Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Colorimetric detection of lipopolysaccharides based on a lipopolysaccharide-binding peptide and AuNPs†
Chunyang
Lei
a,
Zhaohui
Qiao
a,
Yingchun
Fu
 a and
Yanbin
Li
*ab
a and
Yanbin
Li
*ab
aCollege of Biosystems Engineering and Food Science, Zhejiang University, Hangzhou 310058, China. E-mail: yanbinli@zju.edu.cn
bDepartment of Biological and Agricultural Engineering, University of Arkansas, Fayetteville, Arkansas 72701, USA
First published on 14th October 2016
Abstract
In this study, a novel label-free colorimetric assay for the detection of lipopolysaccharides (LPSs) was developed based on a LPS-binding peptide (LBP) and unmodified gold nanoparticles (AuNPs). Cationic LBP probes with a C-terminal cysteine thiol are used to anchor the surface of unmodified AuNPs with anionic surface charge and cause the aggregation of AuNPs, resulting in the red-to-purple color change and the appearance of a new absorbance peak at 586 nm in the absorption spectrum of AuNPs. In the presence of an LPS, specific binding between the LPS and the LBP leads to the formation of LPS–LBP complexes and subsequently prohibits the aggregation of unmodified AuNPs, which exhibits distinct differences in both the color and the absorption spectrum of AuNPs. On this basis, the concentration of the LPS can be visualized through the purple-to-red color change of the AuNPs and quantitatively determined by UV-vis spectroscopy. Under the optimized conditions, the proposed assay is capable of sensitively and selectively measuring LPSs in the range of 10–1000 nM with a detection limit of 2.0 nM.
1. Introduction
Lipopolysaccharides (LPSs), known as bacterial endotoxins, are the major structural constituent of the outer membrane of Gram-negative bacteria, contributing greatly to the integrity of the bacteria, and protecting the membrane from certain kinds of chemical attack.1,2 LPSs are comprised of a variable polysaccharide and a conserved glucosamine-based phospholipid called lipid A, which anchors the LPS into the membrane.3,4 The release of the LPS from bacteria can elicit host monocytes and macrophages to secrete a broad range of inflammatory cytokines, such as tumor necrosis factor-α, interleukin-1, and interleukin-8.5,6 Massive secretion of these cytokines can lead to diarrhea, septic shock and even death. Due to the extreme toxicity of bacterial LPSs, several research efforts have been initiated toward the development of specific and sensitive assays for LPSs.Currently, the enzymatic limulus amebocyte lysate (LAL) assay is clinically used for the quantification of the LPS through the gel formation of the LAL in the presence of the LPS.7 However, the LAL assay is very sensitive and highly susceptible to changes in temperature and pH, as well as potential interference from other carbohydrate derivatives, such as β-glucans.8 Therefore, numerous efforts have been devoted to the development of alternative assays for LPSs. Owing to the highly negative charges of the LPS endowed with its phosphate and carboxylic groups, the utilization of the electrostatic interaction between the LPS and positively charged probes to develop sensors for LPSs has attracted much interest.9 Chromo- and fluorogenic sensors based on positively charged small molecules, liposomes, polymers, and nanoparticles have been reported for the sensitive determination of bacterial LPS in aqueous solutions.10–13 Despite these, innovative attempts have shown great promise with their individual limit of detection ranging from the micromolar to the low nanomolar level, the construction of the sensors usually involves very sophisticated organic synthesis.
Recently, peptide-based recognition elements have been exploited to construct biosensors in a number of applications due to their high affinity and specificity towards target molecules.14 Generally, peptide-based recognition elements are oligopeptides developed from binding domains of proteins, or screened by phage display, possessing several merits over antibodies, including low molecular weight, ease of synthesis and modification, and good chemical/thermal stability.15 Due to these unique properties, peptide-based biosensors have been developed for various analytes including metallic ions, proteins, proteases, kinases and Bacillus species.16–22 By comparison, applications of LPS-binding peptides in biosensing have been much less explored and only a few reports have been found so far. Brock's group developed a peptide beacon derived from the LPS-binding domain of the LPS receptor CD14 for selective detection of LPSs.23 Xing's group designed a LBP-binding peptide-perylene diimide functionalized magnetic nanoparticles for bacterial LPS detection and clearance.24 Liedberg's group reported the sensitive detection of the LPS using a LBP-binding peptide/graphene-oxide platform.25 However, fluorophore conjugations are essentially required in order to convert the binding event into a measurable signal in these studies, which involves complicated synthesis and purification procedures. Hence, in order to simplify the detection process of LPSs, a label-free method based on LPS-binding peptides is highly desirable.
Gold nanoparticles (AuNPs), with unique optical properties associated with distance-dependent surface plasmon resonance (SPR) absorption, are ideal materials for developing high sensitive colorimetric biosensors.26–28 Herein, we have developed a simple, sensitive and label-free colorimetric assay for LPSs based on unmodified AuNPs and a LPS-binding peptide. The positively charged peptide probe with a C-terminal cysteine thiol can be anchored on AuNPs by the Au–S bond. The binding of peptides decreases the negative charge density on the AuNP surface, resulting in AuNP aggregation. AuNPs remain monodispersed when the LPS, a negatively charged glycolipid, binds to the peptide, and neutralizes the positive charges of the LBP. Therefore, the determination of LPSs can be achieved according to the SPR absorption changes of AuNPs.
2. Experimental section
2.1 Materials and apparatus
The LBP (sequence: KYSSSISSIRC), and a control peptide PKR (sequence: SYSSSISSISAC) with purity >95%, were synthesized commercially by KareBay Biochem, Inc. (Ningbo, China). The Lipopolysaccharide (LPS) from Escherichia coli 055: B5 was purchased from Sigma-Aldrich (Shanghai, China). 2-(N-Morpholino)ethanesulfonic acid (MES), sodium citrate, EDTA, ampicillin and bovine serum albumin (BSA) were obtained from Sangon (Shanghai, China). HAuCl4 and dextran were obtain from Sinopharm Chemical Reagent Co., Ltd (Shanghai, China). 10% dextrose injections were purchased from CR Double-Crane Pharmaceuticals Co., Ltd (Anhui, China). Tachypleus amebocyte lysate for the endotoxin detection kit was purchased from Chinese Horseshoe Crab Reagent Manufactory Co., Ltd. (Xiamen, China). All the commercially available reagents were used without further purification. All solutions were prepared using fresh ultrapure water (18.2 MΩ cm) from a Milli-Q automatic ultrapure water system. The absorption spectra of AuNPs were recorded on a Synergy™ H1 Multi-Mode Reader (Biotek, USA).2.2 Synthesis of gold nanoparticles (AuNPs)
AuNPs were synthesized as reported previously.29 In brief, 50 mL of 1 mM HAuCl4 solution was added into a 250 mL round flask, and was heated to boiling. Then, 5 mL of 38.8 mM sodium citrate solution was added rapidly under vigorous stirring. The solution was maintained at the boiling state for 10 min, during which time the color changed from yellow to deep wine red. A stable and monodispersed gold nanoparticle colloidal solution was obtained and stored at 4 °C. The AuNPs were characterized by transmission electron microscopy (JEM1230, JOEL) and dynamic light scattering (Zetasizer Nano ZS-90, Malvern).2.3 Peptide-induced aggregation of AuNPs
In a 200 μL PCR tube, the sample consists of 10 μL of peptide solution (10 μM, LBP/PKR) and 50 μL of AuNP dispersion (10 nM) in 10 mM MES buffer, pH 6.5. After incubation at room temperature (25 °C) for 10 min, the sample was pipetted to the well of a 96-well plate, and the absorption spectrum from 450 nm to 700 nm was recorded using a Synergy™ H1 Multi-Mode Reader.For the investigation of the effect of the concentration of the LPS on the aggregation of AuNPs, a series of samples containing 10 μL of peptide solution with different concentrations and 50 μL of AuNP dispersion (10 nM) in 10 mM MES buffer, pH 6.5 were taken. After incubation for 6 min at room temperature, the sample was pipetted to the well of a 96-well plate, and the absorption spectrum from 450 nm to 700 nm was recorded using a Synergy™ H1 Multi-Mode Reader.
2.4 Colorimetric assay of LPSs
For LPS detection, 10 μL of peptide solution (10 μM) was added to 10 μL of 10 × buffer (1 × buffer: 10 mM MES, pH 6.5), and the sample was replenished to 40 μL with H2O. Then, 10 μL of LPS solution with different concentrations was added and incubated for 15 min. Finally, 50 μL of AuNPs was added and incubated at room temperature for 6 min, and the absorption spectrum from 450 nm to 700 nm was recorded using a Synergy™ H1 Multi-Mode Reader. For a real sample assay, 10 μL of the LPS spiked in 10% dextrose injections was used instead of 10 μL of H2O, and the absorbance measurement protocol was set as that of the above-mentioned procedure. At the same time, the spiked LPS samples were quantified using the tachypleus amebocyte lysate for the endotoxin detection kit.3. Results and discussion
3.1 Mechanism of the colorimetric assay for LPSs
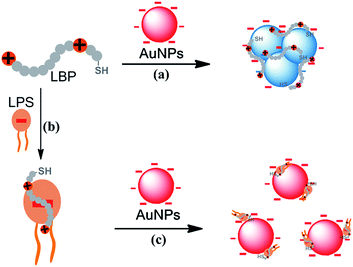
The mechanism of the colorimetric assay for the LPS based on the LBP and AuNPs is shown in Scheme 1. A LPS-binding peptide (LBP, sequence: KYSSSISSIRAC) selected by the phage display method with a high affinity (Kd, 0.01 nM) for the LPS, was employed as the probe for the assay of the LPS.30,31 The peptide contains two basic amino acid residues (Lys and Arg), giving the peptide a net charge of +2 at neutral pH. Moreover, the cysteine residue at the C-terminal of the LBP makes the peptide anchor on the surface of citrate-capped AuNPs via the Au–S bond. The binding of peptides reduces the negative charge density on the AuNP surface and breaks the electrostatic stability of AuNPs, resulting in aggregation of AuNPs and a corresponding red-to-purple color change (route a). With the existence of LPSs, the peptide probe selectively binds the LPS, leading to the formation of LPS–LBP complexes (route b). The amount of negative charges in the LPS is much more than that of the positive charges in the LBP, and as a result, the net charge of the LPS–LBP complex remains negative. The binding of LPS–LBP complexes does not induce the aggregation of AuNPs because the complexes are negatively charged, and the solution remains red (route c). Therefore, the presence of the LPS can be determined by the color changes of AuNPs.The proof-of-concept experiment was carried out to study the effect of the LBP on the stability of unmodified AuNPs. Another peptide, PKR (SYSSSISSISAC) with the two basic amino acid residues (Lys and Arg) replaced by neutral amino acid residues (e.g. Ser), was used as the control probe in the experiments. The absorption spectra and color of AuNPs and that in the presence of the LBP or PKR are shown in Fig. S1.† The addition of the LBP induced a significant change in the absorption spectrum of AuNPs as well as a red-to-purple color change. Thus, the LBP can disrupt the stability of AuNPs and cause the aggregation of AuNPs. Conversely, the influence of PKR on the stability of AuNPs is negligible, implying that the positively charged amino acid residues are essential for the LBP-mediated AuNP destabilization.
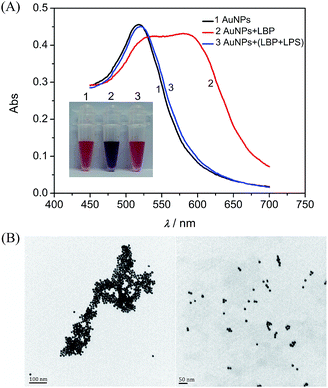
Following the design, the feasibility of the proposed method was investigated. Fig. 1A depicts a typical colorimetric response of this assay for the LPS. Citrate-capped 13 nm AuNPs with λmax at 518 nm in the absorption spectrum were used in this study. When the AuNPs were mixed with the LBP, the absorption peak at 518 nm decreased and one distinct absorption peak appeared at about 586 nm, and the color of the mixture was purple. The corresponding Transmission Electron Microscope (TEM) image (Fig. 1B, left) and the average hydrodynamic diameter (129.7 ± 44.6 nm) measured by dynamic light scattering (DLS, Fig. S2†), demonstrated the formation of aggregates upon addition of the peptide probe to the AuNP solution. In contrast, when the AuNPs were added to the LBP solution treated with the LPS, the absorption peak at 586 nm decreased sharply with a major peak at about 521 nm, and a red color was observed, which was confirmed by the TEM image (Fig. 1B, right) and a smaller average hydrodynamic diameter of 23.1 ± 7.1 nm. DLS and absorption spectroscopic results revealed that the addition of the LBP and LPS to the initially prepared AuNPs slightly increased the average hydrodynamic diameter of AuNPs from 14.2 ± 3.1 nm to 23.1 ± 7.1 nm, and shifted the surface plasmon band from 518 to 521 nm in the absorption spectrum, presumably due to the damping effect of the anchoring LPS–LBP complexes on the gold surface.32 Therefore, the binding of the LPS to the LBP prevents the aggregation of AuNPs induced by the LBP, resulting in a remarkable change in color and providing the colorimetric detection of the LPS.
3.2 Conditional optimization
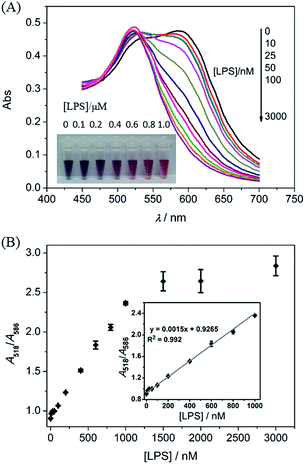
In order to obtain a better response to LPSs, some important factors were optimized. As shown in Fig. S3,† the absorbance at 586 nm drastically increased in 1 min and reached a plateau value within 6 min after addition of the LBP into the AuNP solution. Next, the aggregation of AuNPs induced by different concentrations of the LBP were investigated, and the results are shown in Fig. S4.† Upon the addition of the LBP, the absorbance at 518 nm (A518) decreased, while that at 586 nm (A586) increased (Fig. S4A†). As demonstrated previously, the ratio of A518 to A586 (A518/A586) can be considered as an indicator for the degree of the dispersion/aggregation state of AuNPs.33,34 The value of A518/A586 gradually decreased with increasing concentrations of the LBP from 0 to 1.5 μM, and reached a minimum as the concentrations of the LBP were higher than 1.0 μM (Fig. S4B†). In addition, the incubation time between the LPS and the LBP was optimized as 15 min (Fig. S5†).3.3 Colorimetric assay for the LPS
Under the pre-optimized conditions, the dynamic response range of the LPS was recorded through the absorbance changes of the AuNPs. When the concentration of the LPS increased from 0 to 3.0 μM, the absorbance at 586 nm gradually decreased, and the value of A518/A586 continually increased (Fig. 2). The value of A518/A586 is linearly related to the concentration of the LPS within the range from 0.01 to 1.0 μM (Fig. 2B inset). The limit of detection (LOD) for the LPS was 2.0 nM estimated from 3S/N, where S is the standard deviation of the blank solution obtained in three individual measurements, and N is the slope of the calibration curve in the linear region. The LOD of the proposed assay is better than many previously reported LPS sensors (details in Table S1†).13,23,24 Furthermore, the response of the LPS at a concentration of 0.1 μM can be distinguished by the naked eye.3.4 Selectivity assay
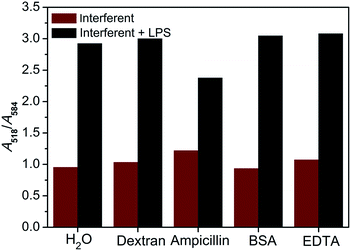
To evaluate the specificity of the proposed colorimetric assay, the responses towards several potential coexisting interferences were examined. As shown in Fig. 3, potential interferents such as dextran, ampicillin, bovine serum albumin (BSA) and EDTA caused little influence on the colorimetric responses to the LPS, indicating good selectivity of the proposed assay. These results clearly demonstrated that this LBP-based assay could be selective for the targeted detection of LPSs.3.5 The real sample assay
Endotoxins can elicit inflammatory responses in animal studies, and the presence of endotoxins in the products of injections can result in pyrogenic responses, ranging from fever and chills to irreversible and fatal septic shock.35 Therefore, the practical application of the proposed method in the real sample assay was estimated by spiking different concentrations of the LPS into 10% dextrose injections. In parallel, the concentrations of the LPS in the samples were also detected by the LAL assay. The experimental results are shown in Table 1. Using the results of the LAL assay as the standard, the recovery of this method varied from 94.8% to 102.7% and the RSDs ranged from 1.7% to 3.3%, indicating that the proposed assay possessed a good application prospect in clinical research.| Samples | Spiked (nM) | LAL assay (nM) | This method (nM) | RSD (%) | Recovery (%) |
|---|---|---|---|---|---|
| 1 | 200 | 194.9 | 200.2 | 3.3 | 102.7 |
| 2 | 400 | 407.7 | 386.9 | 1.7 | 94.8 |
| 3 | 800 | 792.3 | 787.2 | 2.1 | 99.4 |
4. Conclusions
In summary, we have developed a simple and novel strategy using an unmodified LBP probe and bare AuNPs for the colorimetric detection of the LPS. The selective binding of the peptide probe with the LPS prohibits the peptide-mediated AuNP aggregation, switching the solution color from blue to purple red as a visualized read-out. The proposed peptide-based AuNP assay possesses several advantages: (I) high sensitivity and good selectivity are achieved for the LPS assay; (II) the results of LPS detection can be simply visualized without the requirement of any instruments; (III) labor-intensive modification of AuNPs and expensive peptide labeling are avoided. Furthermore, this peptide/nanomaterial-based assay could serve as a universal biosensing strategy for the detection of different bio-macromolecules by adopting the corresponding binding peptides.Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 21505120) and the China Postdoctoral Science Foundation Funded Project (No. 2015M570499 and 2016T90536).References
- R. J. Ulevitch and P. S. Tobias, Curr. Opin. Immunol., 1994, 6, 125–130 CrossRef CAS PubMed.
- L. S. Young, W. J. Martin, R. D. Meyer, R. J. Weinstein and E. T. Anderson, Ann. Intern. Med., 1977, 86, 456–471 CrossRef CAS PubMed.
- J. Qian, T. A. Garrett and C. R. Raetz, Biochemistry, 2014, 53, 1250–1262 CrossRef CAS PubMed.
- S. Singh, M. Kalle, P. Papareddy, A. Schmidtchen and M. Malmsten, Biomacromolecules, 2013, 14, 1482–1492 CrossRef CAS PubMed.
- R. S. Munford, Annu. Rev. Pathol.: Mech. Dis., 2006, 1, 467–496 CrossRef CAS PubMed.
- M. Tidswell and S. P. LaRosa, Expert Rev. Anti-Infect. Ther., 2011, 9, 507–520 CrossRef CAS PubMed.
- P. F. Roslansky and T. J. Novitsky, J. Clin. Microbiol., 1991, 29, 2477–2483 CAS.
- F. R. Lourenco, S. Botelho Tde and J. Pinto Tde, PDA J. Pharm. Sci. Technol., 2012, 66, 542–546 CrossRef CAS PubMed.
- A. P. Das, P. S. Kumar and S. Swain, Biosens. Bioelectron., 2014, 51, 62–75 CrossRef CAS PubMed.
- G. Jiang, J. Wang, Y. Yang, G. Zhang, Y. Liu, H. Lin, G. Zhang, Y. Li and X. Fan, Biosens. Bioelectron., 2016, 85, 62–67 CrossRef CAS PubMed.
- M. Lan, J. Wu, W. Liu, W. Zhang, J. Ge, H. Zhang, J. Sun, W. Zhao and P. Wang, J. Am. Chem. Soc., 2012, 134, 6685–6694 CrossRef CAS PubMed.
- M. Rangin and A. Basu, J. Am. Chem. Soc., 2004, 126, 5038–5039 CrossRef CAS PubMed.
- L. Zeng, J. Wu, Q. Dai, W. Liu, P. Wang and C. S. Lee, Org. Lett., 2010, 12, 4014–4017 CrossRef CAS PubMed.
- Y. Cui, S. N. Kim, R. R. Naik and M. C. McAlpine, Acc. Chem. Res., 2012, 45, 696–704 CrossRef CAS PubMed.
- Q. Liu, J. Wang and B. J. Boyd, Talanta, 2015, 136, 114–127 CrossRef CAS PubMed.
- W. Li, Z. Nie, K. He, X. Xu, Y. Li, Y. Huang and S. Yao, Chem. Commun., 2011, 47, 4412–4414 RSC.
- L. Li, H. Lin, C. Lei, Z. Nie, Y. Huang and S. Yao, Biosens. Bioelectron., 2014, 54, 42–47 CrossRef CAS PubMed.
- C. Lei, X. Xu, J. Zhou, X. Liu, Z. Nie, M. Qing, P. Li, Y. Huang and S. Yao, Chem.–Asian J., 2014, 9, 2560–2567 CrossRef CAS PubMed.
- M. S. Mannoor, S. Zhang, A. J. Link and M. C. McAlpine, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 19207–19212 CrossRef CAS PubMed.
- I. L. Medintz, A. R. Clapp, F. M. Brunel, T. Tiefenbrunn, H. T. Uyeda, E. L. Chang, J. R. Deschamps, P. E. Dawson and H. Mattoussi, Nat. Mater., 2006, 5, 581–589 CrossRef CAS PubMed.
- G. Chen, Y. Xie, H. Zhang, P. Wang, H. Cheung, M. Yang and H. Sun, RSC Adv., 2014, 4, 6560–6563 RSC.
- H. Sun, R. C. Panicker and S. Q. Yao, Biopolymers, 2007, 88, 141–149 CrossRef CAS PubMed.
- S. Voss, R. Fischer, G. Jung, K. H. Wiesmuller and R. Brock, J. Am. Chem. Soc., 2007, 129, 554–561 CrossRef CAS PubMed.
- F. Liu, J. Mu, X. Wu, S. Bhattacharjya, E. K. Yeow and B. Xing, Chem. Commun., 2014, 50, 6200–6203 RSC.
- S. K. Lim, P. Chen, F. L. Lee, S. Moochhala and B. Liedberg, Anal. Chem., 2015, 87, 9408–9412 CrossRef CAS PubMed.
- J. N. Anker, W. P. Hall, O. Lyandres, N. C. Shah, J. Zhao and R. P. Van Duyne, Nat. Mater., 2008, 7, 442–453 CrossRef CAS PubMed.
- R. A. Sperling, P. Rivera Gil, F. Zhang, M. Zanella and W. J. Parak, Chem. Soc. Rev., 2008, 37, 1896–1908 RSC.
- J. Deng, P. Yu, L. Yang and L. Mao, Anal. Chem., 2013, 85, 2516–2522 CrossRef CAS PubMed.
- R. Jin, G. Wu, Z. Li, C. A. Mirkin and G. C. Schatz, J. Am. Chem. Soc., 2003, 125, 1643–1654 CrossRef CAS PubMed.
- M. Matsumoto, Y. Horiuchi, A. Yamamoto, M. Ochiai, M. Niwa, T. Takagi, H. Omi, T. Kobayashi and M. M. Suzuki, J. Microbiol. Methods, 2010, 82, 54–58 CrossRef CAS PubMed.
- M. M. Suzuki, M. Matsumoto, A. Yamamoto, M. Ochiai, Y. Horiuchi, M. Niwa, H. Omi, T. Kobayashi and T. Takagi, J. Microbiol. Methods, 2010, 83, 153–155 CrossRef CAS PubMed.
- J. Nam, N. Won, H. Jin, H. Chung and S. Kim, J. Am. Chem. Soc., 2009, 131, 13639–13645 CrossRef CAS PubMed.
- Y. Jiang, H. Zhao, N. Zhu, Y. Lin, P. Yu and L. Mao, Angew. Chem., Int. Ed., 2008, 47, 8601–8604 CrossRef CAS PubMed.
- D. Liu, W. Chen, K. Sun, K. Deng, W. Zhang, Z. Wang and X. Jiang, Angew. Chem., Int. Ed., 2011, 50, 4103–4107 CrossRef CAS PubMed.
- M. B. Gorbet and M. V. Sefton, Biomaterials, 2005, 26, 6811–6817 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6ay02371a |
| This journal is © The Royal Society of Chemistry 2016 |