Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
A highly selective fluorogenic probe for the detection and in vivo imaging of Cu/Zn superoxide dismutase†
Liyun
Zhang‡
*a,
Jun Cheng
Er‡
bc,
Hao
Jiang
d,
Xin
Li
a,
Zhaofeng
Luo
d,
Thomas
Ramezani
e,
Yi
Feng
e,
Mui Kee
Tang
b,
Young-Tae
Chang
b and
Marc
Vendrell
*e
aInstitute of Technical Biology and Agriculture Engineering, Key Laboratory of Ion Beam Bioengineering, Hefei Institutes of Physical Science, Chinese Academy of Sciences, Hefei, Anhui 230031, P. R. China. E-mail: zly0605@ustc.edu.cn
bDepartment of Chemistry, National University of Singapore, 3 Science Drive 2, 117543, Singapore
cGraduate School for Integrative Sciences and Engineering, National University of Singapore, Centre for Life Sciences, #05-01, 28 Medical Drive, 117456 Singapore
dSchool of Life Sciences, University of Science and Technology of China, Hefei, Anhui 230027, P. R. China
eMRC Centre for Inflammation Research, Queen's Medical Research Institute, University of Edinburgh, EH16 4TJ Edinburgh, UK. E-mail: mvendrel@staffmail.ed.ac.uk
First published on 25th February 2016
Abstract
Copper/zinc superoxide dismutase (Cu/Zn SOD) is an essential enzyme that protects tissue from oxidative damage. Herein we report the first fluorogenic probe (SODO) for the detection and in vivo imaging of Cu/Zn SOD. SODO represents a unique chemical probe for translational imaging studies of Cu/Zn SOD in inflammatory disorders.
Superoxide dismutases (SOD, EC 1.15.1.1) are metalloenzymes that protect tissue from the oxidative stress caused by reactive oxygen species (ROS).1 The main function of SODs is to catalyse the dismutation of superoxide radicals (O2˙−) to hydrogen peroxide (H2O2) and oxygen. There are several isoforms of SODs, which can be distinguished by their metal cofactors and their distribution in cells.2 Among the different isoforms of SODs, copper/zinc superoxide dismutase (Cu/Zn SOD or SOD1) is widely distributed and comprises around 90% of the total SODs. Alterations in the expression and activity of Cu/Zn SOD have been associated with the onset of a number of diseases. Mutations in human Cu/Zn SOD are implicated in the development of neurological disorders, such as familial amyotrophic lateral sclerosis (fALS), Alzheimer's disease and Parkinson's disease.3–5 Furthermore, elevated activities of Cu/Zn SOD have been reported in cancer (e.g. acute myelogenous leukaemia, Hodgkin's lymphoma) and chronic inflammatory diseases (e.g. rheumatoid arthritis, ischemic injury).5–7 On the contrary, decreased levels of Cu/Zn SOD have been associated with an inhibition of the immune response and the promotion of oxidative stress in age-related disorders.8,9
Despite the importance of Cu/Zn SOD in regulating the balance between healthy and disease states, the exact mechanism that correlates Cu/Zn SOD to the progression of different pathologies remains largely unknown.5 Current probes to visualize SODs mainly rely on the intrinsic fluorescence of Tyr or Trp residues10,11 or the use of non-specific metal chelators, such as bathocuproine.12 These methods have very limited practical use in vivo, due to spectral shortcomings (e.g. short excitation/emission wavelengths) and their poor selectivity between SODs and other ROS-related enzymes.
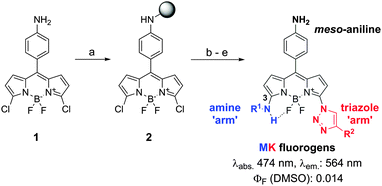
Fluorogenic probes are advantageous for in vivo imaging since they provide high signal-to-noise ratios without the need for washing steps.13,14 Our group and others have reported the preparation of fluorogenic probes based on the 4,4-difluoro-4-bora-3a,4a-diaza-s-indacene (BODIPY) scaffold,15,16 one of the most exploited fluorophores for cell imaging due to its photostability and permeability properties.17,18 BODIPY fluorogens can be synthesized by direct conjugation of electron-rich groups (e.g. substituted benzene rings) to the BODIPY core, leading to photoinduced electron transfer (PeT) quenching and subsequent turn-on fluorescence emission in hydrophobic environments. In order to enhance the fluorogenic response of probes binding to Cu/Zn SOD, we designed a new class of BODIPY fluorogens combining PeT-quenching substituents and chemical groups restricting the rotational flexibility of the BODIPY core. The restriction of torsional motion has proven an effective strategy to generate turn-on fluorescent probes,19,20 and previous studies have shown that “–NH” groups directly linked to the position C3 of BODIPY can form intramolecular hydrogen bonds with the fluorine atoms.21 We synthesized MK fluorogens by modifying a 3,5-dichloro-BODIPY scaffold (1) with benzylamines (M) forming intramolecular hydrogen bonds with the fluorine atoms, and triazole groups (K) as PeT-quenchers (Scheme 1). MK fluorogens were prepared by loading 1 onto 2-chlorotrityl chloride polystyrene (CTC-PS) resin, followed by nucleophilic substitution and copper-catalysed azide–alkyne cycloaddition. A total of 40 MK compounds with diverse amine and alkyne groups were isolated in moderate to high yields with very high purities using mild acidic cleavage conditions22 (for detailed chemical structures and characterisation data, see ESI†).
The spectral characterisation of the MK derivatives confirmed their strong fluorogenic behaviour with minimal fluorescence in aqueous media, long Stokes shifts (i.e. around 90 nm) and red-shifted emission wavelengths when compared to the BODIPY core (Table S1 in ESI†). As expected, the incorporation of benzylamines at the position C3 of the BODIPY scaffold restricted the torsional motion of the fluorophore, leading to an increase in the quantum yields in non-polar solvents. We also observed that the fluorescence emission of MK fluorogens correlated with solvent viscosity as a result of the decreased rotation of both triazole and aniline substituents (Fig. S1 in ESI†). Moreover, the fluorogenic response of MK derivatives was stronger in non-polar solvents due to the reduced PeT quenching effect from the meso-aniline group in non-polar environments (Fig. S2, S3 and Table S2 in ESI†). Altogether, these results assert MK derivatives as BODIPY fluorogens with excellent spectral properties to detect polarity changes associated to the binding at large macromolecules.
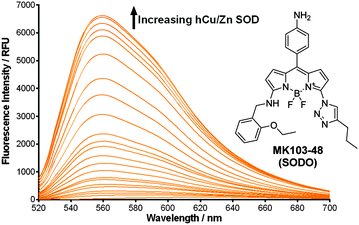
In view of these properties, we assessed our MK fluorogens in vitro to bind at the macromolecular dimeric structure of Cu/Zn SOD. The screening of diversity-oriented fluorescence libraries has become an effective strategy to identify highly selective molecular probes,23 and we observed that compounds with 2-ethoxybenzylamine (M103) as the amine group displayed high fluorescence emission after incubation with human Cu/Zn SOD (hCu/Zn SOD) (Fig. S4 in ESI†). MK103-48 showed the strongest response among all compounds and was selected for further studies (hereinafter named as SODO (SOD Orange), Fig. 1). SODO displayed up to 150-fold increase in fluorescence emission after binding at hCu/Zn SOD, with a limit of detection of 10 μg mL−1 (Fig. S5 in ESI†). Notably, SODO reached quantum yields around 45% and displayed a remarkable 60 nm hypsochromic shift in the fluorescence spectrum after binding (Fig. 1 and Fig. S5, S6 in ESI†).
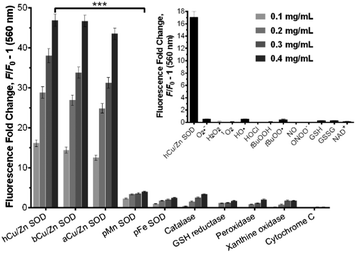
We examined the binding of SODO at Cu/Zn SODs from different species. As with hCu/Zn-SOD, SODO displayed a concentration-dependent response in Cu/Zn SODs from bovine blood (bCu/Zn SOD) and from Arabidopsis thaliana (aCu/Zn SOD) (Fig. 2). These results suggest that the binding of SODO is species-independent and occurs at a conserved hydrophobic region of Cu/Zn SOD. While Cu/Zn SOD stands for the majority of SOD in tissue, high selectivity for the Cu/Zn SOD isoform is essential for imaging studies. We assessed the fluorescence response of SODO in the other two SOD isoforms (i.e. Mn-SOD and Fe-SOD) and observed high selectivity for Cu/Zn SOD over Mn-SOD and Fe-SOD, where minimal binding was detected (Fig. 2). To the best of our knowledge, SODO is the first small fluorophore able to detect Cu/Zn SOD with high specificity over other SODs. We also assessed the fluorescence emission of SODO in other ROS-related enzymes (e.g. catalase, peroxidase) (Fig. 2) and metabolites (e.g. H2O2, O2˙−, 1O2, OH˙) (Fig. 2, inset). SODO exhibited very high specificity for Cu/Zn-SOD, showing minimal fluorescence in other enzymes and metabolites involved in oxidative damage and inflammatory processes. Finally we studied whether the binding of SODO affected the catalytic activity of Cu/Zn SOD (Fig. S7 in ESI†). SODO did not significantly perturb enzymatic function; hence being an excellent reporter of Cu/Zn SOD without altering the normal physiology of cells. Altogether, these results confirm SODO as the first fluorogenic probe to detect Cu/Zn SOD without cross-reacting with other SOD isoforms, enzymes or ROS.
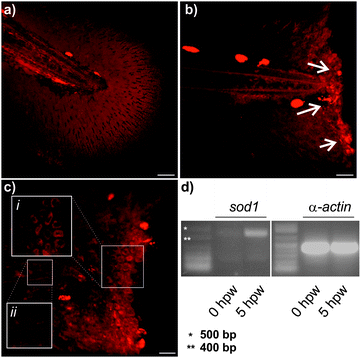
In view of the high selectivity and fluorogenic properties of SODO, we employed it to visualise changes in the expression of Cu/Zn SOD in vivo. We used SODO to image Cu/Zn SOD during the onset of inflammatory processes in zebrafish embryos.9 We employed a zebrafish tail fin injury model of inflammation by amputating the tail fin of embryos at 3 days post fertilization (dpf),24 which allowed us to examine the in vivo fluorogenic response of SODO in the inflammatory milieu. As shown in Fig. 3b, zebrafish undergoing inflammation displayed bright fluorescence in the wound margins (white arrows), which correspond to inflamed areas where Cu/Zn SOD is highly expressed. High magnification images corroborated the expression of Cu/Zn SOD in the cytoplasm of epithelial cells (Fig. 3c). We further confirmed these results by measuring the levels of the sod1 gene before and after wounding using semi-quantitative reverse transcription-polymerase chain reaction (RT-PCR). As shown in Fig. 3d, the sod1 gene was highly upregulated 5 h after wounding, in agreement with the fluorescence emission profile of SODOin vivo (Fig. S8 in ESI†). We also observed that SODO brightly stained oxidatively-stressed fibroblasts (Fig. S9 in ESI†), containing high levels of Cu/Zn SOD.25 Cell viability assays in fibroblasts also corroborated the marginal cytotoxicity of SODO within the working concentration range (Fig. S10 in ESI†).
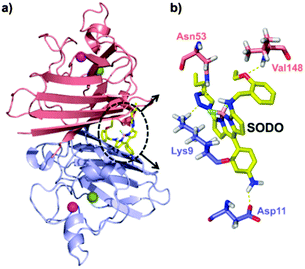
In order to determine the binding mode of SODO in Cu/Zn SOD, we performed docking calculations to analyse the interaction between SODO at hCu/Zn SOD. Cu/Zn-SOD is found in all eukaryotic species as a homodimeric enzyme of ∼32 kDa containing one Cu and one Zn ion in each of the subunits, which are stabilized by an intra-chain disulfide bond.26 Our model predicted the interaction of SODO at the interface of the two subunits of Cu/Zn SOD (Fig. 4a). The binding at this conserved hydrophobic pocket, which is away from the catalytic site of the enzyme, is consistent with the previously observed species-independent response of SODO (Fig. 2) and the fact that the enzymatic activity of Cu/Zn SOD remained unaffected by SODO (Fig. S7 in ESI†). A closer examination of the binding revealed four hydrogen bonds between SODO and hCu/Zn SOD: one hydrogen bond between the oxygen atom of the ethoxy group and Val148, two hydrogen bonds between the nitrogen atoms of the triazole ring and the residues Lys9 and Asn53, and a final hydrogen bond between the meso-aniline group and Asp11 (Fig. 4b). The binding analysis suggests that the fluorogenic response of SODO is the result of combining the restriction in the rotation of the fluorophore by forming four hydrogen bonds and the deactivation of the quenching PeT due to the migration to a hydrophobic environment, as observed in our results from the in vitro characterisation assays.
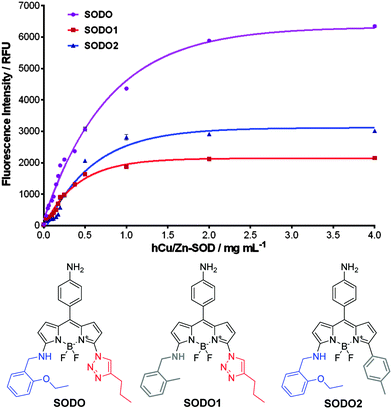
In order to corroborate this hypothesis, we prepared two derivatives of SODO lacking the chemical groups involved in the interaction with hCu/Zn SOD (Fig. 5). We synthesized SODO1 as the derivative without the ethoxy group in the amine ‘arm’ and SODO2 as the derivative lacking the triazole nitrogen atoms (ESI† for synthetic details and characterisation), and compared their fluorogenic response to SODO after binding to hCu/Zn SOD. SODO1 and SODO2 showed remarkably lower fluorescence emission than SODO, confirming the relevance of both ethoxy and triazole groups for binding at hCu/Zn SOD (Fig. 5). These results confirmed the need of four hydrogen bonds, which are missing in the analogues SODO1 and SODO2 (Fig. S11 in ESI†), to restrict the torsional motion of SODO and induce its maximal fluorogenic response.
In summary, we have designed a new class of BODIPY fluorogens with enhanced spectral properties by incorporating both rotational restriction and PeT-quenching groups. These new BODIPY fluorogens show excellent properties as polarity probes with minimal background emission in aqueous media and long Stokes shifts upon fluorescence activation. In vitro studies identified one derivative (SODO) as a highly selective fluorogenic probe for Cu/Zn SOD. SODO shows remarkable fluorescence emission only after binding to Cu/Zn SOD with very high selectivity over ROS-related enzymes and metabolites as well as the other SOD isoforms (i.e. Mn-SOD and Fe-SOD). The high selectivity of SODO enabled its use for imaging Cu/Zn SOD in vivo during the onset of an inflammatory response in a zebrafish tail fin injury model. Furthermore, we performed computational modelling to analyse the binding of SODO at Cu/Zn SOD. Structure–activity studies suggest that the binding occurs at the interface of the two enzymatic subunits and involves four residues to restrict the torsional motion of the BODIPY fluorophore and deactivate its PeT-quenching groups. SODO is the first fluorogenic probe for Cu/Zn SOD and represents a unique probe for the detection and in vivo imaging of Cu/Zn SOD during the progression of inflammatory disorders.
L. Z. acknowledges the ‘973’ program (2014CB932002), the Natural Science Foundation of China (11105150) and the Special Financial Grant from China Postdoctoral Science Foundation (2013T60613). J. C. E. acknowledges a NGS scholarship. Y. F. is a Wellcome Trust Sir Henry Dale Fellow (WT 100104/Z/12/Z). Y.-T. C. acknowledges funding from the National Medical Research Council (NMRC/CBRG/0015/2012). M. V. acknowledges funding from Medical Research Council, Marie Curie Career Integration Grant (333487) and the WT Institutional Strategic Support Fund.
References
- I. Fridovich, Annu. Rev. Biochem., 1995, 64, 97 CrossRef CAS PubMed.
- I. N. Zelko, T. J. Mariani and R. J. Folz, Free Radical Biol. Med., 2002, 33, 337 CrossRef CAS PubMed.
- H. J. Lee, K. J. Korshavn, A. Kochi, J. S. Derrick and M. H. Lim, Chem. Soc. Rev., 2014, 43, 6672 RSC.
- M. A. Hough, J. G. Grossmann, S. V. Antonyuk, R. W. Strange, P. A. Doucette, J. A. Rodriguez, L. J. Whitson, P. J. Hart, L. J. Hayward, J. S. Valentine and S. S. Hasnain, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 5976 CrossRef CAS PubMed.
- R. Noor, S. Mittal and J. Iqbal, Med. Sci. Monit., 2002, 8, 210 Search PubMed.
- R. Gonzales, C. Auclair, E. Voisin, H. Gautero, D. Dhermy and P. Boivin, Cancer Res., 1984, 44, 4137 CAS.
- L. Papa, G. Manfredi and D. Germain, Genes Cancer, 2014, 5, 15 CAS.
- A. Vasilaki and M. J. Jackson, Free Radical Biol. Med., 2013, 65, 317 CrossRef CAS PubMed.
- M. Marikovsky, V. Ziv, N. Nevo, C. Harris-Cerruti and O. Mahler, J. Immunol., 2003, 170, 2993 CrossRef CAS.
- S. T. Ferreira, L. Stella and E. Gratton, Biophys. J., 1994, 66, 1185 CrossRef CAS PubMed.
- E. Luchinat, A. Gianoncelli, T. Mello, A. Galli and L. Banci, Chem. Commun., 2015, 51, 584 RSC.
- D. V. Martyshkin, S. B. Mirov, Y. X. Zhuang, J. P. Crow, V. Ermilov and J. S. Beckman, Spectrochim. Acta, Part A, 2003, 59, 3165 CrossRef CAS.
- P. Shieh, V. T. Dien, B. J. Beahm, J. M. Castellano, T. Wyss-Coray and C. R. Bertozzi, J. Am. Chem. Soc., 2015, 137, 7145 CrossRef CAS PubMed.
- Y. Hori, T. Norinobu, M. Sato, K. Arita, M. Shirakawa and K. Kikuchi, J. Am. Chem. Soc., 2013, 135, 12360 CrossRef CAS PubMed.
- J. C. Er, M. K. Tang, C. G. Chia, H. Liew, M. Vendrell and Y.-T. Chang, Chem. Sci., 2013, 4, 2168 RSC.
- H. Sunahara, Y. Urano, H. Kojima and T. Nagano, J. Am. Chem. Soc., 2007, 129, 5597 CrossRef CAS PubMed.
- T. Kowada, H. Maeda and K. Kikuchi, Chem. Soc. Rev., 2015, 44, 4953–4972 RSC.
- A. Vázquez-Romero, N. Kielland, M. J. Arévalo, S. Preciado, R. J. Mellanby, Y. Feng, R. Lavilla and M. Vendrell, J. Am. Chem. Soc., 2013, 135, 16018 CrossRef PubMed.
- Y.-H. Ahn, J.-S. Lee and Y.-T. Chang, J. Comb. Chem., 2008, 10, 376 CrossRef CAS PubMed.
- Y. Urano, M. Kamiya, K. Kanda, T. Ueno, K. Hirose and T. Nagano, J. Am. Chem. Soc., 2005, 127, 4888 CrossRef CAS PubMed.
- W. Qin, V. Leen, T. Rohand, W. Dehaen, P. Dedecker, M. van der Auweraer, K. Robeyns, L. van Meervelt, D. Beljonne, B. van Averbeke, J. N. Clifford, K. Driesen, K. Binnemans and N. l. Boens, J. Phys. Chem. A, 2008, 113, 439 CrossRef PubMed.
- M. Vendrell, G. G. Krishna, K. K. Ghosh, D. Zhai, J.-S. Lee, Q. Zhu, Y. H. Yau, S. G. Shochat, H. Kim, J. Chung and Y.-T. Chang, Chem. Commun., 2011, 47, 8424 RSC.
- J.-S. Lee, M. Vendrell and Y.-T. Chang, Curr. Opin. Chem. Biol., 2011, 15, 760 CrossRef CAS PubMed.
- L. Li, B. Yan, Y. Q. Shi, W. Q. Zhang and Z. L. Wen, J. Biol. Chem., 2012, 287, 25353 CrossRef CAS PubMed.
- J. H. Chen, K. Stoeber, S. Kingsbury, S. E. Ozanne, G. H. Williams and C. N. Hales, J. Biol. Chem., 2004, 279, 49439 CrossRef CAS PubMed.
- J. A. Tainer, E. D. Getzoff, K. M. Beem, J. S. Richardson and D. C. Richardson, J. Mol. Biol., 1982, 160, 181 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Structures and characterisation for all MK compounds. Full characterisation data (NMR, HR-MS) for all SODO derivatives. See DOI: 10.1039/c6cc00095a |
| ‡ Both authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2016 |

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
: