Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Efficient total syntheses and biological activities of two teixobactin analogues†
Anish
Parmar
a,
Abhishek
Iyer
ab,
Charlotte S.
Vincent
c,
Dorien
Van Lysebetten
b,
Stephen H.
Prior
d,
Annemieke
Madder
b,
Edward J.
Taylor
*c and
Ishwar
Singh
*a
aSchool of Pharmacy, JBL Building, University of Lincoln, Beevor St. Lincoln LN67DL, UK. E-mail: isingh@lincoln.ac.uk
bOrganic and Biomimetic Chemistry Research Group, Department of Organic Chemistry, Ghent University, Krijgslaan 281 (S4), B-9000 Ghent, Belgium
cSchool of Life Sciences, JBL Building, University of Lincoln, Beevor St. Lincoln LN67DL, UK. E-mail: etaylor@lincoln.ac.uk
dSchool of Chemistry, JBL Building, University of Lincoln, Beevor St. Lincoln LN67DL, UK
First published on 9th March 2016
Abstract
The discovery of the new antibiotic teixobactin has been timely in the race for unearthing novel antibiotics wherein the emergence of drug resistant bacteria poses a serious threat worldwide. Herein, we present the total syntheses and biological activities of two teixobactin analogues. This approach is simple, efficient and has several advantages: it uses commercially available building blocks (except AllocHN-D-Thr-OH), has a single purification step and a good recovery (22%). By using this approach we have synthesised two teixobactin analogues and established that the D-amino acids are critical for the antimicrobial activity of these analogues. With continuing high expectations from teixobactin, this work can be regarded as a stepping stone towards an in depth study of teixobactin, its analogues and the quest for synthesising similar molecules.
The decreased potency of drugs such as penicillin,1 vancomycin2 and oxacillin3 due to their excessive use is a consequence of the emergence of drug resistant bacteria. It has been predicted that antimicrobial resistance (AMR) will have disastrous consequences and it is estimated that by 2050 an additional 10 million people yearly could succumb to drug resistant infections.4 The recently published article in Nature5 describing the discovery of teixobactin has provided a much needed breakthrough in the challenging field of antibiotic peptides. The bacteria against which it is effective have, thus far, not shown any detectable resistance to teixobactin. Moreover, given the multiple mechanisms of attack by teixobactin described by Süssmuth6 resistance is unlikely in the near future. Unfortunately, although teixobactin provides some much needed answers, the problem is far from over. Organic chemists have worked round the clock to synthesise novel antibiotics and although progress has been steady, bacteria have time and again confounded even the best in this field.7 This is where the multichannel device, iChip,8 has made a considerable contribution enabling the identification of molecules like teixobactin, some of which could be active against drug resistant bacteria. However, the iChip device has its limitations. The probability of finding an antibiotic is extremely low (10−7 percent) and even with high-throughput screening, which takes considerable time and resources, there is no guarantee when will be the discovery of the next drug like teixobactin. Undeniably, breakthroughs via organic synthesis have to be made to keep the drug resistance problem under check. The time for this is indeed now and teixobactin is a good starting point. This study is aimed at doing so and provides a general approach through which, not only teixobactin, but also other analogues can be synthesised. The total synthesis of teixobactin will give access to analogues which nature cannot provide us with and therefore an organic synthesis approach to the bigger problem at hand should not be prematurely discarded. It is to be noted that during the preparation of this manuscript a report from the Albericio group appeared describing the synthesis of teixobactin analogue 1 obtained in a 6% yield. Moreover, the role of the D-amino acids have not been proven.9 Our work presents an efficient syntheses of both the D and L versions of the analogues of teixobactin (22% yield for 1) and their role in antimicrobial activity.
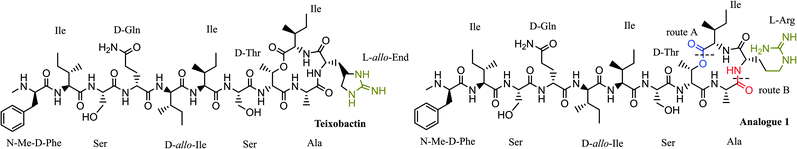
Since teixobactin has been fully characterised by NMR and LC-MS, the structural complexity of teixobactin can be described as moderate to difficult. It has also been described as an unusual depsipeptide due to the presence of the non-natural amino acids L-allo-enduracididine and N-methylphenylalanine along with four D-amino acids (Fig. 1). Despite not possessing a lipid tail, the commercial unavailability of the enduracididine amino acid makes the total synthesis of the molecule more time consuming than expected.10 The commercially available natural amino acid arginine (a linear guanidine) is the closest structural match suitable for the replacement of the L-allo-enduracididine (a cyclic guanidine) as can be seen in Fig. 1 (green). In this work, arginine was selected as a replacement of enduracididine for synthesis of teixobactin analogue 1. A first approach towards tackling this molecule was to synthesise the complete peptide on solid phase and cyclise post-cleavage from the resin via an ester bond (ESI,† Fig. S1). This route has been used with success for the synthesis of the analogues of callipeltin B.11
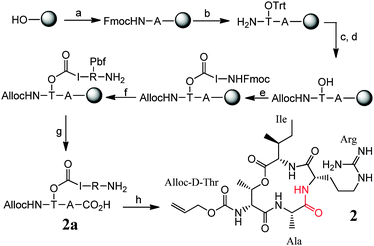
Following this strategy, a fully protected linear peptide was synthesised and cleaved from the resin without cleaving the protecting groups. However, the synthesis failed at the esterification step. Several conditions have been tested for the esterification as described in Table S1 (ESI†). Unfortunately, none of them yielded the esterified product. This could be due to the steric bulk of protecting groups on the amino acids. This led to the conclusion that a direct and linear route is not the way to cyclisation. Therefore a new second synthetic route had been devised which involves cyclisation via amide bond formation (Fig. 1b). For the total synthesis of analogue 1, an optimised pathway for the synthesis of the core ring structure (2) of teixobactin was required. Therefore, the initial efforts were focussed on obtaining this key fragment (Fig. 2).
For optimisation of the synthesis, we chose Wang resin. Fmoc-Ala-OH was loaded onto this resin via ester bond formation. The unreacted resin was capped using 10% acetic anhydride/diisopropylethylamine (Ac2O/DIPEA) followed by (a) the attachment of Fmoc-D-Thr(Trt)-OH via amide bond formation and subsequent (b) Fmoc removal. The orthogonal Alloc protecting group was installed on the amine (c) followed by (d) trityl group removal by 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 96 TFA
96 TFA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) TIS
TIS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DCM and proceeded with (e) the challenging esterification reaction between Ile and Thr. It is to be noted that excess Ile and base were required to drive the reaction to completion.12 This was succeeded by (f) amide bond formation using Arg and subsequent (g) cleavage from the resin using 95
DCM and proceeded with (e) the challenging esterification reaction between Ile and Thr. It is to be noted that excess Ile and base were required to drive the reaction to completion.12 This was succeeded by (f) amide bond formation using Arg and subsequent (g) cleavage from the resin using 95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.5
2.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.5 TFA
2.5 TFA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) TIS
TIS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O. The final step was (h) the amide bond formation between Arg and Ala which proceeds smoothly yielding the desired cyclised fragment. This approach was then slightly modified and used for the total synthesis of the teixobactin analogue 1 as described in Fig. 3.
H2O. The final step was (h) the amide bond formation between Arg and Ala which proceeds smoothly yielding the desired cyclised fragment. This approach was then slightly modified and used for the total synthesis of the teixobactin analogue 1 as described in Fig. 3.
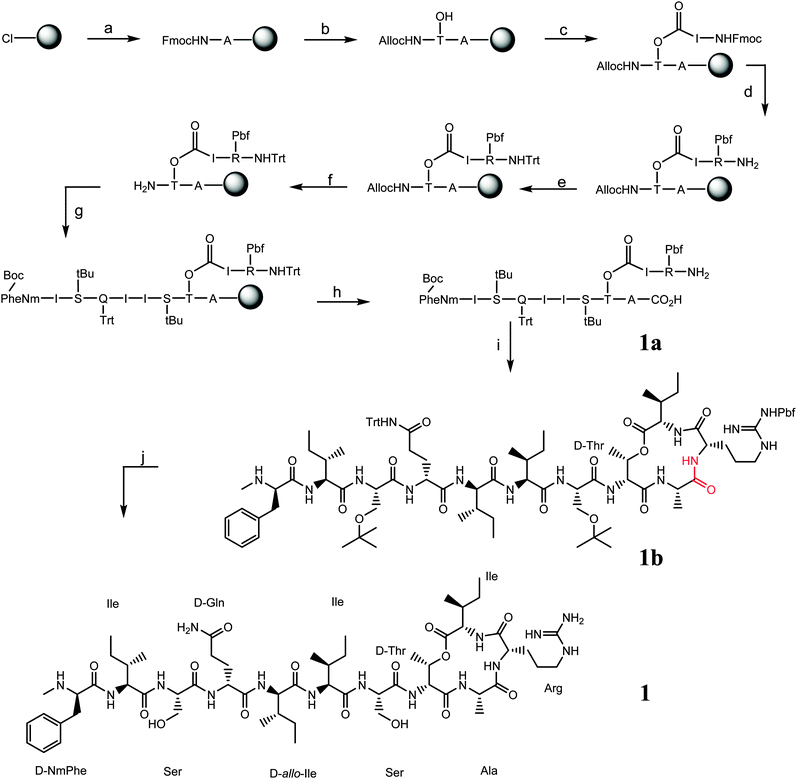
The approach used in this work involves synthesising the peptide completely on solid phase and only a single purification was required after the final cleavage. The synthesis of building block AllocHN-D-Thr-OH (modified from ref. 13) was also reported for first time and which can be directly coupled to the resin without protecting the amino acid side chain. 2-Chlorotritylchloride resin was chosen instead of Wang resin as the peptide could be cleaved off from the resin without removing the side chain protecting groups of the attached amino acids.14
(a) The first amino acid loaded on the resin in this case is Fmoc-Ala-OH followed by (b) an amide bond coupling with Alloc-D-Thr-OH. (c) Fmoc-Ile-OH is then coupled at this stage via an ester bond to the free –OH side chain of threonine. Next, (d) arginine was coupled via an amide bond, the Fmoc protecting group is removed and (e) the N-terminus is protected via a trityl protecting group15 (combining cleavage and deprotection in a single step) to facilitate the cleavage and cyclisation as described in reactions (h and i). (f) The Alloc group protecting the N-terminus of the threonine is then removed16 and (g) the peptide chain is built via standard solid phase peptide synthesis (SPPS). Partial cleavage was performed using 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 93 TFA
93 TFA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) TIS
TIS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DCM followed by cyclisation using 1-[bis(dimethylamino) methylene]-1H-1,2,3-triazolo[4,5-b]pyridinium 3-oxid hexafluorophosphate (HATU) as a coupling reagent and DIPEA as a base in DMF for 1 h. The protecting groups are then cleaved off using 95
DCM followed by cyclisation using 1-[bis(dimethylamino) methylene]-1H-1,2,3-triazolo[4,5-b]pyridinium 3-oxid hexafluorophosphate (HATU) as a coupling reagent and DIPEA as a base in DMF for 1 h. The protecting groups are then cleaved off using 95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.5
2.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.5 TFA
2.5 TFA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) TIS
TIS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
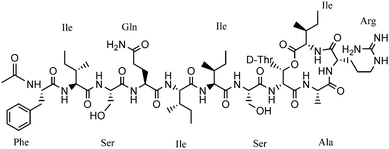
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O yielding the desired peptide (22% recovery, refer ESI,† S10). After successful synthesis of analogue 1, the general applicability of this approach was tested for the synthesis of analogue 3 (Fig. 4). In analogue 3, the three D-amino acids residues (Phe, Gln and Ile) were replaced by L-amino acid residues. The synthesis of analogue 3 also worked efficiently (17% recovery, refer ESI,† S10).
H2O yielding the desired peptide (22% recovery, refer ESI,† S10). After successful synthesis of analogue 1, the general applicability of this approach was tested for the synthesis of analogue 3 (Fig. 4). In analogue 3, the three D-amino acids residues (Phe, Gln and Ile) were replaced by L-amino acid residues. The synthesis of analogue 3 also worked efficiently (17% recovery, refer ESI,† S10).
The detailed characterisation of 1 and 3 were performed using LC-MS and NMR. The NMR spectra of product 1 (refer ESI,† page S15) was shown to be identical as reported previously.5,9 The NOEs of 1 were characteristic of a random coil, however, the NOEs of 3 suggested a considerable degree of structure.
The analogues 1 and 3 were evaluated for their antibacterial activity. MIC results showed a trend similar to teixobactin for analogue 1 against both Gram-positive and Gram-negative bacteria. Analogue 3 was not active against Gram-negative bacteria. Moreover, analogue 1 was 64 times more effective than analogue 3 (Table 1) against Gram-positive bacteria. This difference in antibacterial activity has established that the three D-amino acids residues of 1 are critical for the antibacterial activity.
In conclusion, this work reports efficient total syntheses of two teixobactin analogues (22% yield of teixobactin analogue 1). Analogue 1 is identical to teixobactin in all aspects with the exception of the L-allo-enduracididine amino acid. The methodology described here is not specific to only one molecule, but it can also be used as a general strategy for synthesis of other analogues of teixobactin. The role of three D-amino acids has also been established. The three D-amino acids present in the teixobactin analogue 1 but absent in analogue 3 are critical for antibacterial activity. This work also reports the synthesis of new AllocHN-D-Thr-OH building block and has incorporated it as such in the syntheses of teixobactin analogues. We believe that this work to be pivotal for the synthesis of teixobactin and its analogues and therefore will be helpful to address the current challenges of antimicrobial resistance.
The funding support from the Royal Society and Horizon 2020 is acknowledged. Anish Parmar, Abhishek Iyer and Charlotte Vincent would like to thank University of Lincoln for funding. Jan Goeman from Ghent University and Mick Lee from University of Leicester are thanked for the LC-MS/HRMS analysis. Dorien Van Lysebetten acknowledges the BOF for financial support.
Notes and references
- F. W. Goldstein, L. D. M. Médicale, F. H. Saint-joseph and R. Losserand, J. Antimicrob. Chemother., 1999, 44, 141–144 CrossRef CAS PubMed.
- P. Courvalin, Clin. Infect. Dis., 2006, 42, 25–34 CrossRef PubMed.
- J. H. Lee, Appl. Environ. Microbiol., 2003, 69, 6489–6494 CrossRef CAS PubMed.
- http://amr-review.org/, 2015.
- L. L. Ling, T. Schneider, A. J. Peoples, A. L. Spoering, I. Engels, B. P. Conlon, A. Mueller, D. E. Hughes, S. Epstein, M. Jones, L. Lazarides, V. A. Steadman, D. R. Cohen, C. R. Felix, K. A. Fetterman, W. P. Millett, A. G. Nitti, A. M. Zullo, C. Chen and K. Lewis, Nature, 2015, 517, 455–459 CrossRef CAS PubMed.
- F. von Nussbaum and R. D. Süssmuth, Angew. Chem., Int. Ed., 2015, 54, 6684–6686 CrossRef CAS PubMed.
- L. V. J. Blair, J. M. A. Webber, M. A. Baylay, A. J. Ogbolu and D. O. Piddock, Nat. Rev. Microbiol., 2015, 42, 42–51 Search PubMed.
- D. Nichols, N. Cahoon, E. M. Trakhtenberg, L. Pham, a. Mehta, a. Belanger, T. Kanigan, K. Lewis and S. S. Epstein, Appl. Environ. Microbiol., 2010, 76, 2445–2450 CrossRef CAS PubMed.
- Y. E. Jad, G. A. Acosta, T. Naicker, M. Ramtahal, A. El-Faham, T. Govender, H. G. Kruger, B. G. De La Torre and F. Albericio, Org. Lett., 2015, 17, 6182–6185 CrossRef CAS PubMed.
- W. Craig, J. Chen, D. Richardson, R. Thorpe and Y. Yuan, Org. Lett., 2015, 17, 4620–4623 CrossRef CAS PubMed.
- M. Kikuchi, Y. Watanabe, M. Tanaka, K. Akaji and H. Konno, Bioorg. Med. Chem. Lett., 2011, 21, 4865–4868 CrossRef CAS PubMed.
- M. J. Martin, R. Rodriguez-Acebes, Y. Garcia-Ramos, V. Martinez, C. Murcia, I. Digon, I. Marco, M. Pelay-Gimeno, R. Fernández, F. Reyes, A. M. Francesch, S. Munt, J. Tulla-Puche, F. Albericio and C. Cuevas, J. Am. Chem. Soc., 2014, 136, 6754–6762 CrossRef CAS PubMed.
- M. Acid, L. Acid, N. Shibata, J. E. Baldwin, A. Jacobs and E. Wood, Tetrahedron, 1996, 52, 12839–12852 CrossRef.
- K. Barlos, D. Gatos, J. Kallitsis, G. Papaphotiu, P. Sotiriu, Y. Wenqing and W. Schäfer, Tetrahedron Lett., 1989, 30, 3943–3946 CrossRef CAS.
- C. Gros, C. Boulègue, N. Galeotti, G. Niel and P. Jouin, Tetrahedron, 2002, 58, 2673–2680 CrossRef CAS.
- P. Grieco, P. M. Gitu and V. J. Hruby, J. Pept. Res., 2001, 57, 250–256 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Peptide synthesis, HPLC, LC-MS analysis, NMR spectra. See DOI: 10.1039/c5cc10249a |
| This journal is © The Royal Society of Chemistry 2016 |