Nanoscale dynamics of phospholipids reveals an optimal assembly mechanism of pore-forming proteins in bilayer membranes†
Abstract
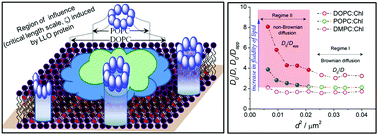
Cell membranes are believed to be highly complex dynamical systems having compositional heterogeneity involving several types of lipids and proteins as the major constituents. This dynamical and compositional heterogeneity is suggested to be critical to the maintenance of active functionality and response to chemical, mechanical, electrical and thermal stresses. However, delineating the various factors responsible for the spatio-temporal response of actual cell membranes to stresses can be quite challenging. In this work we show how biomimetic phospholipid bilayer membranes with variable lipid fluidity determine the optimal assembly mechanism of the pore-forming protein, listeriolysin O (LLO), belonging to the class of cholesterol dependent cytolysins (CDCs). By combining atomic force microscopy (AFM) and super-resolution stimulated emission depletion (STED) microscopy imaging on model membranes, we show that pores formed by LLO in supported lipid bilayers can have variable conformation and morphology depending on the fluidity of the bilayer. At a fixed cholesterol concentration, pores formed in 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) membranes were larger, flexible and more prone to coalescence when compared with the smaller and more compact pores formed in the lower fluidity 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) membranes. In contrast, 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) membranes did not show any evidence of pore formation. Fluorescence correlation spectroscopy (FCS) in STED mode revealed the appearance of a length scale, ξ, below which lipid dynamics, under the influence of LLO protein binding and assembly, becomes anomalous. Interestingly, the magnitude of ξ is found to correlate with both lipid fluidity and pore dimensions (and flexibility) in DOPC and POPC bilayers. However this length scale dependent crossover, signalling the onset of anomalous diffusion, was not observed in DMPC bilayers. Our study highlights the subtle interplay of lipid membrane mediated protein assembly and lipid fluidity in determining proteo-lipidic complexes formed in biomembranes and the significant insight that STED microscopy provides in unraveling critical aspects of nanoscale membrane biophysics.

 Please wait while we load your content...
Please wait while we load your content...