 Open Access Article
Open Access ArticleEmerging investigators series: sewer surveillance for monitoring antibiotic use and prevalence of antibiotic resistance: urban sewer epidemiology†
Nicole
Fahrenfeld
*a and
Kevin J.
Bisceglia
b
aDepartment of Civil & Environmental Engineering, Rutgers, The State University of New Jersey, 96 Frelinghuysen Road, Piscataway, NJ 08854, USA. E-mail: nfahrenf@rutgers.edu; Tel: +(848) 445 8416
bDepartment of Chemistry, Hofstra University, 151 Hofstra University, Hempstead, NY 11549, USA
First published on 31st August 2016
Abstract
Sewer surveillance may be a useful tool for epidemiology that would benefit from improved understanding of the fate of microbial agents and prescription antibiotics during conveyance in sewer systems. The aim of this review is to provide an overview of the factors affecting the loading and fate of antibiotics and antibiotic resistant bacteria (ARB) in sewer systems. A review of surveillance studies for antibiotics and antibiotic resistant bacteria is presented. Then, the role of potentially complicating sewer inputs (e.g., the presence of health care facilities in a sewershed),and evidence for temporal variations in antibiotics and ARB are reviewed. Recommendations for best practices for sampling are made. Finally, evidence is presented for in-sewer attenuation of antibiotics and attenuation, growth and gene transfer for ARB. There is potential for, but limited evidence of, sewers serving as a reservoir for ARB growth and horizontal gene transfer. This review highlights the need for better understanding of ARB carriage in the general population and the impact of in-sewer processes on the fate of antibiotics and ARB.
| Nicole Fahrenfeld Nicole Fahrenfeld is an Assistant Professor in Civil and Environmental Engineering at Rutgers, The State University of New Jersey. She received her Ph.D. in Civil Engineering with a concentration in Environmental and Water Resources Engineering from Virginia Tech. She earned her B.S. in Environmental Engineering from Johns Hopkins University and M.S. in Environmental Engineering and Science from Clemson University. Her research interests include pathogen fate and transport, microbial source tracking, bioremediation, and emerging contaminants. |
| Kevin Bisceglia Kevin Bisceglia is an Assistant Professor of Chemistry at Hofstra University. He earned a B.S. and M.E. in Environmental Engineering from Manhattan College, and a Ph.D. in Environmental Engineering and Chemistry from Johns Hopkins University. His research interests include environmental analytical chemistry, water quality, and chemical fate and transport in the built environment. |
Water impactSewer surveillance may be a useful tool for epidemiology that would benefit from improved understanding of the fate of microbial agents and prescription antibiotics during conveyance in sewer systems. Improving our understanding of these contaminants in sewer systems may also help in characterization of risk in sanitary and combined sewer overflows and the general potential exposure hazard for public works employees. |
Manuscript
An executive order recently called for improved antibiotic stewardship and monitoring of antibiotic resistance in the U.S.1 Sewer surveillance may be a useful tool for epidemiologists wishing to monitor antibiotic resistance in sewered communities,2 that would benefit from improved understanding of the fate of microbial agents and prescription antibiotics during conveyance in sewer systems. Monitoring antibiotic resistance at the community level often focuses on aggregating results from patients seeking treatment for infections, which limits our understanding of resistance carriage in non-health care associated populations. Alternatively, municipal sewage can serve as an all-inclusive reservoir of community-level public health data. Indeed, using sewage rather than individual fecal samples has been suggested as a “practical target” for antibiotic resistance screening to improve detection while reducing workload.3,4 To date, the main focus of using sewer surveillance has been in the illicit drug monitoring community,5 but it has also been successfully applied to other markers of health (e.g., Tamiflu consumption,6 polio virus,7,8 incidence of obesity9). Knowledge of antibiotic consumption may facilitate tracking the incidence of infections, prescription compliance, and use of antibiotics of last resort. It is likely that a broad range of useful data may be collected in sewer systems by including the monitoring of biological agents.Understanding the fate of antibiotic resistant bacteria (ARB) in sewer systems may help in characterization of the potential exposure hazard for public works employees and risk in sanitary sewer and combined sewer overflows, which are poorly characterized sources of ARB in the environment. Alarmingly, but not surprisingly, high levels of antibiotic resistant bacteria were found in surface waters affected by combined sewer overflows.10,11 Because sewer deposits represent a major portion of the solids and biological loading in combined sewer overflows,12 understanding their relative loading of ARB is critical.
The aim of this review is to provide an overview of the factors affecting the loading and fate of antibiotics and antibiotic resistant bacteria in sewer systems, make recommendations about best practices in the monitoring of AR and ARB in sewer systems, compile evidence for the role of sewers in the survival and growth of ARB, and highlight areas of uncertainty that merit additional study.
1. Potential for surveillance of antibiotic use
Sewer surveillance may prove especially useful for monitoring community-level public health where there is a lack of alternative means of tracking (e.g., illicit drug use13) or where it is exclusively associated with specific behaviors (e.g., tracking Tamiflu6 and antiviral compliance,14 or cotinine and nicotine use15). The potential benefits of tracking antibiotics directly include (1) to understand their potential role in selection for and survival of antibiotic resistant organisms and genetic elements in sewers (section 5), (2) to estimate rates of prescription compliance, and (3) to provide an alternative record for monitoring trends in the use of specific drug classes, especially in locations where prescription/sales records are not available. Methods for quantifying antibiotics and their metabolites in sewage generally involve one or more cleanup steps (such as filtration), then pre-concentration (e.g., solid phase extraction) followed by liquid chromatography-(tandem) mass spectrometry. Necessary analytical conditions are dictated by the structure of the desired biomarker. A recent review of the analytical approaches in agriculture, which are similar to those employed in wastewater analysis, is available elsewhere.16 This review also highlights the important consideration that measured concentrations do not necessarily represent bioavailability.Antibiotics entering sewage collection systems should reflect excreted concentrations because patients are no longer directed to dispose of unused antibiotics in sewerage systems.17 Sewer surveillance could represent an improvement over monitoring via sales18 or prescription rates.19 Where prescriptions rates were available, researchers found a positive but insignificant correlation between prescription rates for four fluoroquinolones and detection rates in hospital sewage.20 Outside of medical settings, prescription rates may be a less accurate measure of antibiotic use given that there is a 9.9–44% worldwide admitted non-compliance rate for antibiotics prescribed for acute infections.21 Thus, using sewage-based monitoring could be beneficial to understanding compliance outside of hospitals which is especially important because non-compliance may result in antibiotic resistance in patients [e.g., Thomas et al.22].
Sewage monitoring for antibiotics requires understanding of excretion rates which will differ from administered doses of antibiotics due to compound specific differences in adsorption and metabolism. For example, 90% of tetracycline and only 15% sulfamethoxazole is excreted in urine and feces (as reviewed by Jjemba23). Excretion has been noted as a source of uncertainty in sewage studies tracking illicit drug use.13 There are varying reports for the agreement between predicted environmental concentrations (PEC, μg L−1) and observed antibiotic concentrations in raw sewage. Several researchers have calculated PEC for antibiotics in raw sewage:
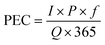 | (1) |
1.1 Evidence for fate and transport affecting antibiotic levels in sewers
Lower levels of antibiotics may reach sewage system intakes than are excreted if sorption and/or losses to biodegradation occur during conveyance. The redox conditions, hydraulics, sewer type, sediment type, and degradation rate may impact the fate of antibiotics during conveyance. While sewers are designed to convey wastewater and any suspended solids they contain, sediment deposition in sewers is ubiquitous.28 Sewer sediments and biofilms develop in sanitary, storm, and combined sewer systems.29 Sorption of antibiotics to biofilm has been demonstrated in annular biofilm reactors30 and should be occurring in sewer lines. In a simulated sewer system collecting wastewater from a hospital, fluoroquinolone was found to accumulate in sewer sediments.24 Jarnheimer et al.24 concluded that sewer sediments would provide a “time-integrated” measure of antibiotic use and can be expected to attenuate flux of antibiotics in sewer systems. Whether this attenuation results in an environment driving selection for resistance is important to understand (section 5).The potential for biodegradation of other high molecular weight compounds in sewers has been demonstrated.31,32 Biodegradation of antibiotics in sewers can be expected to be a function of antibiotic class and redox conditions. Gravity sewers will be re-aerated while pressurized (rising main) sewers will be anaerobic33 and this difference in redox state will affect biodegradation. While there is a growing body of knowledge on the biodegradation of antibiotics in WWTP and soils, no known research has assessed the biodegradation of antibiotics in sewer lines. However, studies comparing PEC and influent concentrations provide some insight. For example, researchers comparing consumption rates to WWTP influent concentrations noted that the administered ratio of sulfamethoxazole and trimethoprim was consistent with the ratio observed in the treatment plant influent26 suggesting if excretion of these compounds is the same that either a lack of transformation or similar rates of degradation/sorption is occurring for these compounds during conveyance. More information is needed to determine the importance of biotransformation of antibiotics during conveyance.
2. Potential for surveillance of antibiotic resistance in sewers
Programs for tracking antibiotic resistance in humans in health care settings are underway on several continents [North America: Canadian Antimicrobial Resistance Surveillance System (CARSSR) in Canada, National Antimicrobial Resistance Monitoring System for Enteric Bacteria (NARMS) in the US; Australia (administered through the Australian Commission on Safety and Quality in Health Care); European Antimicrobial Resistance Surveillance Network (EARS-Net), and Central Asian and Eastern European Surveillance of Antimicrobial Resistance (CASEAR)]. These surveillance programs for antibiotic resistance target sick populations and researchers have noted that resistance rates in healthy populations are not well monitored.34 In contrast, results of sewer surveillance have potential for revealing population level variations in resistance for both healthy and sick human populations.Using sewage rather than individual fecal samples has been suggested as a “practical target” for antibiotic resistance screening to improve detection while reducing workload.3,4 Sewer surveillance would primarily target antibiotic resistant bacteria (ARB) excreted in human feces and urine, but may also contain ARB washed off skin and in saliva, mucus, sputum, etc., entering waste collection systems. The majority of the excreted ARB may be expected to be commensal ARB, given that resistance can persist in intestinal flora following antibiotic treatment [e.g., Jakobsson et al.35]. In contrast to individual gut microbiomes, which vary to the point that no single core set of bacterial species has been found in all guts, a growing body of research indicates that human fecal bacteria that survive in sewers are highly conserved across municipalities in the US36–38 and that variations in this community are related to public health (e.g., obesity).9 It has been suggested that the averaging that is accomplished by sewage sampling would allow for broader analysis of demographic influences (e.g., comparing cities, countries).
Currently applied methods to detect antibiotic resistant bacteria in sewage fall into two general categories: cultivation-based phenotypic (e.g., disc diffusion or microdilution) and genotypic approaches (e.g., PCR, quantitative PCR, metagenomics). The cultivation based techniques provide proof of viable ARB and standard methods are available (e.g., disc diffusion methods). Genotypic methods provide evidence of the presence of ARG and can be used in mixed microbial communities. Genotypic results may be reported as presence/absence (PCR), quantitative (qPCR), or semi-quantitative (metagenomics, which have not yet been reported for sewer surveillance). For a more complete review of current methods and the advantages and disadvantages, the reader is referred to a recent review.39
3. Role and relative importance of other sources of antibiotics and ARB in sewers
Sewersheds containing hospitals can be expected to have different antibiotic profiles from urban sewage. Certain antibiotics reserved for difficult to treat infections (‘antibiotics of last-resort,’ examples provided in Table S1†) may only be present in hospital sewage. In a study in Portugal, the antibiotic profiles detected in hospital sewage compared to residential sewage matched prescription patterns: ciprofloxacin, ofloxacin were higher in hospital sewage and sulfamethoxazole, tetracycline, and penicillin G were higher in raw urban influent.43 Therefore, hospital inputs in a sewershed must be delineated before interpreting surveillance results for antibiotics. The relative role of hospital sewage as a source of antibiotics and other pharmaceutical micropollutants has received considerable attention and the reader is referred to other studies and reviews for further information on this topic and state of science regarding the potential necessity for pre-treatment.19,44–46 Several studies have measured antibiotics in urban and/or hospital sewage and we have summarized the levels observed in (Fig. 1 adapted from Verlicchi et al.46). Our literature review confirms several cephalosporin, tetracyclines, macrolide, and quinolone antibiotics are present in municipal and hospital sewage above the predicted no-effect level [previously estimated by Kümmerer and Henninger by dividing the minimum inhibitory concentration for 50% of the population (MIC50) by 10].47 Whether these concentrations are selecting for ARB in sewers will be in part a function of their bioavailability (section 1.1).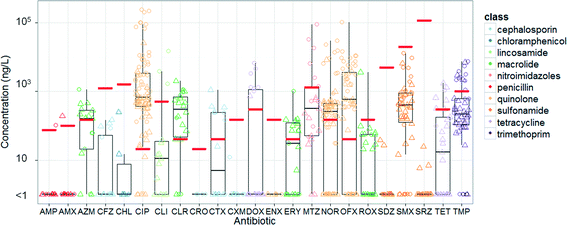 | ||
| Fig. 1 Box and jitter plot of hospital sewage (triangles) and municipal sewage (squares) antibiotic concentrations for select antibiotics. Red lines represent predicted no-effects concentrations (MIC/100) reported by Kümmerer and Henninger.47 Color coding for jitter based on antibiotic class. Antibiotic data from ref. 20, 24–26, 43, 63, 80–103. AMP = ampicilin, AMX = amoxicillin, AZM = azithromycin, CFZ = cefazolin, CHL = chloramphenicol, CIP = ciprofloxacin, CLI = clindamycin, CLR = clarithromycin, CRO = ceftriaxone, CTX = cefotaxime, CXM = cefuroxime, DOX = doxycycline, ENX = enoxacin, ERY = erythromycin, MTZ = metronidazole, NOR = norfloxacin, OFX = ofloxacin, ROX = roxithromycin, SDZ = sulfadiazine, SMX = sulfamethoxazole, SRZ = sulphamerazine, TET = tetracycline, TMP = trimethoprim. | ||
3.1 Hospital and health care associated ARB
Hospital effluents contribute ARB to sewers: a significantly higher proportion of sulfamethoxazole resistant E. coli was observed downstream of hospital inputs in municipal sewers compared to upstream of the hospital.51 This type of in-sewer study is rare and more often comparisons are made between municipal influent and hospital effluents. Generally, hospital effluents have been found to have higher rates of AR and select ARG compared to municipal effluent (Table 1). There have been reports of exceptions to this trend. Higher incidence of resistance in isolates from municipal influent than hospital effluent was reported for (1) E. coli with tetracycline in Denmark,52 ceftazidime in Poland,53 ciprofloxacin in Ireland,51 (2) Enterococci with nitrofurantoin in Portugal,54 vancomycin in Sweden, Spain, and the UK,3,41 and (3) coliform with amoxicillin in Portugal55 and cephaloridine in South Africa.56 The matrix with the highest incidence of resistant isolates was not necessarily consistent for the same organism with different antibiotics. For example, while higher incidence of erythromycin resistant Enterococci were observed in hospital effluent, higher incidence of vancomycin resistant Enterococci were observed in municipal sewage in Sweden.3 Of particular interest is the prevalence of VRE in municipal influents which was especially surprising in Sweden, given that a reduction in clinical VRE infections occurred after use of the growth promoter avoparcin ceased. The VRE observed in municipal influents may represent high level of sustained resistance due to co-selecting factors.41 This finding was not consistent across geographies, with higher incidence of VRE in municipal influent in Sweden and the UK3,41 and higher incidence of VRE in hospital effluent in Spain and Portugal.3,57 Clonal analysis and typing confirmed the clinical origin of the VRE in sewage in Portugal.54 Whether hospital ARB loads affect municipal influent will depend in part on the proportion of the municipal influent that comes from hospitals (i.e., dilution of hospital inputs) and may be obscured by high levels of community resistance. However, these inputs could be amplified if ARB are able to preferentially grow or engage in horizontal gene transfer (HGT) in sewer systems (see section 5).
| Location | Isolate | Antibiotic resistance phenotype | ARG | Comp. to human database or clinical samples | Sampling Schemed | Ref. | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Anti-biotica | Resistant counts, samples, isolates, or strains in sewageb (% or CFU mL−1) | Resistant counts, samples, isolates or strains in hospital sewageb (% or CFU mL−1) | ARG | ARG sewagec | ARG hospital sewagec | |||||
| a Subscript on Antibiotic denotes mg L−1 tested for resistance. AMK = amikacin, AMP = ampicillin, AMX = amoxicillin, CAZ = ceftazidime, CER = cefaloridine, CHL = chloramphenicol, CIP = ciprofloxacin, CPD = cefpodoxime, CTX = cefotaxime, ERY = erythromycin, FOX = cefoxitin, GEN = gentamycin, IPM = imipenem, KAN = kanamycin, LIN = linezolid, NAL = nalidixic acid, NEO = neomycin, NIT = nitrofurantoin, OXY = oxytetracycline, S = sulfonamide, SMX = sulfamethoxazole, STR = streptomycin, SXT = sulfamethoxazole/trimethoprim, TET = tetracycline, TMP = trimethoprim, TZP = piperacillin-tazobactam, VAN = vancomycin. b Phenotype resistance is shown as resistant CFU mL−1 or as a percentage reported using study units. Percentage values listed represent percentages of samples with resistance (s), percent resistant colony forming units (c) in a study with a given number of samples (s), percent resistant most probable number per mL (MPN mL−1), percent resistant isolates (i), or percent gentamicin resistant PFGE typed isolates (GENRpi). c ARG were reported as percentages of vancomycin resistant Enterococci isolates (VRE), isolates (i), gentamicin resistant strains (GENRst), or extended-spectrum beta-lactamase producing isolates (ESBL), gene copies/16S rRNA gene copies, or presence/absence (+/−). d Hospital sewage = HS, raw urban sewage = RS. + Urban sewage downstream of major hospital input. ** Estimated from manuscript figures using WebPlotDigitizer. | ||||||||||
| South Africa | Coliform | AMP25 | 20 (c, N = 9s) | 38 (c, N = 9s) | 56 | |||||
| CER15 | 19 | 15 | ||||||||
| CHL25 | 45 | 62 | ||||||||
| KAN25 | 3 | 39 | ||||||||
| NEO25 | 2 | 37 | ||||||||
| OXY25 | 7 | 48 | ||||||||
| STR25 | 22 | 76 | ||||||||
| S25 | 35 | 73 | ||||||||
| TET25 | 4 | 31 | ||||||||
| Stockholm and Uppsala, Sweden | Enterococci | VAN8 | 60 (N = 35s) | 36 (N = 14s) | vanA | 96% (N = 23 VRE) | 20% (N = 5 VRE) | Grab or flow integrated RS, grab for HS | 41 | |
| vanB | 4% | 80% | ||||||||
| Stockholm, Sweden | E. coli | AMP32CT | 16 (N = 1326i) | 36 (N = 451i)** | Y | 24 h composite for RS, 4 h composite for HS | 40 | |||
| X2 | 3 | 15 | ||||||||
| CAZ4 | 3 | 16 | ||||||||
| CHL32 | 3 | 10 | ||||||||
| CIP4 | 3 | 7 | ||||||||
| GEN16 | 2 | 7 | ||||||||
| NAL32 | 17 | 25 | ||||||||
| CPD3 | 6 | 19 | ||||||||
| TET16 | 13 | 28 | ||||||||
| TMP16 | 11 | 26 | ||||||||
| Portugal | Heterotrophs | AMX32 | 28.3 (c, N = 20s) | 37.8 (c, N = 7s) | 24 h composite for RS, grab HS | 43 | ||||
| CIP4 | 7.6 | 19.5 | ||||||||
| Aeromonads/Pseudomonads | AMX32 | 41.3 (N = 20s, c) | 48.8 (c, N = 7s) | |||||||
| CIP4 | 3.7 | 10.7 | ||||||||
| Enterobacteria | AMX32 | 45.7 (c, N = 20s) | 52.7 (c, N = 7s) | |||||||
| CIP4 | 3.5 | 15.8 | ||||||||
| Portugal | Enterococci | CIP1 | 4 (c, N = 4)1 | 14 (c, N = 3)8 | vanA | 32% (N = 19i) | 58% (N = 33i) | Y | 24 h composite for RS, grab for HS | 57 |
| VAN16 | ||||||||||
| Portugal | Enterococci | AMP | 11 (N = 37i) | 45 (N = 93i)+ | aac(6′)- | (N = 0 VRE) | 40% (N = 25 VRE) | Grab | 54 | |
| CIP | 27 | 75 | aph(2′′) | |||||||
| CHL | 11 | 17 | aph(3′)-IIIa | 84% | ||||||
| ERY | 59 | 69 | ermB | 84% | ||||||
| GEN | 30 | 40 | vanA | 84% | ||||||
| KAN | 49 | 61 | vanB | 16% | ||||||
| LIN | 0 | 0 | ||||||||
| NIT | 14 | 6 | ||||||||
| STR | 32 | 48 | ||||||||
| TET | 41 | 37 | ||||||||
| VAN | 0 | 27 | ||||||||
| Portugal | Coliform | AMP32 | 48 (c, N = 4s) | 11 (c, N = 3s) | int1** | 10–1.5/16S rRNA | 10–1.7/16S rRNA | 24 h composite for RS, grab for HS | 55 | |
| CIP4 | 1 | 3 | bla TEM** | 10–2.4/16S rRNA | 10–1.9/16S rRNA | |||||
| marA** | 10–3.4/16S rRNA | 10–2.6/16S rRNA | ||||||||
| vanA** | 10–5.1/16S rRNA | 10–3.1/16S rRNA | ||||||||
| Girona, Spain | bla TEM** | 10–4.5–10−3.3/16S rRNA | 10–3.2–10−2.2/16S rRNA | Grab | 95 | |||||
| ermB** | 10–2.2–10−1.3/16S rRNA | 10–1.9–10−1.4/16S rRNAs | ||||||||
| qnrS** | 10–3.1–10−1.9/16S rRNA | 10–3.0–10−1.9/16S rRNA | ||||||||
| sul1** | 10–3.0–10−1.8/16S rRNA | 10–2.2–10−1.2/16S rRNA | ||||||||
| tet(W)** | 10–1.8–10−1.5/16S rRNA | 10–1.6–10−1.4/16S rRNA | ||||||||
| Denmark | E. coli | GEN | 11 CFU mL−1 | 36 CFU mL−1 | aac (3)-II | 33% (N = 21 GENRst) | 66% (N = 15 GENRst) | Y | Flow weighted 24 h RS, 2-sample composites for HS | 52 |
| AMP | 65 (N = 17 GENRpi) | 93 (N = 15 GENRpi) | aac (3)-IV | 38% | 27% | |||||
| 76 | 100 | ant (2′′)-I | 29% | 7% | ||||||
| SMX | 88 | 100 | ||||||||
| STR | 88 | 33 | ||||||||
| TET | 53 | 73s | ||||||||
| TMP | ||||||||||
| Sweden | Enterococci | ERY8 | 26 (N = 35s) | 86 (N = 14s) | Grab | 3 | ||||
| VAN8 | 57 | 43 | ||||||||
| VAN20 | 57 | 36 | ||||||||
| Spain | ERY8 | 100 (N = 49s) | 83 (N = 23s) | |||||||
| VAN8 | 98 | 30 | ||||||||
| VAN20 | 90 | 22 | ||||||||
| UK | VAN8 | 18 (N = 22s) | ||||||||
| VAN20 | 67 (N = 21s) | 5 | ||||||||
| 52 | ||||||||||
| Olsztyn, Poland | E. coli | CTX5 | 95 (N = 56i) | 97 (N = 167i) | bla CTX−M-1 | 18% (N = 11 ESBL i) | 6% (N = 62 ESBL i) | Grab | 53 | |
| CAZ10 | 79 | 73 | bla CTX−M-3 | 27% | 18% | |||||
| CPD10 | 61 | 66 | bla CTX−M-9 | 18% | 32% | |||||
| TZP30/6 | 7 | 13 | bla TEM−1 | 73% | 87% | |||||
| GEN10 | 61 | 69 | bla TEM−47 | 9 | 0 | |||||
| AMK30 | 48 | 55 | bla TEM−49 | 18 | 8 | |||||
| IPM10 | 4 | 4 | bla TEM−52 | 0 | 2 | |||||
| CHL30 | 13 | 29 | bla OXA−1 | 0 | 18 | |||||
| SXT1.25/23.8 | 11 | 40 | bla SHV−5 | 27 | 10 | |||||
| Ireland | E. coli | AMP32 | 24.5 (N = 8.8 × 106 MPN mL−1) | 39.8 (N = 5.38 × 106 MPN mL−1) | bla CTX−M-1 | + | + | Grab | 51 | |
| bla CTX−M-9 | − | − | ||||||||
| STR32 | 16.5 | 18.5 | bla TEM | + | + | |||||
| SXT25/6 | 11.1 | 22.6 | bla SHV | − | + | |||||
| TET4 | 12.4 | 17 | ||||||||
| CTX2 | 0 | 2.2 | ||||||||
| CIP4 | 7.15 | 5.4 | ||||||||
| FOX32 | 0.11 | 2.45 | ||||||||
The impact of other another health care settings is less well studied. Bäumlisberger et al.58 investigated ARGs and mobile genetic elements (MGE) arising from German nursing home wastewater effluent by sampling sewage upstream and downstream in different seasons. Although they found seasonal differences in both microbial community structure and the ARG and MGE abundance, they concluded that nursing homes are not significant sources of ARG or MGE, because upstream and downstream samples were not significantly different.
3.2 Non-human fecal sources of antibiotics and ARB
Other non-human sources of ARB may be present in sewers that could complicate interpretation of sewer surveillance. Slaughterhouse wastewater contains ARB.59 However, a study comparing slaughterhouse and municipal wastewater found higher levels of extended spectrum beta-lactamse (ESBL)-producing E. coli in the municipal wastewater.60 Combined and separate sanitary sewers may both contain feces from domestic pets from intentional disposal or livestock or domestic pets from runoff. Domestic pets have been shown to carry ARB, as reviewed by Guardabassi et al.61 Food waste may also be a source of ARB in sewers given that foods may carry ARB.62 The relative importance of these sources of ARB would need to be better understood to correctly interpret sewer surveillance data.4. Temporal variations and sampling design
Best practices for sampling sewage for sewer surveillance may be taken from reviews of the illicit drug literature. However, some considerations for antibiotics and ARB supported by the literature are included here. Antibiotic concentrations often vary across the day in sewage. The variation of antibiotic concentrations in sewage will be partly a function of the timing and number of doses of an antibiotic, half-life in the body, sewage flow rate, discharge timing, and conveyance system residence time. In two separate studies of hospital sewage, considerable variation in antibiotic concentrations in sewage was observed over a 24 h period.20,63 Adjusting for variation in flow rate did not explain the variation in ofloxacin, ciprofloxacin, sulfamethoxazole, trimethoprim, metronidazole, and doxycycline in hospital sewage.63 It is possible that accounting for dosing and half-lives could help explain these results. The fairly consistent concentrations of sulfonamides and trimethoprim across a day in municipal sewage was explained by the twice daily dosing and ∼10 h half-life.26 In contrast, the variable daily profile of macrolides in sewage was explained by the once a day dosing and 10–14 h half-lives in the body.26 Likewise, certain antibiotics (e.g., macrolides) have shown seasonal variation in sewage that corresponded to seasonal variation in sales.26 The potential for daily and seasonal variation must be considered when designing sampling schemes for antibiotic surveillance in sewers.Likewise, sampling of sewage has revealed temporal variation in ARB and ARG that a surveillance program must take into account. Seasonal variation in resistance was observed in Enterobacteriaceae isolated from hospital sewage: a higher prevalence of ESBL-positive and beta-lactamase producing isolates were observed in summer and spring corresponding to greater antibiotic consumption.64 Variation in mecA (conferring beta-lactam resistance) concentrations were observed across a year of grab sampling municipal wastewater influent in Sweden, but no seasonality was observed.65 Detecting seasonal variations may depend on sampling design. As with other contaminants, transient, diurnal, and weather related variations in flow may not be adequately described without targeted sampling, compositing, and correction for flow variations. Kwak et al.40 discuss the challenges with cultivating E. coli in wastewater with respect to knowing what small samples represent and how solids can impact counts. Therefore, best practices for sampling wastewater must be employed for surveillance studies. Despite this, many studies comparing hospital and municipal influent used different sampling methods for the two matrices given the difficulty of obtaining access to hospital sewage often limiting researchers to grab samples, while WWTP were prepared for 24 h composite sampling. Ideas for how to normalize observations to the population level can be gleaned from the illicit drug literature [e.g., Lai et al.66].
5. Evidence for the survival and selection for ARB in sewers?
Using sewage for surveillance relies upon understanding the potential for growth and decay of ARB during conveyance, and the exchange of resistance genes in sewer systems. Sewers act as bioreactors67 and sewer microbial communities are distinct from microbial communities in wastewater treatment plants.68 The microbial community in sewage arriving at the treatment plant changes from that present in fecal inputs with a majority of sewage bacteria not associated with fecal sources.37,69 This is due in part to other non-fecal microbial inputs. For example, the presence of certain freshwater protists in sewage was considered evidence of infiltration impacting the microbial community.70 The difference in the temperature of the sewer and gut environment may also impact the microbial community. Typical chemical characteristics in sewers are included as Table S2.† There is evidence of not only survival of indicator organisms but also multiplication of fecal coliform in storm water sewer sediments.71Biological activity in sewer sediments is also apparent from the changes in redox,72 bulk density,73 organic carbon,74 and extrapolymeric substances in the deposits.75 Low dissolved oxygen conditions could help facultative aerobes and anaerobic fecal microbes survive in sewer deposits. Sewers deposits have also been shown to attenuate the flux of microbial agents. A release of polio virus in a sewer was detectable at the wastewater treatment plant inlet for more than four days after the release.76 Thus, the factors effecting the prevalence of ARB in sewer systems may be expected to include dilution,57 attenuation in biofilms or sewer sediments, growth/decay, and gene exchange. Seasonal variations are not expected to be due to changes in sewer water temperature, which was not shown to impact sewer microbial community structures.37 Rather, VandeWalle et al.37 demonstrated that water quality parameters in sewers were found to drive temporal patterns in sewer microbial communities. The impact of pH is evident from efforts to control sulfate reducing and methanogenic communities in sewer biofilms.77 Likewise, dissolved oxygen and nitrate were found to be associated with shifts in microbial communities in sewer biofilm sampled at different locations within a collection system.78 Sewers are warm (10–30 °C, see Table S2†), moist, nutrient rich, and contain biofilm, all of which may support survival, HGT, or growth of ARB. Identification of PFGE typed isolates of gentamicin resistant E. coli from hospitals in wastewater treatment plant inflow was evidence of the survival of these ARB in sewers.52
The potential for the spread of antibiotic resistance in sewers was proposed based on the observation that levels of antibiotics in sewage can exceed the semi-maximum inhibitory concentration and that sewers have high concentrations of bacteria.47,49 While there is potential for gene exchange and growth of ARB based on comparison of predicted no-effect concentrations and observed sewage concentrations (Fig. 1), there is limited evidence demonstrating these phenomena in sewers/sewage beyond the measurement of high concentrations of ARB and ARG in sewage (Table 1). A study of hospital wastewater in a simulated sewer system did not find evidence for selection for fluoroquinolone resistance despite accumulation of fluoroquinolone in sewer sediments.24 Antibiotic residuals were associated with different bacterial communities in hospital and urban sewage via correspondence analysis and the percentage of ARB and the concentrations of penicillin G, tetracycline, and ciprofloxacin were correlated.43 However, the relative importance of selection for resistance after excretion into sewers or prior to excretion is unknown. Other compounds present in sewers may co-select for resistance including heavy metals and disinfectants,79 which could explain persistence of resistance in observed sewers. Correlations between ARB prevalence and arsenic were also observed in sewers.43
Limited evidence is available for determining the role of HGT in sewers. HGT of gentamicin resistance plasmids in Staphylococcus aureus has been observed in dewatered sewage and sewage bioreactors.80 But, the antibiotic concentrations observed in hospital sewage for this study were reported to be below the levels needed to increase plasmid transfer frequencies indicating that HGT was likely not able to occur in sewers. However, HGT is one possible explanation for the observation of the identical resistance profiles for ESBL producers from different Phene Plate (PhP) typed E. coli isolates in hospital sewage.40 Many other factors may affect the survival of ARB and potential for HGT in sewers including sewer type (combined versus separate sanitary sewers), pipe material (a variety of original and liner materials may be used that could affect pH and the chemical microenvironment), flow type (open channel versus forced main), amount of sewer deposits, and sewer maintenance (i.e., frequency of cleaning, amount of infiltration and inflow).
6. Conclusions
Sewers systems may serve as an important role in understanding population level trends as interest increases in monitoring antibiotic resistance and mitigating its spread in the environment. Sewage contains measurable levels of antibiotics that may be reasonably estimated via PEC calculations. But, comparisons to PEC are complicated by the variability in excretion rates, attenuation, and degradation in sewer lines. Sewage is also a source of ARB. Some correspondence exists between ARB found in sewage and in human waste and sera, but the paucity of data on ARB carriage in non-health care associated populations limits our ability to perform comparisons. Hospitals are a concentrated source of both ARB and ARG that may need to be accounted for in sewage surveillance studies. However, higher levels of certain ARB have been observed in municipal sewage that does not contain exceptional amounts of hospital waste, which highlights the gap in knowledge that exists about ARB in general (i.e., healthy) populations. In general trends can be expected to vary by antibiotic identity, ARB identity, and geography. Several pertinent factors (e.g., sewer pipe/liner material, system type) likely influencing the transmission of antibiotics and ARB in sewers have not been studied. Most notably, there is potential for, but limited data demonstrating, the growth/decay/HGT for ARB in sewer systems. Improved understanding of the processes occurring in our sewer infrastructure is needed to understand the health risk to public works employees and with sewer overflows, and to aid in interpretation of sewer surveillance data. This may be achieved through the application of techniques to better understand the resistome of the sewage microbial community (i.e., metagenomics) and the selective pressure applied by mixtures of antibiotics, their degradation products, and other co-selecting chemicals in sewers.Acknowledgements
This work was supported in part by a grant from the National Science Foundation (#1510461).References
- B. Obama, Executive Order, 2014 Search PubMed.
- R. G. Sinclair, C. Y. Choi, M. R. Riley and C. P. Gerba, Adv. Appl. Microbiol., 2008, 65, 249–269 CrossRef CAS PubMed.
- A. Blanch, J. Caplin, A. Iversen, I. Kühn, A. Manero, H. Taylor and X. Vilanova, J. Appl. Microbiol., 2003, 94, 994–1002 CrossRef CAS PubMed.
- K. B. Linton, M. H. Richmond, R. Bevan and W. A. Gillespie, J. Med. Microbiol., 1974, 7, 91–103 CrossRef CAS PubMed.
- E. Zuccato, C. Chiabrando, S. Castiglioni, D. Calamari, R. Bagnati, S. Schiarea and R. Fanelli, Environ. Health, 2005, 4, 1–7 CrossRef PubMed.
- A. C. Singer, J. D. Järhult, R. Grabic, G. A. Khan, G. Fedorova, J. Fick, R. H. Lindberg, M. J. Bowes, B. Olsen and H. Söderström, PLoS One, 2013, 8, e60221 CAS.
- Y. Manor, R. Handsher, T. Halmut, M. Neuman, A. Bobrov, H. Rudich, A. Vonsover, L. Shulman, O. Kew and E. Mendelson, J. Clin. Microbiol., 1999, 37, 1670–1675 CAS.
- T. Pöyry, M. Stenvik and T. Hovi, Appl. Environ. Microbiol., 1988, 54, 371–374 Search PubMed.
- R. J. Newton, S. L. McLellan, D. K. Dila, J. H. Vineis, H. G. Morrison, A. M. Eren and M. L. Sogin, mBio, 2015, 6, e02574–14 CrossRef PubMed.
- Y. Suzanne, J. Andrew and D. O'Mullan Gregory, J. Water Health, 2013, 11, 297–310 CrossRef PubMed.
- S. L. McLellan, E. J. Hollis, M. M. Depas, M. Van Dyke, J. Harris, C. O. Scopel and J. Gt, Lakes Reservoirs, 2007, 33, 566–580 Search PubMed.
- A. Hannouche, G. Chebbo and C. Joannis, Environ. Sci. Pollut. Res., 2014, 21, 5311–5317 CrossRef CAS PubMed.
- K. J. Bisceglia and K. A. Lippa, Environ. Sci. Pollut. Res., 2014, 21, 4453–4460 CrossRef CAS PubMed.
- T. Azuma, N. Nakada, N. Yamashita and H. Tanaka, Chemosphere, 2013, 93, 1672–1677 CrossRef CAS PubMed.
- T. Rodríguez-Álvarez, R. Rodil, M. Rico, R. Cela and J. B. Quintana, Anal. Chem., 2014, 86, 10274–10281 CrossRef PubMed.
- D. S. Aga, M. Lenczewski, D. Snow, J. Muurinen, J. B. Sallach and J. S. Wallace, J. Environ. Qual., 2016, 45, 407–419 CrossRef CAS PubMed.
- P. Ortner and M. McCullagh, J. Hosp. Palliat. Nurs., 2010, 12, 15–26 CrossRef.
- T. P. Van Boeckel, S. Gandra, A. Ashok, Q. Caudron, B. T. Grenfell, S. A. Levin and R. Laxminarayan, Lancet Infect. Dis., 2014, 14, 742–750 CrossRef PubMed.
- A. Schuster, C. Hädrich and K. Kümmerer, Water, Air, Soil Pollut.: Focus, 2008, 8, 457–471 CrossRef.
- V. Diwan, A. J. Tamhankar, R. K. Khandal, S. Sen, M. Aggarwal, Y. Marothi, R. V. Iyer, K. Sundblad-Tonderski and C. Stålsby-Lundborg, BMC Public Health, 2010, 10, 414 CrossRef PubMed.
- J.-C. Pechère, D. Hughes, P. Kardas and G. Cornaglia, Int. J. Antimicrob. Agents, 2007, 29, 245–253 CrossRef PubMed.
- J. K. Thomas, A. Forrest, S. M. Bhavnani, J. M. Hyatt, A. Cheng, C. H. Ballow and J. J. Schentag, Antimicrob. Agents Chemother., 1998, 42, 521–527 CAS.
- P. K. Jjemba, Ecotoxicol. Environ. Saf., 2006, 63, 113–130 CrossRef CAS PubMed.
- P.-Å. Jarnheimer, J. Ottoson, R. Lindberg, T.-A. Stenström, M. Johansson, M. Tysklind, M.-M. Winner and B. Olsen, Scand. J. Infect. Dis., 2004, 36, 752–755 CrossRef CAS PubMed.
- M. Carballa, F. Omil and J. M. Lema, Chemosphere, 2008, 72, 1118–1123 CrossRef CAS PubMed.
- A. Göbel, A. Thomsen, C. S. McArdell, A. Joss and W. Giger, Environ. Sci. Technol., 2005, 39, 3981–3989 CrossRef.
- A. L. van Nuijs, A. Covaci, H. Beyers, L. Bervoets, R. Blust, G. Verpooten, H. Neels and P. G. Jorens, Environ. Sci. Pollut. Res., 2015, 22, 9110–9118 CrossRef CAS PubMed.
- R. M. Ashley and T. Hvitved-Jacobsen, in Wet-weather flow in the urban watershed: Technology and management, ed. R. Field and D. Sullivan, Lewis Publishers, Boca Raton, FL, 2003 Search PubMed.
- R. Crabtree, Water Environ. J., 1989, 3, 569–578 CrossRef CAS.
- D. B. Wunder, V. A. Bosscher, R. C. Cok and R. M. Hozalski, Water Res., 2011, 45, 2270–2280 CrossRef CAS PubMed.
- P. Jin, B. Wang, D. Jiao, G. Sun, B. Wang and X. C. Wang, Water Res., 2015, 84, 112–119 CrossRef CAS PubMed.
- L. A. Rodenburg, S. Du, H. Lui, J. Guo, N. Oseagulu and D. E. Fennell, Environ. Sci. Technol., 2012, 46, 6612–6620 CrossRef CAS PubMed.
- Y. Liu, B.-J. Ni, R. Ganigué, U. Werner, K. R. Sharma and Z. Yuan, Water Res., 2015, 70, 350–359 CrossRef CAS PubMed.
- F. F. Reinthaler, G. Herbert, F. Gebhard, H. Doris, L. Eva, M. Franz, M. Angelika, P. Josefa, P. Brigitte and W. Ingrid, J. Water Health, 2013, 11, 13–20 CrossRef PubMed.
- H. E. Jakobsson, C. Jernberg, A. F. Andersson, M. Sjölund-Karlsson, J. K. Jansson and L. Engstrand, PLoS One, 2010, 5, e9836 Search PubMed.
- O. C. Shanks, R. J. Newton, C. A. Kelty, S. M. Huse, M. L. Sogin and S. L. McLellan, Appl. Environ. Microbiol., 2013, 79, 2906–2913 CrossRef CAS PubMed.
- J. L. VandeWalle, G. W. Goetz, S. M. Huse, H. G. Morrison, M. L. Sogin, R. G. Hoffmann, K. Yan and S. L. McLellan, Environ. Microbiol., 2012, 14, 2538–2552 CrossRef CAS PubMed.
- S. L. McLellan, R. J. Newton, J. L. Vandewalle, O. C. Shanks, S. M. Huse, A. M. Eren and M. L. Sogin, Environ. Microbiol., 2013, 15, 2213–2227 CrossRef CAS PubMed.
- L. Rizzo, C. Manaia, C. Merlin, T. Schwartz, C. Dagot, M. C. Ploy, I. Michael and D. Fatta-Kassinos, Sci. Total Environ., 2013, 447, 345–360 CrossRef CAS PubMed.
- Y.-K. Kwak, P. Colque, S. Byfors, C. G. Giske, R. Möllby and I. Kühn, Int. J. Antimicrob. Agents, 2015, 45, 25–32 CrossRef CAS PubMed.
- A. Iversen, I. Kühn, A. Franklin and R. Möllby, Appl. Environ. Microbiol., 2002, 68, 2838–2842 CrossRef CAS PubMed.
- C. M. Yang, M. F. Lin, P. C. Liao, H. W. Yeh, B. V. Chang, T. K. Tang, C. Cheng, C. H. Sung and M. L. Liou, Lett. Appl. Microbiol., 2009, 48, 560–565 CrossRef CAS PubMed.
- A. R. Varela, S. André, O. C. Nunes and C. M. Manaia, Water Res., 2014, 54, 327–336 CrossRef CAS PubMed.
- K. Kümmerer, Chemosphere, 2009, 75, 417–434 CrossRef PubMed.
- K. S. Le Corre, C. Ort, D. Kateley, B. Allen, B. I. Escher and J. Keller, Environ. Int., 2012, 45, 99–111 CrossRef CAS PubMed.
- P. Verlicchi, A. Galletti, M. Petrovic and D. Barceló, J. Hydrol., 2010, 389, 416–428 CrossRef CAS.
- K. Kümmerer and A. Henninger, Clin. Microbiol. Infect., 2003, 9, 1203–1214 CrossRef.
- CDC, Interim guidance for environmental infection control in hospitals for Ebola virus, http://www.cdc.gov/vhf/ebola/hcp/environmental-infection-control-in-hospitals.html#eight).
- K. Kümmerer, Chemosphere, 2001, 45, 957–969 CrossRef.
- T. P. G. Chagas, L. M. Seki, D. M. da Silva and M. D. Asensi, J. Hosp. Infect., 2011, 77, 281 CrossRef CAS PubMed.
- S. Galvin, F. Boyle, P. Hickey, A. Vellinga, D. Morris and M. Cormican, Appl. Environ. Microbiol., 2010, 76, 4772–4779 CrossRef CAS PubMed.
- L. Jakobsen, D. Sandvang, L. H. Hansen, L. Bagger-Skjøt, H. Westh, C. Jørgensen, D. S. Hansen, B. M. Pedersen, D. L. Monnet, N. Frimodt-Møller, S. J. Sørensen and A. M. Hammerum, Environ. Int., 2008, 34, 108–115 CrossRef CAS PubMed.
- E. Korzeniewska, A. Korzeniewska and M. Harnisz, Ecotoxicol. Environ. Saf., 2013, 91, 96–102 CrossRef CAS PubMed.
- C. Novais, T. M. Coque, H. Ferreira, J. C. Sousa and L. Peixe, Appl. Environ. Microbiol., 2005, 71, 3364–3368 CrossRef CAS PubMed.
- C. Narciso-da-Rocha, A. R. Varela, T. Schwartz, O. C. Nunes and C. M. Manaia, J. Glob. Antimicrob. Resist., 2014, 2, 309–315 CrossRef.
- W. O. K. Grabow and O. W. Prozesky, Antimicrob. Agents Chemother., 1973, 3, 175–180 CrossRef CAS PubMed.
- A. R. Varela, G. Ferro, J. Vredenburg, M. Yanık, L. Vieira, L. Rizzo, C. Lameiras and C. M. Manaia, Sci. Total Environ., 2013, 450–451, 155–161 CrossRef CAS PubMed.
- M. Bäumlisberger, L. Youssar, M. B. Schilhabel and D. Jonas, PLoS One, 2015, 10, e0122635 Search PubMed.
- N. Ayaz, Y. Gencay and I. Erol, Ann. Microbiol., 2014, 1–8, DOI:10.1007/s13213-014-0961-5.
- A. A. Diallo, H. Brugère, M. Kérourédan, V. Dupouy, P.-L. Toutain, A. Bousquet-Mélou, E. Oswald and D. Bibbal, Water Res., 2013, 47, 4719–4729 CrossRef CAS PubMed.
- L. Guardabassi, S. Schwarz and D. H. Lloyd, J. Antimicrob. Chemother., 2004, 54, 321–332 CrossRef CAS PubMed.
- H. Sørum and T. M. L'Abée-Lund, Int. J. Food Microbiol., 2002, 78, 43–56 CrossRef.
- R. Lindberg, P.-Å. Jarnheimer, B. Olsen, M. Johansson and M. Tysklind, Chemosphere, 2004, 57, 1479–1488 CrossRef CAS PubMed.
- E. Korzeniewska and M. Harnisz, J. Environ. Manage., 2013, 123, 1–7 CrossRef CAS PubMed.
- S. Börjesson, S. Melin, A. Matussek and P.-E. Lindgren, Water Res., 2009, 43, 925–932 CrossRef PubMed.
- F. Y. Lai, S. Anuj, R. Bruno, S. Carter, C. Gartner, W. Hall, K. P. Kirkbride, J. F. Mueller, J. W. O'Brien, J. Prichard, P. K. Thai and C. Ort, Environ. Sci. Technol., 2015, 49, 999–1008 CrossRef CAS PubMed.
- T. Hvitved-Jacobsen, J. Vollertsen and P. H. Nielsen, Water Sci. Technol., 1998, 37, 233–241 CrossRef CAS.
- T. Hvitved-Jacobsen, J. Vollertsen and A. H. Nielsen, Sewer processes: microbial and chemical process engineering of sewer networks, CRC press, 2013 Search PubMed.
- S. L. McLellan, S. M. Huse, S. R. Mueller-Spitz, E. N. Andreishcheva and M. L. Sogin, Environ. Microbiol., 2010, 12, 378–392 CrossRef CAS PubMed.
- A. Korajkic, L. W. Parfrey, B. R. McMinn, Y. V. Baeza, W. VanTeuren, R. Knight and O. C. Shanks, Water Res., 2015, 69, 30–39 CrossRef CAS PubMed.
- R. P. Marino and J. J. Gannon, Water Res., 1991, 25, 1089–1098 CrossRef.
- S. Clegg, C. F. Forster and R. W. Crabtree, Environ. Technol., 1992, 13, 561–569 CrossRef CAS.
- R. Banasiak, R. Verhoeven, R. De Sutter and S. Tait, Water Res., 2005, 39, 5221–5231 CrossRef CAS PubMed.
- J. Vollertsen and T. Hvitved-Jacobsen, Water Res., 1999, 33, 3127–3141 CrossRef CAS.
- J. Cooke and C. F. Forster, Environ. Technol., 1994, 15, 175–181 CrossRef CAS.
- T. Hovi, M. Stenvik, H. Partanen and A. Kangas, Epidemiol. Infect., 2001, 127, 101–106 CAS.
- O. Gutierrez, D. Park, K. R. Sharma and Z. Yuan, Water Res., 2009, 43, 2549–2557 CrossRef CAS PubMed.
- J. Luo, H. Liang, L. Yan, J. Ma, Y. Yang and G. Li, Bioresour. Technol., 2013, 148, 189–195 CrossRef CAS PubMed.
- M. Ferreira da Silva, I. Vaz-Moreira, M. Gonzalez-Pajuelo, O. C. Nunes and C. M. Manaia, FEMS Microbiol. Ecol., 2007, 60, 166–176 CrossRef CAS PubMed.
- K. Ohlsen, T. Ternes, G. Werner, U. Wallner, D. Loffler, W. Ziebuhr, W. Witte and J. Hacker, Environ. Microbiol., 2003, 5, 711–716 CrossRef CAS PubMed.
- A. L. Batt, S. Kim and D. S. Aga, Chemosphere, 2007, 68, 428–435 CrossRef CAS PubMed.
- H. A. Duong, N. H. Pham, H. T. Nguyen, T. T. Hoang, H. V. Pham, V. C. Pham, M. Berg, W. Giger and A. C. Alder, Chemosphere, 2008, 72, 968–973 CrossRef CAS PubMed.
- P. Gao, Y. Ding, H. Li and I. Xagoraraki, Chemosphere, 2012, 88, 17–24 CrossRef CAS PubMed.
- G. C. Ghosh, T. Okuda, N. Yamashita and H. Tanaka, Water Sci. Technol., 2009, 59, 779–786 CrossRef CAS PubMed.
- E. M. Golet, A. C. Alder and W. Giger, Environ. Sci. Technol., 2002, 36, 3645–3651 CrossRef CAS PubMed.
- E. M. Golet, I. Xifra, H. Siegrist, A. C. Alder and W. Giger, Environ. Sci. Technol., 2003, 37, 3243–3249 CrossRef CAS PubMed.
- A. Gulkowska, H. W. Leung, M. K. So, S. Taniyasu, N. Yamashita, L. W. Y. Yeung, B. J. Richardson, A. P. Lei, J. P. Giesy and P. K. S. Lam, Water Res., 2008, 42, 395–403 CrossRef CAS PubMed.
- A. Jelic, M. Gros, A. Ginebreda, R. Cespedes-Sánchez, F. Ventura, M. Petrovic and D. Barcelo, Water Res., 2011, 45, 1165–1176 CrossRef CAS PubMed.
- A. Joss, E. Keller, A. C. Alder, A. Göbel, C. S. McArdell, T. Ternes and H. Siegrist, Water Res., 2005, 39, 3139–3152 CrossRef CAS PubMed.
- B. Kasprzyk-Hordern, R. M. Dinsdale and A. J. Guwy, Water Res., 2009, 43, 363–380 CrossRef CAS PubMed.
- D. E. Koch, A. Bhandari, L. Close and R. P. Hunter, J. Chromatogr. A, 2005, 1074, 17–22 CrossRef CAS PubMed.
- A. Y.-C. Lin and Y.-T. Tsai, Sci. Total Environ., 2009, 407, 3793–3802 CrossRef CAS PubMed.
- J. Radjenovic, M. Petrovic and D. Barceló, Anal. Bioanal. Chem., 2007, 387, 1365–1377 CrossRef CAS PubMed.
- P. H. Roberts and K. V. Thomas, Sci. Total Environ., 2006, 356, 143–153 CrossRef CAS PubMed.
- S. Rodriguez-Mozaz, S. Chamorro, E. Marti, B. Huerta, M. Gros, A. Sànchez-Melsió, C. M. Borrego, D. Barceló and J. L. Balcázar, Water Res., 2015, 69, 234–242 CrossRef CAS PubMed.
- M. Seifrtová, A. Pena, C. M. Lino and P. Solich, Anal. Bioanal. Chem., 2008, 391, 799–805 CrossRef PubMed.
- W.-J. Sim, J.-W. Lee, E.-S. Lee, S.-K. Shin, S.-R. Hwang and J.-E. Oh, Chemosphere, 2011, 82, 179–186 CrossRef CAS PubMed.
- A. L. Spongberg and J. D. Witter, Sci. Total Environ., 2008, 397, 148–157 CrossRef CAS PubMed.
- S. Suarez, J. M. Lema and F. Omil, Bioresour. Technol., 2009, 100, 2138–2146 CrossRef CAS PubMed.
- Q. Sui, J. Huang, S. Deng, G. Yu and Q. Fan, Water Res., 2010, 44, 417–426 CrossRef CAS PubMed.
- S. Terzić, I. Senta, M. Ahel, M. Gros, M. Petrović, D. Barcelo, J. Müller, T. Knepper, I. Martí, F. Ventura, P. Jovančić and D. Jabučar, Sci. Total Environ., 2008, 399, 66–77 CrossRef PubMed.
- A. J. Watkinson, E. J. Murby and S. D. Costanzo, Water Res., 2007, 41, 4164–4176 CrossRef CAS PubMed.
- J. Xu, Y. Xu, H. Wang, C. Guo, H. Qiu, Y. He, Y. Zhang, X. Li and W. Meng, Chemosphere, 2015, 119, 1379–1385 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6ew00158k |
| This journal is © The Royal Society of Chemistry 2016 |
