Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
G-Quadruplex formation using fluorescent oligonucleotides as a detection method for discriminating AGG trinucleotide repeats†
Yoojin
Park
,
Ki Tae
Kim
and
Byeang Hyean
Kim
 *
*
Department of Chemistry, Division of Advanced Materials Science, Pohang University of Science and Technology (POSTEH), Pohang 37673, Republic of Korea. E-mail: bhkim@postech.ac.kr
First published on 3rd October 2016
Abstract
We have developed a simple and sensitive system for detecting AGG trinucleotide repeats through the formation of intermolecular G-quadruplexes using a fluorescent oligonucleotide. The fluorescence signal increased rapidly and dramatically by 44.7-fold with respect to the low background signal in the presence of RNA agg repeats and by 35.0-fold in the presence of DNA AGG repeats.
G-Quadruplexes are intra- and intermolecular secondary structures that contain guanine (G)-rich DNA and RNA sequences. Recently, many G-quadruplexes have been reported to include homo-tetrads [adenine (A), cytosine (C), thymine (T), or uracil (U) tetrads], mixed tetrads (G:C:G:C, G:T:G:T, or A:T:A:T), pentads, hexads, heptads or octads as well as G-tetrads.1,2 In both DNA and RNA, trinucleotide repeats (TNRs) based on AGG units can form ordered atypical G-quadruplex structures consisting of hexad or heptad planes as well as G-tetrads.3,4
AGG trinucleotide repeats are located in biologically important regions in genes. The RNA agg repeats are represented at high frequency in the exonic sequence in human transcripts. They are stabilized on the mRNA and decreased gene expression because of pausing of the ribosomes during translation.1 RNA r(agg)4 sequences have been reported to form an intramolecular parallel G-quadruplex from one G-tetrad and one hexad plane (Fig. 1A).1 DNA AGG repeats can form a similar G-quadruplex structure to that of RNA agg repeats, including a G-tetrad and a heptad plane(Fig. 1B).4 DNA AGG repeats are abundant in gene regulatory regions known to modulate gene expression at the transcription level (e.g., the gene promoter region).5 AGG repeats are found most notably in the human c-myb proto-oncogene region. The c-myb gene is overexpressed in many leukemias and tumor cells and is a potential therapeutic target.6,7 An antisense oligonucleotide of the c-myb gene and c-myb mutants disrupts gene function; its use can, therefore, be a therapeutically effective strategy.6 The Myc-associated zinc finger protein (MAZ) can bind to the c-myb G-quadruplex and restrict the c-myb promoter activity.5 Furthermore, the G-quadruplexes formed in c-myb repeats also play critical roles in regulating gene expression.8 The Yuan group used a flexible cyclic polyamide and electrospray ionization mass spectroscopy (ESI-MS) to probe the G-quadruplex formed from the c-myb oncogene promoter.9 These results suggested that induced formation of G-quadruplexes and their detection have the potential to modulate c-myb gene expression. Although AGG repeats have biological significance, no fluorescence-based methods have been developed previously to probe them in genes. Therefore, employing G-quadruplex formation as a means for the detection of both RNA agg and DNA AGG repeats using highly discriminating fluorescent probes remains a challenge. In a previous study, metal complexes were found to bind the G-quadruplex structures and produce enhanced fluorescence signals.10 We also developed a system to monitor the formation of a (3+1) intermolecular G-quadruplex and to detect G-quadruplex-forming sequences through fluorescence enhancement.11 Inspired by a probing system based on competition between two interstrand G-quadruplexes, here we report a fluorescence probe for the AGG repeats, which can form a very stable intermolecular G-quadruplex including hexad and heptad planes as well as a G-tetrad plane. This system has three properties that facilitate the recognition of the AGG repeats. First, the background signal has a very low intensity when 5-(1-pyrenylethynyl)-2′-deoxyuridine (PyU) is used instead of BODIPY-phenyl-2′-deoxyuridine (BodU) in the absence of the target sequence.11,12 Second, the system exhibits dramatic discrimination, with strong fluorescence intensity response only to the formation of an intermolecular G-quadruplex with the AGG repeats, especially RNA agg repeats, even though other XGG (X = T, DNA C, RNA C, U) repeats might also possibly form a G-quadruplex. Third, the probe rapidly folds into a stable parallel intermolecular G-quadruplex and the fluorescence intensity also rapidly increases in the presence of the target sequences. Accordingly, induced G-quadruplex formation between the probe and AGG repeats appears to be a simple, selective, and sensitive method for the detection of AGG repeats, and is a particularly distinguished method for probing trinucleotide repeats (Fig. 1C).
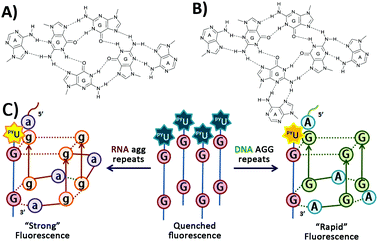 | ||
| Fig. 1 (A) Hexad structure1 by RNA (agg)4, (B) heptad structure4 by DNA (AGG)4, and (C) schematic representation of a fluorescent probe for G-quadruplex formation from both RNA agg repeats and DNA AGG repeats, based on the formation of intermolecular G-quadruplexes. | ||
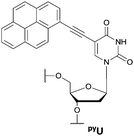
First, we designed tetramolecular G-quadruplex structures having pyU in their sequences. G-Rich oligodeoxynucleotides (ODNs) containing pyrene units at their 5′-ends are assembled together to form a stable G-quadruplex with quenched fluorescence intensity as a result of π-stacking and hydrophobic interactions.11–14 A fluorophore conjugated with deoxyribose can induce a stable G-quadruplex by stacking on the terminal G-tetrad; addition of a T unit at the opposite end of the G-rich sequence also greatly improves the stability of the G-quadruplex.14–17 We synthesized four ODNs (PYG2–PYG5) incorporating PyU units and possessing G sequences of various lengths (Table 1). We used MALDI-TOF mass spectroscopy to confirm the structures of PYG2–PYG5 oligonucleotides. PYG2–PYG5 exhibited fluorescence quenching in the presence of KCl as a result of the formation of parallel tetramolecular G-quadruplexes (Fig. 2 and Fig. S1, ESI†). The fluorescence emission intensity was lower than that obtained previously using BodU.11 We presume that the fluorescence quenching of PYG2–PYG5 resulted from hydrophobic interactions of their pyrene units that stabilized their assemblies. The formation of secondary structures from PYG2–PYG5 in the presence of KCl resulted in red-shifts in the UV-Vis spectra in the pyrene absorption region (Fig. S2, ESI†).13
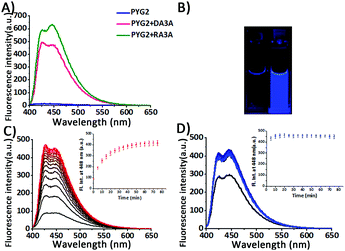
Next, we measured the fluorescence emission of PYG2–PYG5 to evaluate their potential for use in the sensing of AGG repeats through the formation of intermolecular G-quadruplexes in the presence of added K+ ions. All samples were annealed prior to measurement. When PYG2–PYG5 interacted with RA3A, the RNA target sequence, we observed very strong fluorescence enhancements: 44.7-, 20.5-, 8.6-, and 5.5-fold increases in fluorescence, respectively. In the presence of DA3A as the DNA target sequence, PYG2–PYG5 also exhibited large increases in fluorescence: 35.0-, 6.9-, 2.3-, and 1.5-fold, respectively (Fig. 2A and Fig. S1, ESI†). The fluorescence intensity of PYG2, which provided the largest increases in fluorescence intensity for both targets, did not change in the absence of KCl (Fig. S3, ESI†). As expected, the fluorescence of PYG2–PYG5 recovered upon formation of secondary structures with the target sequences. The UV absorbance wavelength of PYG2 shifted in the pyrene region upon addition of the target sequences (Fig. S4, ESI†). Notably, both PYG4 and PYG5 preferred to form a tetramolecular G-quadruplex in competition with the fluorescent (3+1) intermolecular G-quadruplex. It is known that longer G-rich sequences can form secondary structures with higher thermal stability and more rapid association; the addition of other bases at the 5′ or 3′ end can also increase the thermal stability of the G-quadruplexes.14,17 The ODN featuring a pyrene unit as well as a consecutive G-rich sequence forms a stable G-quadruplex even in the absence of K+ ions and does not dissociate upon increasing the temperature.13,15 In the presence of KCl, PYG3 forms a parallel G-quadruplex structure that is stable at room temperature, but dissociates at a temperature lower than that of the PYG4 and PYG5 sequences. PYG3 participates in the competition between the tetramolecular G-quadruplex and the (3+1) intermolecular G-quadruplex in the presence of G-rich target sequences.11,15 Nevertheless, the increases of fluorescence intensity with PYG3 were lower than those with PYG2 in the presence of AGG repeats. For both DA3A and RA3A, probe PYG2—featuring two G units, two T units, and a pyU unit at the 5′-end—exhibited the highest efficiency, in terms of its good brightness and fluorescence enhancement. Remarkably, this enhancement in fluorescence was visible to the naked eye (Fig. 2B). PYG2 also exhibited strong fluorescence intensity enhancements at 20 °C after 80 min: 29.1-fold upon addition of single-stranded DA3A, and 31.4-fold upon addition of single-stranded RA3A without annealing (Fig. 2C and D). Here, the fluorescence intensity in the presence of DA3A almost reached the maximum fluorescence intensity of DA3A and PYG2 after annealing. This result suggests that these products, (3+1) intermolecular G-quadruplexes, are thermodynamically favoured products (Fig. 2C; Fig. S5, ESI†). The fluorescence enhancement was also exhibited upon addition of single-stranded RA3A, with the maximum fluorescence intensity maintained after 20 min (Fig. 2D). Because K+ ions induce rapid formation of parallel G-quadruplexes, including hetero-quadruplexes,17,18 the construction of intermolecular G-quadruplexes from PYG2 and target sequences also occurred with rapid responses and resulted in an enhanced fluorescence signal. The Tm value of the (3+1) intermolecular G-quadruplexes with PYG2 and targets is higher than that of the secondary structure of PYG2 (Fig. S6, ESI†). Accordingly, we speculated that PYG2 might be a suitable candidate for probing AGG repeats with a very low background signal. To examine the effect of the T units in PYG2 on the detection of the AGG repeats, we replaced it with A, C, and G units obtaining PYG2A, PYG2C and PYG2G, respectively. The fluorescence intensities of these sequences were all quenched well in the presence of KCl; they also provided distinguishable fluorescence emission intensities in the presence of DA3A and RA3A (Fig. S7, ESI†). Nevertheless, the enhancements in their fluorescence intensity were lower than those of PYG2 in the presence of the target AGG repeats. We suspect that an interaction between the T unit of the PYG2 probe and the A unit of the AGG repeat sequence stabilized the secondary structure and was responsible for this difference in fluorescence intensity.
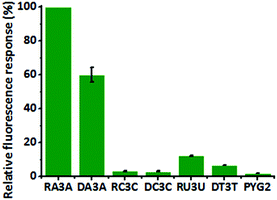
We investigated the structure of PYG2 through circular dichroism (CD) spectroscopy in the absence and presence of KCl. Although PYG2 displayed quenched fluorescence intensity in both cases, the CD spectra featured different patterns. Probe PYG2 formed the secondary structure in the presence of KCl (Fig. S8, ESI†). The CD spectrum of DA3A revealed that the peak near 260 nm increased while that near 240 nm decreased in the presence of PYG2, exhibiting the typical pattern of a parallel G-quadruplex. These characteristic peaks also appeared for DA4 in its G-quadruplexes (Fig. 3A; Fig. S9, ESI†).19,20 The patterns of the CD bands were similar in the cases of the RNA agg repeats, but with some distinctions. The mixture of PYG2 and RA3A also exhibited bands of a parallel G-quadruplex at 260 and 240 nm. An additional positive band of low intensity appeared near 300 nm, attributed to a G-quadruplex dimer formed through hexad–hexad stacking. The spectrum of the intramolecular parallel G-quadruplex of RNA RA4 also featured these significant peaks (Fig. 3B; Fig. S9, ESI†).21,22 In the absence of KCl, the spectrum of the mixture of DA3A/PYG2 and RA3A/PYG2 did not feature any significant peaks (Fig. S9, ESI†). In order to obtain the binding stoichiometry of PYG2 to DA3A and RA3A, the fluorescence Job plots were recorded. The inflection point in each graph is observed at PYG2 fractions of 0.53 and 0.51, which correspond to a 1:1 binding stoichiometry to DA3A and RA3A (Fig. S10, ESI†). Taken together, it appeared that the AGG repeat sequences associated with PYG2 to form a bimolecular parallel G-quadruplex (Fig. 3; Fig. S11, ESI†).23 Thus, we applied PYG2 to detect longer AGG repeats that could form intramolecular G-quadruplexes. We monitored the fluorescence changes of DA4 and RA4 in the presence of PYG2. Although these sequences themselves can form very stable G-quadruplexes, the presence of PYG2 increased the fluorescence as soon as it was added at the same concentration as the target sequences, without annealing; in addition, enhanced fluorescence occurred in the presence of the targets annealed with the probe (Fig. S12, ESI†). The enhanced fluorescence intensity was less discriminated than those for the DA3A and RA3A sequences containing three repeat AGG units, because of greater competition between the formation of an intermolecular G-quadruplex and an intramolecular G-quadruplex.11 The CD spectrum of DA4 with PYG2 was similar to that of DA4 as parallel G-quadruplexes, and the CD spectrum of RA4 with PYG2 displayed comparable patterns with that of RA4. Therefore, PYG2 binds to DA4 and RA4 by forming intermolecular G-quadruplexes (Fig. S13, ESI†). The fluorescence Job's plot suggested 1:1 binding stoichiometry in both PYG2/DA4 and PYG2/RA4 (Fig. S14, ESI†). To demonstrate the feasibility of using PYG2 as a probe for AGG repeats, we examined its response toward varying concentrations of the AGG repeat sequence. The fluorescence intensities of PYG2 were enhanced upon increasing the concentration of both targets DA4 and RA4. We also evaluated the sensitivity of PYG2 in terms of its detection threshold. When we annealed with the probe, the detection thresholds of DA3A and RA3A were 0.02 and 0.01 μM, respectively; the targets added to the probe without annealing exhibited detection thresholds of 0.05 and 0.01 μM, respectively (Fig. S15, ESI†). As the length of the target is increased, the probe exhibited higher fluorescence intensity and a rapid response (Fig. S16, ESI†). We also observed thermodynamic control in the system. Despite the high thermal stability of the AGG G-quadruplex, we monitored enhanced fluorescence intensity when we added PYG2 to the intramolecular G-quadruplex of both DA4 and RA4 (Fig. S17, ESI†). Thus, the probe also shows fast response with the G-quadruplex of target sequences under thermodynamic control. Therefore, this system appeared to have the potential for use as a fast, simple, and sensitive method for the detection of G-quadruplexes as well as long sequences of AGG repeats. Based on these findings, we evaluate the selectivity of PYG2 for probing the AGG repeat sequences. We investigated the enhancements in its fluorescence intensity in the presence of RC3C, RU3U, DC3C and DT3T, which are trinucleotide repeats having the sequence (XGG)3X (X = C, U and T) (Fig. 4). Although the CGG repeat has G dinucleotides, it forms a stable hairpin or coexists as a hairpin and a G-quadruplex in both DNA and RNA.24,25 Much like G-tetrads, the TGG and UGG repeats are known as tetramolecular G-quadruplex-forming sequences containing T- and U-tetrads, respectively.1,26 PYG2 dramatically displayed enhanced fluorescence intensity for only the AGG repeats. The conformation and configuration of the G-quadruplex affected the interactions of PYG2 with the XGG trinucleotide sequences (Fig. S18, ESI†). PYG2 also exhibited relatively low fluorescence emission with other G-quadruplex-forming sequences (Fig. S19, ESI†). Thus, this strategy provided high selectivity toward the AGG repeats over other G-quadruplex-forming sequences, especially XGG repeats, based on the formation of the intermolecular G-quadruplex, distinguishing this method from those reported previously.
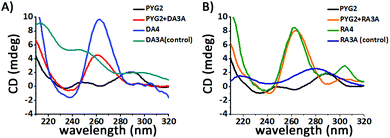 | ||
| Fig. 3 CD spectra of PYG2 in the presence of (A) DA3A and (B) RA3A repeat sequences (2.0 μM of annealed sample, 1 mL; 10 mM Tris–HCl buffer, pH 7.2; 500 mM KCl; 20 °C). | ||
In conclusion, a 5-mer fluorescent oligonucleotide probe induces (3+1) intermolecular G-quadruplex formation allowing detection of AGG repeats from a very low background signal. Our tested strategy of probing AGG repeats based on a G-quadruplex structure is distinct from other systems for the detection of trinucleotide repeats. It provides highly selective, sensitive, and rapid recognition of both RNA agg repeats and DNA AGG repeats with exceptional fluorescence properties. Because PYG2 is a remarkable probe for the detection of AGG repeat sequences and induced formation of G-quadruplexes, it makes these AGG repeats a potential therapeutic target.
We thank the NRF project (2014R1A2A1A12067404 and 2015M3A9B8029067) for financial support.
Notes and references
- M. Malgowska, D. Gudanis, R. Kierzek, E. Wyszko, V. Gabelica and Z. Gdaniec, Nucleic Acids Res., 2014, 42, 10196 CrossRef CAS PubMed.
- N. Borbone, J. Amato, G. Oliviero, V. D'Atri, V. Gavelica, E. De Pauw, G. Piccialli and L. Mayol, Nucleic Acids Res., 2011, 39, 7848 CrossRef CAS PubMed.
- (a) K. Sobczak, G. Michlewski, M. Mezer, E. Kierzek, J. Korl, M. Olejniczak, R. Kierzek and W. J. Krzyzosiak, J. Biol. Chem., 2010, 285, 12755 CrossRef CAS PubMed; (b) P. Galka-Marciniak, M. O. Urbanek and W. J. Krzyzosiak, Biol. Chem., 2012, 393, 1299 CrossRef CAS PubMed.
- A. Matsugami, T. Okuizumi, S. Uesugi and M. Katahira, J. Biol. Chem., 2003, 278, 28147 CrossRef CAS PubMed.
- S. L. Palumbo, R. M. Memmott, D. J. Uribe, Y. Krotova-Khan, L. H. Hurley and S. W. Ebbinghaus, Nucleic Acids Res., 2008, 36, 1755 CrossRef CAS PubMed.
- (a) A. M. Gewirtz and B. Calabretta, Science, 1988, 242, 1303 CAS; (b) A. M. Gewirtz, Oncogene, 1999, 18, 3056 CrossRef CAS PubMed; (c) H. K. Yi, S. Y. Nam, J. C. Kim, J. S. Kim, D. Y. Lee and P. H. Hwang, Exp. Hematol., 2002, 30, 1139 CrossRef CAS PubMed.
- (a) D. J. Patel, A. T. Phan and V. Kuryavyi, Nucleic Acids Res., 2007, 35, 7429 CrossRef CAS PubMed; (b) G. W. Collie and G. N. Parkinson, Chem. Soc. Rev., 2011, 40, 5867 RSC.
- C. Broxson, J. Beckett and S. Tornaletti, Biochemistry, 2011, 50, 4162 CrossRef CAS PubMed.
- (a) Q. Zhang, X. Cui, S. Lin, J. Zhou and G. Yuan, Org. Lett., 2012, 14, 6126 CrossRef CAS PubMed; (b) X. Cui, Q. Zhang, H. Chen, J. Zhou and G. Yuan, J. Am. Soc. Mass Spectrom., 2014, 25, 684 CrossRef CAS PubMed.
- (a) L. Lu, D. S.-H. Chan, D. W. J. Kwong, H.-Z. He, C.-H. Leung and D.-L. Ma, Chem. Sci., 2014, 5, 4561 RSC; (b) M. Wang, Z. Mao, T.-S. Kang, C.-Y. Wong, J.-L. Mergny, C.-H. Leung and D.-L. Ma, Chem. Sci., 2016, 7, 2516 RSC.
- K. T. Kim and B. H. Kim, Chem. Commun., 2013, 49, 1717 RSC.
- Y. J. Seo, G. T. Hwang and B. H. Kim, Tetrahedron Lett., 2006, 47, 4037 CrossRef CAS.
- O. Doluca, J. M. Withers, T. S. Loo, P. J. B. Edwards, C. Gonzalez and V. V. Filichev, Org. Biomol. Chem., 2015, 13, 3742 CAS.
- J. D'Onofrio, L. Petraccone, E. Erra, L. Martino, G. Di Fabio, L. De Napoli, C. Giancola and D. Montesarchio, Bioconjugate Chem., 2007, 18, 1194 CrossRef PubMed.
- E. E. Merkina and K. R. Fox, Biophys. J., 2005, 89, 365 CrossRef CAS PubMed.
- H. Zhu and F. D. Lewis, Bioconjugate Chem., 2007, 18, 1213 CrossRef CAS PubMed.
- P. L. Tran, A. D. Cian, J. Gros, R. Moriyama and J.-L. Mergny, Top. Curr. Chem., 2013, 330, 243 CrossRef CAS PubMed.
- S. Roy, K. J. Zanotti, C. T. Murphy, F. A. Tanious, W. D. Wilson, D. H. Ly and B. A. Armitage, Chem. Commun., 2011, 47, 8524 RSC.
- A. Matsugami, K. Ouhashi, M. Kanagawa, H. Liu, S. Kanagawa, S. Uesugi and M. Katahira, J. Mol. Biol., 2001, 313, 255 CrossRef CAS PubMed.
- I. Kejnovska, J. Kypr and M. Vorickova, Chirality, 2003, 15, 584 CrossRef CAS PubMed.
- F. Nishikawa, K. Murakami, A. Matsugami, M. Katahira and S. Nishikawa, Oligonucleotides, 2009, 19, 179 CrossRef CAS PubMed.
- J. M. Lipay and M.-R. Mihailescu, Mol. BioSyst., 2009, 5, 1347 RSC.
- A. Paul, P. Sengupta, Y. Krishnan and S. Ladame, Chem. – Eur. J., 2008, 14, 8682 CrossRef CAS PubMed.
- K. Usdin, Nucleic Acids Res., 1998, 26, 4078 CrossRef CAS PubMed.
- K. Sobczak, M. Mezer, G. Michlewski, J. Krol and W. J. Krzyzosiak, Nucleic Acids Res., 2003, 31, 5469 CrossRef CAS PubMed.
- F. M. Chen, Biophys. J., 1997, 73, 348 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6cc06566j |
| This journal is © The Royal Society of Chemistry 2016 |