AOB driven N2O models
Initially, single-pathway models were proposed describing either the NN or the ND pathway. The main difference between models is with regards to the stoichiometric coefficients, the number of considered substrates, the identity of the direct electron donor, and the inclusion or absence of substrate inhibition. Initial models described NO and N2O production as directly dependent on NH4+, DO and NO2− levels.49,50 In subsequent models, NH2OH was considered an intermediate of NH3 oxidation, allowing the NN pathway to be modelled as a fraction of NH2OH oxidation to NO2−, either via NOH51 or via NO.52 In the ND pathway, NH2OH acts as an electron donor for the consecutive reduction of NO2− to N2O via NO.53 To increase their predicting capabilities, newer models consider unionized species as the true substrates (NH3–HNO2vs. NH4+–NO2−) and more complex functions are included in the process rates, resulting in more model parameters.54 However, N2O dynamics cannot be captured with single-pathway models, and recent models that combine the NN and ND pathways provide better descriptions of N2O production than single-pathway models.6,7The two-pathway AOB model by Pocquet et al.7 considers NH3 and HNO2 as substrates and NH2OH as the electron donor for both NO and HNO2 reduction to N2O in the NN and ND pathways, respectively. NO is formed from NH2OH oxidation, and HNO2 is formed from subsequent NO oxidation: in other words, all NH2OH is first converted to NO, which is considered as a substrate for subsequent oxidation to HNO2. In this model, NH2OH oxidation to NO is modelled as consuming oxygen to maintain COD mass balance continuity, but this is in contradiction with the fact that no oxygen is actually consumed in this reaction.11,16 Hence, the Pocquet model implies that NH2OH oxidation is only feasible under aerobic conditions. The ND pathway is described as a one-step process wherein HNO2 is reduced directly to N2O, and the intermediate NO is ignored. Ignoring NO is necessary in the Pocquet model for mathematical reasons: the formed NO in the ND pathway would be a substrate in the NN pathway and be oxidized to HNO2, which in turn could be reduced to NO in the ND pathway. Ignoring NO as an intermediate in the ND pathway is not in agreement with reality but avoids a futile NO cycling between NN and ND pathways.
In a different approach, global cellular oxidation (electron generating) and reduction (electron consuming) reactions in AOB are linked by a common pool of electron carriers, represented by one model component.6 This model aggregates all intracellular electron carriers into one component, which cannot be experimentally quantified. In this model, NH2OH and NO oxidation compete for oxidized electron carriers as cosubstrates and produce reduced electron carriers. The reduction reactions of O2, O2/NH4+, NO and NO2− compete for the reduced carriers, which are transformed back to their oxidized forms.6 Oxidative and reductive processes are uncoupled, and competition is described with specific kinetic parameters. Similarly to the previously described two-pathway model, in the ND pathway, a one-step reduction of NO2− to N2O is included.
The two-pathway AOB models are adequate in predicting a shift in NN and ND contributions to the total N2O production at different DO and NO2− concentrations. However, these models would not describe the increased NO emissions at low DO and high NO2− levels as observed in several nitrifying systems.18,19,25,55,56 Hence, ND-associated NO production would be wrongly attributed to the NN pathway, overestimating the NN contribution to total N2O production. As NO is the direct precursor of N2O, and its emissions can be measured, it would seem preferable to retain NO in any model expressions. Experimental data on NO could then help assess and validate proposed mechanisms and model structures.
HB driven N2O models
Two approaches have been widely used to model heterotrophic denitrification. In the electron competition approach, a model component describing a common pool of electrons, originating from carbon oxidation, exists for which the four enzymes in the denitrification respiratory pathway compete.39 In the direct approach, no internal pool of electrons are considered, as carbon oxidation is assumed to provide a non-limiting supply of electrons to all denitrification enzymes.57 Both approaches describe the electron donor and acceptor limitations with a specific Monod dependency for each denitrification step.57,58 The known oxygen inhibition of the HD pathway has been described by either a single inhibition constant or a specific oxygen inhibition constant for each denitrification step.53,57Even though the indirect approach has been heralded as superior as it can potentially describe more data sets, information about newly proposed reaction kinetics is not available in the literature.59
The direct HD modelling approach adequately predicts COD and nitrogen removal for systems with low intermediate accumulation (NO2−, N2O)48 but might be inadequate for systems with high intermediate accumulation levels.
Abiotic N2O models
Systems treating high-strength wastewaters are particularly prone to chemical production of N2O due to high AOB activity and associated high NH2OH concentrations.60 However, only one model has considered abiotic contribution together with biologically-driven N2O production (ND and HD pathways).47 The abiotic contribution was modelled with no pH dependency as a second order reaction for NH2OH and NO2−, limiting the applicability to conditions of constant pH (eqn (2)).4. Model development (NDHA)
An improved and consilient model including all the relevant mechanisms responsible for N2O production during biological N removal is proposed (Table S2†). This model is consilient in its agreement between the mathematical abstraction and the chemical, biochemical and microbiological observations. The NDHA model considers N2O production from the three known biological pathways (![[N with combining low line]](https://www.rsc.org/images/entities/char_004e_0332.gif)
![[D with combining low line]](https://www.rsc.org/images/entities/char_0044_0332.gif)
![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) A) as well as abiotic production (NDH
A) as well as abiotic production (NDH![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ) (Fig. 1). By explicitly considering NO as the direct precursor of N2O production, three distinct biological NO production pathways can be identified while only including quantifiable state variables.
) (Fig. 1). By explicitly considering NO as the direct precursor of N2O production, three distinct biological NO production pathways can be identified while only including quantifiable state variables.
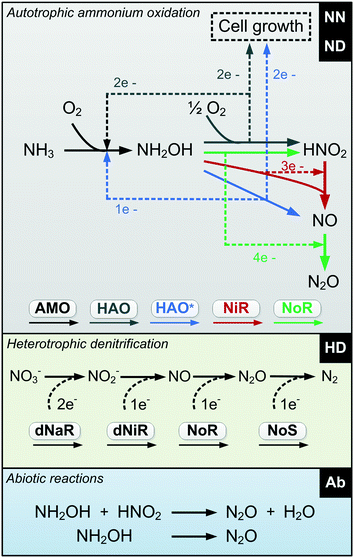 | ||
| Fig. 1 Diagram of the proposed N2O-producing mechanisms occurring during N removal: nitrifier nitrification, nitrifier denitrification, heterotrophic denitrification and abiotic pathways (NDHA). | ||
Different from current AOB driven models, the two autotrophic pathways are distinguished by two NO-producing processes with different DO and HNO2 dependencies. The simplification of current AOB models that ignore NO as an intermediate during ND-driven N2O production is solved: NO is an intermediate of both the NN and the ND pathways. A single autotrophic N2O-producing process accounts for the combined NO reduction. Heterotrophic denitrification is described as a 4-step process, and two chemical reactions, which involve NH2OH and HNO2, describe the abiotic N2O production.
Nitrifier nitrification (NN)
The first process considers NH3 oxidation to NH2OH (P1) (Table S3†). NH2OH can be oxidized incompletely to NONN (P2) – a secondary catalyzed reaction of HAO – or completely to HNO2 – the primary catalysed reaction of HAO – in the presence of DO (P3). The effect of IC limitation on NH3 oxidation is described by a Monod dependency.61 In the NN pathway (P2), NH2OH reacts with H2O;62 the NN process is, therefore, indirectly dependent on the NH3 oxidation rate, reducing the DO dependency only to P1. The fraction of NH2OH oxidized via the NN pathway is described by the factor ε.P1 – AMO: NH3 + O2 → NH2OH
P2 – HAO*: NH2OH → NONN
P3 – HAO: NH2OH + 0.5O2 → HNO2 + H2O
Nitrifier denitrification (ND)
In the ND pathway, HNO2 denitrification to NOND is negatively affected by DO (P4).Different from other two-pathway AOB models, N2O production from its precursor (NO) is described by one process (P5) as there is no evidence of different NO reduction mechanisms within individual cells.23 The NN and ND pathways are, therefore, mainly described by two NO-producing processes with different DO and HNO2 dependencies. These dependencies govern the shift between pathways.24,25 N2ONN production is enhanced at high NH3 and DO levels, while N2OND increases at low DO and high HNO2 levels. By considering NH2OH as an electron donor of both NO and HNO2 reduction, the model minimizes the number of model components and fewer parameters are necessary to describe the electron competition (Table S4†).
The NO/N2O ratio can be used to help elucidate the individual contribution of each pathway during model calibration.7 An advantage of the proposed model is the uncoupling of the NN- and ND-driven NO production, which allows for a more biologically congruent estimate of NO/N2O.
P4 – NIR: 3HNO2 + NH2OH → 4NOND
P5 – NOR: 2(NOND + NONN) + NH2OH → 1.5N2O
Heterotrophic denitrification (HD)
A four-step complete denitrification is considered following the ASM-N model.57 Individual reaction kinetics (pH-dependent), inhibition and substrate affinities are considered for every step as recently suggested for systems with low intermediate accumulation.48 Moreover, because of its wide applicability, the direct approach has been extended to new denitrification models coupled with phosphorus removal.63P6 – HD: NOx,oxidized + COD → NOx,reduced
Heterotrophic consumption and autotrophic production of N2O can occur simultaneously, at different rates, throughout wastewater treatment operations. Ignoring heterotrophic N2O consumption can underestimate the autotrophic production. Thus, an N2O model should always include compatible structures for both the autotrophic and the heterotrophic pathways.64
Abiotic (Ab)
Two biologically-driven abiotic N2O production processes are considered (P7). Nitrification produces NH2OH which can form HNO.65 HNO dimerizes via H2N2O2 to N2O and H2O (eqn (1)). Nitrosation of NH2OH (eqn (2)) with HNO2 has also been postulated as a relevant reaction in partial nitrification reactors.46 Reaction rates are modelled with pH dependent second order kinetics.P7 – Abiotic: NH2OH → N2O; NH2OH + HNO2 → N2O
| (kAbiotic_1·SNH2OH·f(pH)); (kAbiotic_2·SNH2OH·SHNO2) |
Model predictions for every pathway are pH-dependent, due to either substrate speciation or an enzymatic effect on the maximum specific growth rate. Implicit pH calculations also allow for estimations of IC and therefore limitations on AOB growth.66 Aerobic growth of nitrite oxidizing bacteria on FNA and that of heterotrophs on soluble COD are also included.
5. Discussion
Current two-pathway AOB models share the same nitrogenous substrates and reactions to describe one NO and two N2O-producing processes.6,7 The proposed NDHA model adds the denitrification contribution to the NO pool that could not be considered in current models (Table 1).| Pocquet et al. (2016) | Ni et al. (2014) | NDHA | |
|---|---|---|---|
| NH2OH oxidation: steps | 2-step process to HNO2via NO | 2-step process to HNO2via NO | 2 processes: to NO and to HNO2 |
| NH2OH oxidation: e-acceptor | NH2OH and NO oxidation require O2 | Requires O2, NO2− or NO reduction | HNO2 production requires O2, NO does not |
| NH2OH oxidation: anoxic | Not possible | Possible (produces HNO2) | Possible (produces N2O) |
| Direct substrate for HNO2 production | NO | NO | NH2OH |
| Denitrifying NO production | Not considered | Not considered | Considered |
| NO-producing pathways | NN | NN | NN and ND |
| N2O-producing pathways | NN and ND | NN and ND | NN and ND |
| pH-dependent substrate | Yes | No | Yes |
| Additional state variables | No | Yes | No |
| Model parameters (processes) | 13(5) | 18(6) | 13(5) |
The NN pathway is based on NO produced during NH2OH oxidation. Differently from Pocquet et al.7 and in agreement with Ni et al.,6 the production of NONN in the NDHA model does not require the presence of oxygen.
Until now, models have considered HNO2, coupled with an electron donor, as the direct precursor of N2O for the ND pathway.6,7 However, ND-associated NO reduction is not always faster than that of HNO2, leading to NO accumulation.18,19,25,55,56 In the NDHA model, this assumption is resolved and NOND is produced from HNO2 reduction (Fig. 2). Whether the source of NO is NH2OH oxidation or HNO2 reduction will determine the contribution of each autotrophic pathway to N2O production, NN or ND, respectively.
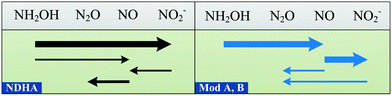 | ||
| Fig. 2 Schematic comparison of the reactions involved in two-pathway AOB models for N2O production. The arrow widths represent typical reaction rates. Model A7 and Model B.6 | ||
Although oxidation and reduction processes are not uncoupled in the NDHA model, the competition for electrons is represented by NH2OH, the common electron donor: HNO2, NO and DO compete for NH2OH instead of reduced electron carriers.
Because of the structural assumption of the current AOB models, NO-associated N2O production is only related to the NN pathway. As well as for the ND pathway, this assumption should be extrapolated to the HD pathway to avoid the NO exchange (simultaneous oxidation–reduction). Consequently, during model calibration, any possible ND or HD contributions to total NO would be falsely associated with the NN pathway. The NDHA model can describe more NO/N2O pathways with the same or fewer parameters than the other models (Table 1).
The same N2O net production rate can result from different individual N2O production/consumption rates. Thus, together with total N2O production, correctly predicting the individual contribution of each pathway is key for N2O models. For example, the mitigation strategy of an autotrophic system with a small N2O sink capacity will differ from that of mixed liquor with a higher N2O consuming capacity.
Advances on N2O models have led to more complete structures that can potentially describe any N2O dynamics data set. However, the structural identifiability of none of the N2O models has ever been assessed, and parameter identifiability analysis, if conducted, is limited to confidence interval depiction. Not all the model parameters are usually estimated from the available data as practical identifiability problems arise due to overparameterization of activated sludge models (ASM).67 Model discrimination studies should therefore critically address calibration results as well as structural limitations. Best-fit parameter estimates provide little information and need to be supplemented with additional metrics (correlation matrix, sensitivity functions, analysis of residuals, estimation biases, etc.) in future model comparisons.
Additional complexity could be added, if necessary, to capture transient phenomena, relevant for systems with dynamic conditions. For example, the physiological state of the biomass can directly affect cellular activity and has been included in denitrifying models.38,68 The high modularity of heterotrophic organisms, lumped into individual parameters for each denitrifying step, could be described by distinct microbial subpopulations and would yield more accurate kinetic parameters.69 However, it is typically out of the scope of ASM models.
6. Conclusions
A consilient mathematical model structure that describes N2O production during biological nitrogen removal is proposed. Three biological pathways, two autotrophic and one heterotrophic, are coupled with abiotic processes. Consistent with experimental studies, the model considers NO as the direct precursor of N2O in all three biologically-driven pathways. This model can describe all relevant NO and N2O production pathways with fewer parameters than other proposed models. A simplified and biologically congruent model will help develop mitigation strategies during wastewater treatment operations.Acknowledgements
This research was funded by the Danish Agency for Science, Technology and Innovation through the Research Project LaGas (12-132633). The authors have no conflict of interest to declare. The fruitful discussions with Mr. Jan-Michael Blum are gratefully acknowledged.References
- D. J. I. Gustavsson and S. Tumlin, Water Sci. Technol., 2013, 68, 887 CrossRef CAS PubMed.
- J. H. Ahn, S. Kim, H. Park, B. Rahm, K. Pagilla and K. Chandran, Environ. Sci. Technol., 2010, 44, 4505–4511 CrossRef CAS PubMed.
- M. J. Kampschreur, H. Temmink, R. Kleerebezem, M. S. M. Jetten and M. C. M. van Loosdrecht, Water Res., 2009, 43, 4093–4103 CrossRef CAS PubMed.
- B.-J. Ni, Z. Yuan, K. Chandran, P. A. Vanrolleghem and S. Murthy, Biotechnol. Bioeng., 2013, 110, 153–163 CrossRef CAS PubMed.
- L. Peng, B.-J. Ni, L. Ye and Z. Yuan, Chem. Eng. J., 2015, 281, 661–668 CrossRef CAS.
- B.-J. Ni, L. Peng, Y. Law, J. Guo and Z. Yuan, Environ. Sci. Technol., 2014, 48, 3916–3924 CrossRef CAS PubMed.
- M. Pocquet, Z. Wu, I. Queinnec and M. Spérandio, Water Res., 2016, 88, 948–959 CrossRef CAS PubMed.
- F. Garcia-Ochoa and E. Gomez, Biotechnol. Adv., 2009, 27, 153–176 CrossRef CAS PubMed.
- D. Dochain and P. A. Vanrolleghem, Dynamic Modelling and Estimation in Wastewater Treatment Processes, IWA Publishing, London, UK, 2001 Search PubMed.
- B. Böttcher and H. P. Koops, FEMS Microbiol. Lett., 1994, 122, 263–266 CrossRef.
- P. de Bruijn, A. A. van de Graaf, M. S. M. Jetten, L. A. Robertson and J. G. Kuenen, FEMS Microbiol. Lett., 1995, 125, 179–184 CrossRef CAS.
- A. B. Hooper and K. R. Terry, Biochim. Biophys. Acta, Enzymol., 1979, 571, 12–20 CrossRef CAS.
- X. Zhu, M. Burger, T. A. Doane and W. R. Horwath, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 6328–6333 CrossRef CAS PubMed.
- J. A. Kozlowski, M. Stieglmeier, C. Schleper, M. G. Klotz and L. Y. Stein, ISME J., 2016, 10, 1836–1845 CrossRef CAS PubMed.
- P. Chain, J. Lamerdin, F. Larimer, W. Regala, V. Lao, M. Land, L. Hauser, A. Hooper, M. Klotz, J. Norton, L. A. Sayavedra-Soto, D. Arciero, N. Hommes, M. Whittaker and D. Arp, J. Bacteriol., 2003, 185, 2759–2773 CrossRef CAS PubMed.
- M. Poth and D. D. Focht, Appl. Environ. Microbiol., 1985, 49, 1134–1141 CAS.
- L. Kuai and W. Verstraete, Appl. Environ. Microbiol., 1998, 64, 4500–4506 CAS.
- A. Rodriguez-Caballero and M. Pijuan, Water Res., 2013, 47, 3131–3140 CrossRef CAS PubMed.
- R. A. Kester, Appl. Environ. Microbiol., 1997, 63, 3872–3877 CAS.
- O. Perez-Garcia, S. G. Villas-Boas, S. Swift, K. Chandran and N. Singhal, Water Res., 2014, 60C, 267–277 CrossRef PubMed.
- M. J. Kampschreur, N. C. G. Tan, R. Kleerebezem, C. Picioreanu, M. S. M. Jetten and M. C. M. Van Loosdrecht, Environ. Sci. Technol., 2008, 42, 429–435 CrossRef CAS PubMed.
- R. Yu and K. Chandran, BMC Microbiol., 2010, 10, 70 CrossRef PubMed.
- A. K. Upadhyay, A. B. Hooper and M. P. Hendrich, J. Am. Chem. Soc., 2006, 128, 4330–4337 CrossRef CAS PubMed.
- J. A. Kozlowski, J. Price and L. Y. Stein, Appl. Environ. Microbiol., 2014, 80, 4930–4935 CrossRef PubMed.
- K. Chandran, L. Y. Stein, M. G. Klotz and M. C. M. van Loosdrecht, Biochem. Soc. Trans., 2011, 39, 1832–1837 CrossRef CAS PubMed.
- S. Park, W. Bae, J. Chung and S.-C. Baek, Process Biochem., 2007, 42, 1671–1676 CrossRef CAS.
- K. M. Udert, T. A. Larsen and W. Gujer, Environ. Sci. Technol., 2005, 39, 4066–4075 CrossRef CAS PubMed.
- CRC Handbook of Chemistry and Physics, ed. D. R. Lide, CRC Press/Taylor and Francis, Boca Raton, FL, 89th edn, 2009 Search PubMed.
- D. Jiang, W. O. Khunjar, B. Wett, S. N. Murthy and K. Chandran, Environ. Sci. Technol., 2015, 49, 2523–2531 CrossRef CAS PubMed.
- B. L. Mellbye, A. Giguere, F. Chaplen, P. J. Bottomley and L. A. Sayavedra-Soto, Appl. Environ. Microbiol., 2016, 82, 3310–3318 CrossRef PubMed.
- S. Panwivia, S. Sirvithayapakorn, C. Wantawin, P. Noophan and J. Munakata-Marr, J. Environ. Sci. Health, Part A: Toxic/Hazard. Subst. Environ. Eng., 2014, 49, 851–856 CrossRef CAS PubMed.
- D. R. H. Graf, C. M. Jones and S. Hallin, PLoS One, 2014, 9, e114118 Search PubMed.
- C. M. Jones, A. Spor, F. P. Brennan, M.-C. Breuil, D. Bru, P. Lemanceau, B. Griffiths, S. Hallin and L. Philippot, Nat. Clim. Change, 2014, 4, 801–805 CrossRef CAS.
- R. A. Sanford, D. D. Wagner, Q. Wu, J. C. Chee-Sanford, S. H. Thomas, C. Cruz-García, G. Rodríguez, A. Massol-Deyá, K. K. Krishnani, K. M. Ritalahti, S. Nissen, K. T. Konstantinidis and F. E. Löffler, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 19709–19714 CrossRef CAS PubMed.
- I. Kucera, V. Dadak and R. Dobry, Eur. J. Biochem., 1983, 130, 359–364 CrossRef CAS PubMed.
- P. R. Alefounder, A. J. Greenfield, J. E. G. Mccarthy and S. J. Ferguson, Biochim. Biophys. Acta, 1983, 724, 20–39 CrossRef CAS.
- D. Richardson, H. Felgate, N. Watmough, A. Thomson and E. Baggs, Trends Biotechnol., 2009, 27, 388–397 CrossRef CAS PubMed.
- D. Wild, R. Von Schulthess and W. Gujer, Water Sci. Technol., 1994, 30, 113–122 CAS.
- J. K. Thomsen, T. Geest and R. P. Cox, Appl. Environ. Microbiol., 1994, 60, 536–541 CAS.
- Y. Pan, L. Ye, B.-J. Ni and Z. Yuan, Water Res., 2012, 46, 4832–4840 CrossRef CAS PubMed.
- J. Heil, B. Wolf, N. Brüggemann, L. Emmenegger, B. Tuzson, H. Vereecken and J. Mohn, Geochim. Cosmochim. Acta, 2014, 139, 72–82 CrossRef CAS.
- Methods in Nitric Oxide Research, ed. M. Feelisch and J. S. Stamler, J. W. and Sons, Chichester, England, 1996, pp. 71–115 Search PubMed.
- C. Döring and H. Gehlen, Zeitschrift für Anorg. und Allg. Chemie, 1961, 312, 32–44 CrossRef.
- S. Liu, H. Vereecken and N. Brüggemann, Geoderma, 2014, 232-234, 117–122 CrossRef CAS.
- O. Spott, R. Russow and C. F. Stange, Soil Biol. Biochem., 2011, 43, 1995–2011 CrossRef CAS.
- A. Soler-Jofra, B. Stevens, M. Hoekstra, C. Picioreanu, D. Sorokin, M. C. M. van Loosdrecht and J. Pérez, Chem. Eng. J., 2016, 287, 720–726 CrossRef CAS.
- W. F. Harper, Y. Takeuchi, S. Riya, M. Hosomi and A. Terada, Chem. Eng. J., 2015, 281, 1017–1023 CrossRef CAS.
- B.-J. Ni and Z. Yuan, Water Res., 2015, 87, 336–346 CrossRef CAS PubMed.
- M. J. Kampschreur, C. Picioreanu, N. Tan, R. Kleerebezem, M. S. Jetten and M. C. van Loosdrecht, Water Environ. Res., 2007, 79, 2499–2509 CrossRef CAS PubMed.
- F. Schreiber, B. Loeffler, L. Polerecky, M. M. Kuypers and D. de Beer, ISME J., 2009, 3, 1301–1313 CrossRef CAS PubMed.
- Y. Law, B.-J. Ni, P. Lant and Z. Yuan, Water Res., 2012, 46, 3409–3419 CrossRef CAS PubMed.
- B.-J. Ni, L. Ye, Y. Law, C. Byers and Z. Yuan, Environ. Sci. Technol., 2013, 47, 7795–7803 CrossRef CAS PubMed.
- B.-J. Ni, M. Ruscalleda, C. Pellicer-Nàcher and B. F. Smets, Environ. Sci. Technol., 2011, 45, 7768–7776 CrossRef CAS PubMed.
- L. Guo and P. A. Vanrolleghem, Bioprocess Biosyst. Eng., 2014, 37, 151–163 CrossRef CAS PubMed.
- Y. Wang, X. Lin, D. Zhou, L. Ye, H. Han and C. Song, Chem. Eng. J., 2016, 289, 330–340 CrossRef CAS.
- C. Domingo-Félez, A. G. Mutlu, M. M. Jensen and B. F. Smets, Environ. Sci. Technol., 2014, 48, 8679–8687 CrossRef PubMed.
- W. C. Hiatt and C. P. L. Grady, Water Environ. Res., 2008, 80, 2145–2156 CrossRef CAS PubMed.
- R. Von Schulthess, D. Wild and W. Gujer, Water Sci. Technol., 1994, 30, 123–132 Search PubMed.
- Y. Pan, B.-J. Ni, H. Lu, K. Chandran, D. Richardson and Z. Yuan, Water Res., 2014, 71, 21–31 CrossRef PubMed.
- F. Schreiber, P. Wunderlin, K. M. Udert and G. F. Wells, Front. Microbiol., 2012, 3, 372 Search PubMed.
- A. Guisasola, S. Petzet, J. A. Baeza, J. Carrera and J. Lafuente, Water Res., 2007, 41, 277–286 CrossRef CAS PubMed.
- G. A. Ritchie and D. J. Nicholas, Biochem. J., 1972, 126, 1181–1191 CrossRef CAS PubMed.
- Y. Liu, L. Peng, X. Chen and B.-J. Ni, Environ. Sci. Technol., 2015, 49, 8595–8601 CrossRef CAS PubMed.
- C. Domingo-Félez, C. Pellicer-Nàcher, M. S. Petersen, M. M. Jensen, B. G. Plósz and B. F. Smets, Biotechnol. Bioeng., 2016 DOI:10.1002/bit.26062.
- N. Igarashi, H. Moriyama, T. Fujiwara, Y. Fukumuri and N. Tanaka, Nature, 1997, 4, 276–284 CAS.
- B. Wett and W. Rauch, Water Res., 2003, 37, 1100–1110 CrossRef CAS PubMed.
- A. Zhu, J. Guo, B.-J. Ni, S. Wang, Q. Yang and Y. Peng, Sci. Rep., 2015, 5, 8493 CrossRef CAS PubMed.
- J. Zheng and P. V. Doskey, Environ. Sci. Technol., 2015, 49, 2132–2139 CrossRef CAS PubMed.
- N. Adouani, L. Limousy, T. Lendormi, E. O. Voit and O. Sire, Int. J. Chem. React. Eng., 2014, 12, 683–693 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6ew00179c |
| This journal is © The Royal Society of Chemistry 2016 |