Open Access Article
Open Access ArticleHydrodynamic thermal confinement: creating thermo-chemical microenvironments on surfaces†
J. F.
Cors
,
A.
Stucki
and
G. V.
Kaigala
*
IBM Research – Zurich, Säumerstrasse 4, 8803 Rüschlikon, Switzerland. E-mail: gov@zurich.ibm.com
First published on 14th October 2016
Abstract
We present a new, general concept termed Hydrodynamic Thermal Confinement (HTC), and its implementation for the creation of microscale dynamic thermo-chemical microenvironments on biological surfaces. HTC is based on a scanning probe and operates under physiological conditions. The temperature can be regulated between 30° and 80 °C with ±0.2 °C precision and temperature ramps of 5 °C s−1 over a footprint of ∼50 μm × 80 μm in a volume of ∼50 × 80 × 15 μm3 (∼50 pl).
Methods to unravel the heterogeneity in biological entities are increasingly needed in several areas of research in the life-sciences where compartmentalization plays a critical role. Biochemical reactions are efficient at a specific temperature or within a range of temperatures. As an example of enzymatic activity, human trypsin is most active around 37 °C,1 whereas the optimal operating temperature of Taq polymerase is around 85 °C.2 Additionally, elevated temperatures have an effect on the conformation of proteins, which can lead to the unmasking of certain epitopes.3 In pathology, the breaking-down of the cross-linking between formalin and tissue prior to histochemical staining is performed at temperatures as high as 100 °C.4 In cytology, fluorescence in situ hybridization probes and buffers are designed to have increased efficiency around 37 °C.5 In certain cases, there is an additional need to cycle between different set-point temperatures, for example, in polymerase chain reaction (PCR).
Compartmentalization and the ability to address small areas within biological samples enable functions such as pharmacological studies on single cells,6 detachment of single adherent cell from surfaces for genetic testing,7 patterning arrays of protein on surfaces,8 and the generation of local chemical gradients.9 Advantages of microscale-localization of chemical reactions on biological samples combined with the temperature dependence of biochemical reactions identifies a need for new technologies for creating thermo-chemical microenvironments on immersed biological samples.
Current heating approaches used with biological samples include ovens, autoclaves, microwaves, radiofrequencies, lasers, and resistive heaters.10 Ovens and autoclaves do not allow heat localization on a specific area of the sample nor is rapid temperature switching possible because of the large thermal inertia of these instruments. Microwave-based droplet heating in a microfluidic chip has been demonstrated, but is limited to reactions performed in a microchannel at temperatures as high as 42 °C.11 Thermal localization with lasers requires light-absorbing materials and suffers from changes in pH conditions, sample structure and chemical composition inherent in a biological environment. In contrast, resistive heating is a favored implementation for local heating in microfluidics because of facilitated on-chip integration as well as precise temperature control.10,12 However, all aforementioned techniques require the insertion of the sample into a microchannel—what we term closed-space microfluidics—thus not allowing direct interaction with the sample. In contrast, thermal probes are atomic-force microscope (AFM)-based techniques that allow to locally measure the temperature and/or heat an “open” surface with nanometer resolution. These techniques rely on mechanical contact between the tip and the substrate for heat transfer.13,14 For example, scanning thermal microscopy in immersion has been shown for mapping nanoscale thermal properties of surfaces.15 Direct contact between the probe tip and the substrate is, however, not a preferred option, in particular when interacting with biological substrates as it may cause structural damage to the sample. Furthermore, in both dry and wet contact-based techniques, the heat transfer relies on conduction, which is a strongly material-dependent process and can be relatively slow compared to convection.
Several research groups developed scanning technologies for interacting with biological substrates in immersion such as FluidFM16 and dip pen nanolithography (DPN).17,18 Both techniques use a cantilever mounted on piezo stages, which provide nanometer resolution. Such resolution is advantageous when studying sub-cellular entities or single DNA strands. However, when operating at the micrometer length-scale, for example on a few cells or a tissue section, there is a need for a more versatile technique that is not perturbed by high topological variations.
In contrast, the microfluidic probe (MFP) technology relies on hydrodynamic localization of a processing liquid on a surface with a footprint on the micrometer length-scale in both the horizontal and vertical dimensions.19 The MFP and its variants have been used to demonstrate local staining of tissue sections,20,21 RNA profiling of cell blocks,22 single-cell analysis,23 microperfusion of brain slices24 and patterning of antibodies to create protein microarrays.8 Previous work using the MFP for local processing focused exclusively on chemical alterations of a microenvironment and did not include heating approaches and thermal management. Interestingly, Wegrzyn et al. used a variant of an MFP for thermometry to monitor ion-channel activation in a few cells,25 but with no means of temperature control over the flow confinement.
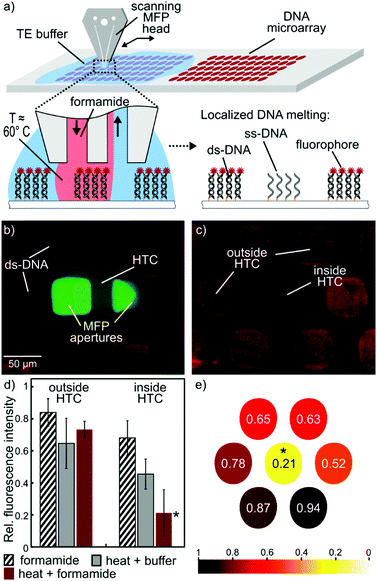
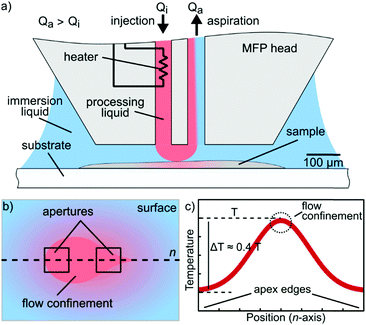
In this paper, we present a new concept and an implementation for simultaneous localization of chemical reagents and heat on immersed surfaces at the microscale without the need for physical contact between the probe and the sample. Our devised concept that we term hydrodynamic thermal confinement (HTC) (Fig. 1) leverages hydrodynamic flow confinement (HFC) for liquid localization generated with an MFP and precise temperature control while minimizing the thermal losses to the surrounding immersion liquid and surface.
The HFC serves to localize the processing liquid on a sample immersed in a liquid while HTC ensures maintenance of a desired temperature within the HFC. Furthermore, HTC provides faster and efficient heat transport to the sample on the surface as it leverages convection by means of continuous liquid flows as opposed to other techniques that rely exclusively on heat conduction. Our implementation of HTC uses a microfluidic probe with an integrated heater and sensing element fabricated using standard microfabrication. We demonstrate temperature control in the HFC using fluorescence thermometry of the confined solution. We further perform local denaturation of DNA on a microarray by using a heated formamide solution, thus localizing thermal and chemical action on an area of ∼80 × 50 μm. To the best of our knowledge, this is the first demonstration of generation of dynamically controlled thermo-chemical microenvironments at the micrometer length scale on immersed surfaces.
The MFP head is a hybrid silicon-glass device with microchannels etched on one side of the silicon using deep reactive-ion etching and resistive elements patterned on the other side using physical vapor deposition (see ESI†). We performed numerical modelling using the finite-element method (FEM) to determine the optimal design and operating parameters, namely, the heater position, channel geometry and flow rates. Simulations showed that there is an operating range between 1 and 10 μl min−1 that provides adequate working conditions for heating the processing liquid (see ESI†).
We further developed an analytical model to describe the heating of the processing liquid flowing through the injection channel with a constant wall temperature (see ESI†).
Power requirements were assessed to implement the conditions considered in the analytical model. Using a restatement of Newton's cooling law:
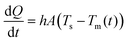 | (1) |
We therefore investigated the temperature variation in the flow confinement along the axis perpendicular to the surface. The cross-sectional and the surface contours show that we can create a microenvironment of ∼50 μm × 80 μm × 15 μm, within which the temperature variation remains below 5 °C (Fig. 2b). This indicates that HTC can be used to alter the microenvironment of samples, such as cell blocks, tissue sections and in certain cases, single isolated cells. The resolution of the confined heat is limited by diffusion from the flow confinement to the surrounding liquid and is approximately 50 μm (see ESI†).
We demonstrate the combined effect of thermal and chemical localization on a biological sample by locally denaturing single spots of ∼60 μm diameter of labelled double-stranded DNA (ds-DNA) on a microarray. The DNA strands on the microarray are 17-base-pair-long (Mycroarray, Ann Arbor, MI). The sequence is typically used to detect a specific point mutation responsible for the sickle cell disease, which leads to a change in the shape of red blood cells (see ESI†). We first incubated complementary fluorescently-labeled DNA in solution on the entire slide, thus all spots of the microarray are comprised of dsDNA. We used a 15% formamide solution in TE buffer (10 mM Tris, 1 mM EDTA, pH 8) as processing liquid to locally denature single DNA spots on a microarray (Fig. 3a and b).
Formamide is a solvent derived from formic acid that lowers the melting temperature of a ds-DNA by forming H-bonds with the nucleotides, thus competing with the H-bonds of the double-helix structure. We demonstrate that the combined use of chemical effect with confined elevated temperatures leads to higher level of DNA denaturation. Fig. 3c shows the denaturation of a single DNA spot, while the neighboring spots remain mostly unaltered.
We performed different experimental sets with various conditions to assess the relative contribution of the chemical and thermal effects (Fig. 3d). In all cases studied, the flow confinement was applied for 3 min on top of a DNA spot, with injection and aspiration flow rates of 2.5 and 7 μl min−1, respectively. These values were set to account for the viscosity of formamide as well as the channel geometry. When using a formamide flow confinement at room temperature, we observed a 30% fluorescence signal decrease. Similarly, the use of TE buffer (pH 8) at 60 °C as processing liquid resulted in a 50% signal decrease. The melting temperature of ds-DNA depends on multiple parameters such as the length of the strand and the G–C base content. Finally, the use of formamide at 60 °C resulted in a dramatic signal decrease, down to less than 20% of the initial value, confirming the synergetic effect of chemistry and elevated temperature. We attribute the remaining fluorescence to partial denaturation of DNA, inherent in thermodynamic reactions. Also, the slight asymmetry in denaturation of the unprocessed DNA spots (Fig. 3e) is attributed to the hybrid silicon-glass composition of the MFP head, silicon being more prone to thermal losses on the account of higher thermal conductivity.
We proposed, modeled, characterized and validated a new concept to perform localized thermo-chemical reactions at the micrometer length scale on immersed biological surfaces. We predict our devised technique to be of significant importance for applications requiring precise control of the temperature over extended periods of time, such as enzymatic reactions, but also in applications requiring rapid temperature changes, such as PCR. With HTC combined with liquid sequencing technique,26 we envision the possibility to perform multi-step biochemical reactions with pH and temperature control. The shape of the microenvironment could be tuned as required by modifying the geometry of the apertures, absolute flow rates and flow rate ratios. Fabrication is currently a limiting factor to localize the heat further as a reduction of the apex size would reduce the heat loss in the immersion liquid. We foresee scaling up the HTC concept to interact with larger samples by creating multiple HTCs operating simultaneously by using MFP heads with arrays of apertures. The technique we present in this paper is versatile and can be used with standard visualization techniques and biological substrates as common as microscope slides and Petri dishes. HTC with a microfluidic probe allows micrometer-scale processing of samples, and will become more prominent as the size of the samples decrease. We strongly believe that the ability to create local, spatio-temporal and thermo-chemical microenvironments on biological samples will enable numerous new investigations in research and diagnostics.
This work was supported by the European Research Council (ERC) Starting Grant, under the 7th Framework Program (Project No. 311122, BioProbe). We thank Aditya Kashyap for assistance in the DNA work, Bernd Gotsmann, Ali Oskooei and Deborah Huber for valuable scientific discussions, Robert Lovchik for technical assistance, and Prof. Bradley Nelson (ETH Zurich), Emmanuel Delamarche, Bruno Michel and Walter Riess are acknowledged for their continuous support.
Notes and references
- D. Figeys, Proteomics, 2013, 13, 1231–1232 CrossRef CAS PubMed.
- F. C. Lawyer, S. Stoffel, R. K. Saiki, S. Y. Chang, P. A. Landre, R. D. Abramson and D. H. Gelfand, Genome Res., 1993, 2, 275–287 CrossRef CAS.
- S. a Pileri, G. Roncador, C. Ceccarelli, M. Piccioli, A. Briskomatis, E. Sabattini, S. Ascani, D. Santini, P. P. Piccaluga, O. Leone, S. Damiani, C. Ercolessi, F. Sandri, F. Pieri, L. Leoncini and B. Falini, J. Pathol., 1997, 183, 116–123 CrossRef CAS PubMed.
- S. R. Shi, R. J. Cote and C. R. Taylor, J. Histochem. Cytochem., 1997, 45, 327–343 CrossRef CAS PubMed.
- D. Huber, J. Autebert and G. V. Kaigala, Biomed. Microdevices, 2016, 18, 40 CrossRef CAS PubMed.
- A. Ainla, E. T. Jansson, N. Stepanyants, O. Orwar and A. Jesorka, Anal. Chem., 2010, 82, 4529–4536 CrossRef CAS PubMed.
- D. Juncker, H. Schmid and E. Delamarche, Nat. Mater., 2005, 4, 622–628 CrossRef CAS PubMed.
- J. Autebert, J. F. Cors, D. P. Taylor and G. V. Kaigala, Anal. Chem., 2016, 88, 3235–3242 CrossRef CAS PubMed.
- M. A. Qasaimeh, T. Gervais and D. Juncker, Nat. Commun., 2011, 2, 464 CrossRef PubMed.
- V. Miralles, A. Huerre, F. Malloggi and M.-C. Jullien, Diagnostics, 2013, 3, 33–67 CrossRef PubMed.
- M. S. Boybay, A. Jiao, T. Glawdel and C. L. Ren, Lab Chip, 2013, 13, 3840 RSC.
- S. M. Shameli, T. Glawdel, V. E. Fernand and C. L. Ren, Electrophoresis, 2012, 33, 2703–2710 CrossRef CAS PubMed.
- R. Garcia, A. W. Knoll and E. Riedo, Nat. Nanotechnol., 2014, 9, 577–587 CrossRef CAS PubMed.
- D. Pires, J. L. Hedrick, A. De Silva, J. Frommer, B. Gotsmann, H. Wolf, M. Despont, U. Duerig and A. W. Knoll, Science, 2010, 328, 732–735 CrossRef CAS PubMed.
- P. D. Tovee and O. V. Kolosov, Nanotechnology, 2013, 24, 465706 CrossRef PubMed.
- P. Dörig, P. Stiefel, P. Behr, E. Sarajlic, D. Bijl, M. Gabi, J. Vörös, J. A. Vorholt and T. Zambelli, Appl. Phys. Lett., 2010, 97, 23701 CrossRef.
- R. D. Piner, J. Zhu, F. Xu, S. Hong and C. A. Mirkin, Science, 1999, 283, 661–663 CrossRef CAS PubMed.
- J. Zhong, G. Sun and D. He, Nanoscale, 2014, 12217–12228 RSC.
- G. V. Kaigala, R. D. Lovchik, U. Drechsler and E. Delamarche, Langmuir, 2011, 27, 5686–5693 CrossRef CAS PubMed.
- R. D. Lovchik, G. V. Kaigala, M. Georgiadis and E. Delamarche, Lab Chip, 2012, 12, 1040–1043 RSC.
- J. F. Cors, R. D. Lovchik, E. Delamarche and G. V. Kaigala, Rev. Sci. Instrum., 2014, 85, 34301 CrossRef CAS PubMed.
- A. Kashyap, J. Autebert, E. Delamarche and G. V. Kaigala, Sci. Rep., 2016, 6, 29579 CrossRef PubMed.
- A. Sarkar, S. Kolitz, D. A. Lauffenburger and J. Han, Nat. Commun., 2014, 5, 3421 Search PubMed.
- A. Queval, N. R. Ghattamaneni, C. M. Perrault, R. Gill, M. Mirzaei, R. A. McKinney and D. Juncker, Lab Chip, 2010, 10, 326 RSC.
- I. Węgrzyn, A. Ainla, G. Jeffries and A. Jesorka, Sensors, 2013, 13, 4289–4302 CrossRef PubMed.
- N. Ostromohov, M. Bercovici and G. V. Kaigala, Lab Chip, 2016, 16, 3015–3023 RSC.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental setup, finite element modelling, analytical model, calibration. See DOI: 10.1039/c6cc08018a |
| This journal is © The Royal Society of Chemistry 2016 |

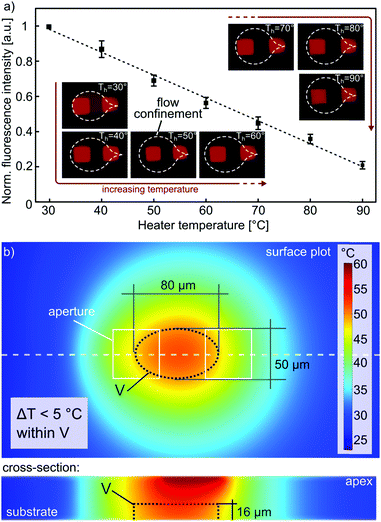
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 μm3 elliptical volume remains below 5 °C.
000 μm3 elliptical volume remains below 5 °C.