Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Differential quantification of SCCA1 and SCCA2 cancer antigens using a hydrogel biochip
Aleksei A.
Tikhonov
*a,
Maria V.
Tsybulskaya
a,
Veronika I.
Butvilovskaya
a,
Elena N.
Savvateeva
a,
Pavel V.
Belousov
bc,
Dmitry V.
Kuprash
bc,
Olga N.
Solopova
d,
Maria A.
Chernichenko
e,
Michail M.
Filushin
f and
Alla Yu.
Rubina
a
aLaboratory of Biological Microchips, Engelhardt Institute of Molecular Biology, Russian Academy of Sciences, Vavilova Str. 32, Moscow 119991, Russia. E-mail: Alex.Tihonoff@gmail.com
bDepartment of Immunology, Faculty of Biology, Lomonosov Moscow State University, 1-12 Leninskie Gory, Moscow 119991, Russia
cLaboratory of Intracellular Signaling in Health and Disease, Engelhardt Institute of Molecular Biology, Russian Academy of Sciences, Vavilova Str. 32, Moscow 119991, Russia
dBiotechnology Laboratory, Russian Research Center for Molecular Diagnostics and Therapy, Sympheropolsky Blvd 8, Moscow 117638, Russia
eAbdominal Department, P.A. Herzen Moscow Cancer Research Institute, Ministry of Healthcare of the Russian Federation, 2nd Botkinskiy Proezd 3, Moscow 125284, Russia
fMicrosurgery Department, P.A. Herzen Moscow Cancer Research Institute, Ministry of Healthcare of the Russian Federation, 2nd Botkinskiy Proezd 3, Moscow 125284, Russia
First published on 7th October 2016
Abstract
Methods employing hydrogel-based microarrays (biochips) allow the simultaneous monitoring of protein interactions with different antibodies immobilized in gel elements. The method was applied for the simultaneous differential quantification of two highly homologous antigens of squamous cell carcinomas (SCCs) SCCA1 and SCCA2 in a single analysis. Two panels of monoclonal antibodies against recombinant SCCA1 and SCCA2 were generated, and two antibodies, C5 (anti-SCCA1) and A11 (anti-SCCA2), were selected for further evaluation based on their ability to specifically interact with their cognate antigens. Using a sandwich analysis, these antibodies were further tested in combination with anti-SCCA antibodies (H31 and SCC107) recognizing both of the SCCA antigens, thus allowing a quantitative independent measurement of both antigens. The intra- and inter-assay coefficients of variation for all resultant tests did not exceed 10% for the range of SCCA concentrations tested and were independent of whether SCCA1 and SCCA2 concentrations were determined simultaneously. The lower limit of detection (LOD) was estimated as 0.006 ng ml−1 for SCCA1 and 0.011 ng ml−1 for SCCA2 using the SCC107-Cy5 developing antibody and 0.014 ng ml−1 and 0.01 ng ml−1 concentrations, respectively, of the H31-Cy5 developing antibody. This assay provides a simple and accurate procedure for the differential quantitation of SCCA1 and SCCA2 using a single analysis of human serum on a biochip.
1. Introduction
SCCA antigen was first isolated by Kato and Torigoe in 1977 from squamous cell carcinoma (SCC) of the uterine cervix and characterized as a glycoprotein with a molecular mass of 48 kDa.1 Later, it was found that SCCA is an inhibitor of serine proteases belonging to the group of ovalbumin-serpins/clade B serpins2,3 and represents a mixture of two highly homologous proteins SCCA1 (serpin B3) and SCCA2 (serpin B4), sharing 92% amino acid identity.2SCCA proteins are normally expressed in various squamous epithelia.4 SCCA expression is increased in human carcinomas, particularly in SCC of various localizations.4–8 Notably, SCCA has been shown to be associated with the tumour size and stage, invasion depth, nodal status, survival rate and response to radiotherapy, making it a valuable tumour marker for cervical SCC.9–13 High pre-treatment SCCA serum levels were reported to be associated with a poor prognosis and high risk of SCC recurrence.12
Currently available in vitro diagnostic systems are overall capable of measuring the total SCCA level.14 For example, Crescenzi et al.15 developed a biochip to determine SCCA-IgM complexes in patients with liver diseases. In certain clinical scenarios, however, it may be important to perform a differential measurement of SCCA1 and SCCA2 in biological fluids. For example, the SCCA2 gene was shown to be overexpressed in lung and prostate cancers and inflammatory bowel diseases,8 as well as in idiopathic fibrosis-associated lung cancer.16
By contrast, SCCA1 overexpression was observed in hepatoblastoma17 and cholesteatoma.18 The monitoring of SCCA1 serum levels was shown to be important in the context of neoadjuvant chemotherapy for non-small-cell lung cancer and breast cancer.19,20
Moreover, the simultaneous measurement of SCCA1 and SCCA2 and the estimation of their ratio in a single analysis may provide more-accurate prognostic results and may be useful for personalizing a treatment strategy. For example, the association of an elevated SCCA2/SCCA1 mRNA level ratio with a poor prognosis was reported for head and neck SCC.21 A high SCCA2/SCCA1 ratio in HPV (human papilloma virus)-positive head and neck SCC patients was associated with a high risk of disease recurrence and with a decrease in survival rate.22 The SCCA2/SCCA1 mRNA level ratio increased during the course of carcinogenesis of uterine cervix SCC, and an elevated SCCA2/SCCA1 ratio at the early disease stage was associated with a higher risk of relapse.23
Importantly, serum levels of SCCA1 and SCCA2 are increased in atopic dermatitis patients and patients with cervical SCC, thus potentially creating many false-positive SCC screening results in patients with (highly prevalent) allergic diseases. However, a high SCCA2/SCCA1 concentration ratio discriminated patients with atopic dermatitis from patients with cervical SCC, providing a strong rationale for the individual measurement of SCCA1 and SCCA2. Moreover, serum concentrations of SCCA1 and SCCA2 were positively correlated with the severity of atopic dermatitis, thus creating a diagnostic niche for SCCA antigens beyond oncology.24
Such issues can be resolved using various multiplex test systems. For example, we previously developed a biochip for quantitating two forms of the prostate-specific antigen (PSAtotal and PSAfree),25 recently certified by the Russian Federal Service for Surveillance in Healthcare.
Previously, we have demonstrated the possibility of determining SCCA1 in the presence of SCCA2.26 The purpose of this study was to develop a method for the simultaneous differentiated measurement of two highly homologous proteins (SCCA1 and SCCA2) within one analysis on a biochip in clinically relevant concentrations. Our aim was also to determine the analytical characteristics of the method and to test it on actual biological samples in comparison to a reference method, used in clinical practice.
2. Experimental section
2.1. Recombinant antigens
The preparation of recombinant SCCA1 and SCCA2 was described previously.26 Additionally, recombinant SCCA1 and SCCA2 were affinity purified on Sepharose 4B (GE Healthcare Bio-Sciences, # 17-0430-01, Piscataway, NJ, USA) with immobilized antibody C5 or A11. The concentrations of SCCA1 and SCCA2 in solutions were determined using an SCC EIA test kit (CanAg-Fujirebio, # 800-10-96, Sweden).2.2. Monoclonal antibody production
Monoclonal antibodies H30, C5, H5, H81 and G9 have been described before (clone H30 was originally described as H3 and subsequently renamed).26 Antibodies F8, A11, H31 and D10 were generated by immunizing BALB/c mice with recombinant SCCA1 and SCCA2. This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals (NRC 2011), the European Convention for the Protection of Vertebrate Animals Used for Experimental and Other Scientific Purposes, Council of Europe (ETS 123), and “The Guidelines for Manipulations with Experimental Animals” (the decree of the Presidium of the Russian Academy of Sciences of April 02, 1980, no. 12000-496). All manipulations with animals were approved by Scientific Council of the Russian Research Center for Molecular Diagnostics and Therapy. Briefly, 2 month-old female BALB/c mice (N. N. Blokhin Russian Cancer Research Center, Moscow, Russia) were immunized in their hind paws with SCCA1 or SCCA2. The procedure was performed twice with an interval of two weeks with a dosage of 50 μg of the protein per immunization. The first immunization was performed using Freund's complete adjuvant and the second through Freund's incomplete adjuvant. The volume ratio of adjuvant–immunogen was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. On the third day following the second immunization, popliteal lymph node cells were hybridized with sp2/0 myeloma cells in accordance with the standard procedure.27 The supernatants of the hybrids were tested by ELISA.28 Positive clones were sub-cloned 2–4 times; monoclonal antibodies were produced in ascitic fluids of mice and purified by affinity chromatography on protein G-Sepharose (LKB, Sweden). The purity of the antibodies was determined by electrophoresis in a 12% polyacrylamide gel. The antibody activity was monitored by solid-phase enzyme immunoassay as described elsewhere.26 Antibodies C5, A11 and H31 were additionally purified by PEG-8000 (Sigma-Aldrich # P-5413, USA) precipitation in 0.01 M sodium phosphate, pH 8.0; then, the precipitates were collected, dissolved in the same buffer, and pelleted for 15 min at 10
1. On the third day following the second immunization, popliteal lymph node cells were hybridized with sp2/0 myeloma cells in accordance with the standard procedure.27 The supernatants of the hybrids were tested by ELISA.28 Positive clones were sub-cloned 2–4 times; monoclonal antibodies were produced in ascitic fluids of mice and purified by affinity chromatography on protein G-Sepharose (LKB, Sweden). The purity of the antibodies was determined by electrophoresis in a 12% polyacrylamide gel. The antibody activity was monitored by solid-phase enzyme immunoassay as described elsewhere.26 Antibodies C5, A11 and H31 were additionally purified by PEG-8000 (Sigma-Aldrich # P-5413, USA) precipitation in 0.01 M sodium phosphate, pH 8.0; then, the precipitates were collected, dissolved in the same buffer, and pelleted for 15 min at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm at +7 °C. The pellets were purified by ion-exchange chromatography on DEAE-cellulose (DE-52, Whatman # 4057-050, UK). The purified antibodies were eluted using a gradient of 0–0.4 M sodium chloride in 0.01 M sodium phosphate, pH 8.0.
000 rpm at +7 °C. The pellets were purified by ion-exchange chromatography on DEAE-cellulose (DE-52, Whatman # 4057-050, UK). The purified antibodies were eluted using a gradient of 0–0.4 M sodium chloride in 0.01 M sodium phosphate, pH 8.0.
2.3. Biochip preparation
Using the copolymerization immobilization technology developed at EIMB RAS (Moscow, Russia),29 a hydrogel biochip with the following probes immobilized in gel elements was created: monoclonal antibodies against recombinant SCCA1 and SCCA2, as well as commercial monoclonal antibody SCC107 (# 801-01 CanAg-Fujerebio, Sweden)30 against natural SCC antigen SCCA. Each molecular probe was immobilized in quadruplicate. We have previously determined that, in the case of hydrogel biochips, the median signal from 4 cells was optimal for reproducibility and increasing the number of cells did not significantly improve the intra-assay coefficient of variation.31A polymerization mixture containing gel-forming monomers based on methacrylamide and the molecular probes to be immobilized were applied by the QArray robot (Genetix, Great Britain) in 0.1 nl microdroplets on an activated substrate surface – Corning 2947 Micro Slides glass supports (Corning, USA).32 Gel elements were polymerized for 50 min at 20 °C in a stream of nitrogen under a UV lamp with a radiation maximum of 350 nm (Sylvania GTE lamp F15T8/350Bl, Great Britain) at a 2 cm distance from the lamp. After the end of polymerization, the biochips were washed for 40 min in PBST (PBS [0.15 M sodium chloride, 0.01 M sodium phosphate, pH 7.2] containing 0.01% Tween-20) and rinsed in distilled water followed by 1 h incubation in a blocking buffer solution (1% polyvinyl alcohol [50 kDa] solution in PBS). A ready-to-use biochip was covered by using an incubation chamber and stored at 2–8 °C.33
2.4. Variability and reproducibility
The quality of the biochips was checked by their imaging in transmitted light in a dedicated biochip analyser using TestChip and QualityControl software packages (EIMB RAS, Moscow, Russia). Biochips were rejected when the deviation of the radii of gel elements exceeded 5% within individual biochips and exceeded 8% among the biochips of a single batch.In addition to geometric screening tests, we also conducted biological control measurements both within each batch and between batches. An intra-assay was performed by measuring control samples in ten replicates. The lot passed the control if the intra-assay coefficient of variation (CV%) did not exceed 10%.
In the inter-assay case, the average values from the biochips in one batch were compared to the same values from biochips in other batches, and if the CV% was greater than 10%, the lot was rejected.
2.5. Biochip for studies of the specificity of the interaction of antibodies with SCCA1 and SCCA2
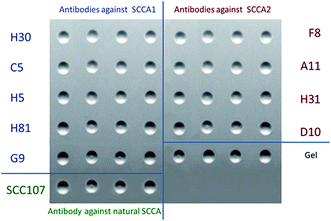
Panels of monoclonal antibodies against SCCA1 (panel A – H30, C5, H5, H81 and G9) and SCCA2 (panel B – F8, A11, H31 and D10), the SCC107 antibody and empty gel elements were immobilized in biochip elements as molecular probes. Each gel element was replicated four times to improve reproducibility (Fig. 1).2.6. Preparation of fluorescent conjugates
SCCA1, SCCA2 and monoclonal antibodies were labelled with fluorescent dye Cyanine 5 (Amersham Cy5 Mono NHS Ester (GE Healthcare, USA)). Cy5 NHS ester solution (2 mg ml−1 in DMF) was added to 75 μl of the protein or antibody solution (0.5 mg ml−1 in 0.05 M bicarbonate buffer, pH 9.5). The reaction was conducted for 1 h at 20 °C. The conjugates were purified by gel filtration on a Micro Bio-Spin column (Bio-Rad Laboratories, France) with Sephadex® G-25 coarse (Sigma-Aldrich, USA) equilibrated with PBS buffer. The degree of modification of serpins and antibodies was estimated spectrophotometrically. The Cy5/protein molar ratio was 1.5–2.0.2.7. Immunoassay
An immunoassay of SCCA1 and SCCA2 on a biochip with monoclonal antibodies immobilized in the elements was performed in two variants: a direct method using fluorescently labelled SCCA1 and SCCA2, and a two-stage sandwich immunoassay of SCCA1 and SCCA2 with development by fluorescently labelled monoclonal antibodies to other antigenic determinants.For direct analysis, 50 μl of a SCCA1-Cy5 or SCCA2-Cy5 solution (concentration range 0.1–400 ng ml−1) in dilution buffer (0.1 M sodium chloride, 0.01 M sodium phosphate, 0.1 M sodium citrate, 0.15% polyvinyl alcohol (50 kDa, Sigma-Aldrich, USA), and 0.15% polyvinylpyrrolidone (360 kDa, Sigma-Aldrich, USA), pH 7.5) was placed into the biochip chamber. Incubation was performed for 1 h and 30 min at 37 °C. The system was washed with PBST for 30 min, then rinsed with distilled water and dried in a stream of air. Then, fluorescence signals from the gel elements were registered.
For the sandwich immunoassay, 50 μl of an SCCA1 or SCCA2 solution in dilution buffer was placed into the biochip chamber. Incubation was performed at 37 °C for 20 h. In our experiments, this extended incubation time was necessary to achieve sufficient sensitivity with slow kinetics at a low analyte concentration.34 The system was washed for 30 min with PBST solution, then rinsed with distilled water and dried in a stream of air. Next, the developing antibodies (10 μg ml−1) in dilution buffer were applied to the biochip, and the system was incubated for 1 h at 37 °C. After the second washing of the biochips (30 min, PBST solution), fluorescence signals from the biochip gel elements were recorded.
2.8. Fluorescence
Fluorescence was induced by laser radiation (650/670 nm filters, excitation/registration) and registered using a fluorescence biochip analyser (EIMB RAS, Moscow, Russia).35 The final fluorescence intensity was calculated as the median value of four identical biochip elements using the ImaGelResearch software (EIMB RAS, Moscow, Russia).2.9. ELISA kit for the SCCA quantitation in serum
We used a commercially available SCC EIA test kit (CanAg-Fujirebio, # 800-10-96, Sweden). Serum concentrations of SCCA were determined according to the manufacturer's instructions.2.10. Statistical methods
Mean and median values, standard deviations (SD) and coefficients of variation (CV) were calculated using Microsoft Excel 2010.2.11. Live subject statement
This study was approved by the Ethical Committee of the P.A. Herzen Moscow Cancer Research Institute, Ministry of Healthcare of the Russian Federation. The samples were collected in accordance with the guidelines issued by the Ethical Committee of the P.A. Herzen Moscow Cancer Research Institute, Ministry of Healthcare of the Russian Federation. Written informed consent was obtained from all participants.3. Results and discussion
3.1. Selection of monoclonal antibodies specific to SCCA1 and/or SCCA2
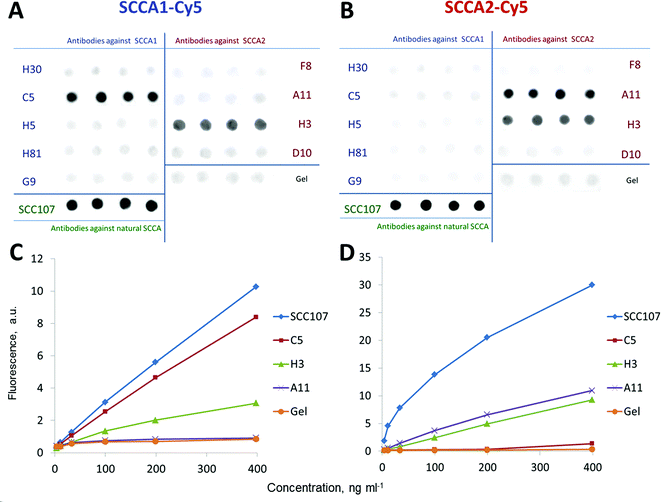
Antibodies for differentially measuring recombinant SCCA1 and SCCA2 were selected by direct immunoassay on a biochip using SCCA1-Cy5 and SCCA2-Cy5 conjugates. Testing of the monoclonal antibodies allowed us to identify two monospecific antibodies, namely, C5 and A11 raised against SCCA1 and SCCA2, respectively. Each of them specifically interacted with only one of the two proteins, and only a background-level signal was observed with non-cognate SCCA conjugates. In the direct analysis, both SCCA1-Cy5 and SCCA2-Cy5 conjugates bound to an SCC107 antibody raised against the natural antigen and recognizing common antigenic determinants shared between SCCA1 and SCCA2.36 This indicates that the tertiary structure of the refolded recombinant proteins matched the structure of the serpins isolated from squamous tumour cells. An oligospecific H31 antibody raised against recombinant SCCA2 is also capable of binding both fluorescently labelled SCCA2 and SCCA1, indicating that it likely recognizes a common antigenic determinant. Antibodies H30, H5, H81 and G9 (anti-SCCA1), as well as F8 and D10 (anti-SCCA2), failed to recognize any of SCCA antigens in this assay (Fig. 2).3.2. Selection of antibody pairs for differentially measuring SCCA1 and SCCA2 using a sandwich immunoassay
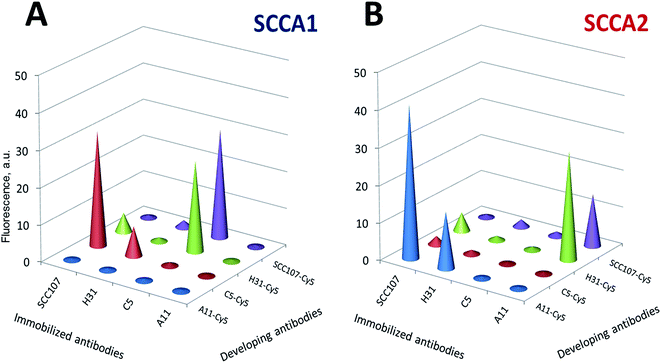
We then proceeded to select antibody pairs for the differential measurement of SCCA1 and SCCA2. Diagrams depicting the levels of fluorescence signals obtained on biochips after sandwich analysis of SCCA1 and SCCA2 using various developing antibodies are displayed in Fig. 3. Cy5 conjugates were obtained for all antibodies that were found to be efficient by direct analysis (SCC107, C5, A11 and H31), and each pair of immobilized/developing antibodies was tested in a sandwich immunoassay on a biochip using the fixed antigen concentration of 25 ng ml−1. Fig. 4 demonstrates a representative set of fluorescence images obtained in the sandwich analysis.Table 1 summarizes the efficiency of the interaction of the monoclonal antibodies with proteins, which was defined as the ratio of the fluorescence signal from a gel element with immobilized antibodies to the signal from the element containing no immobilized antibodies.26,37 The fluorescence signals from the non-specific interaction of the gel elements with the immobilized antibodies did not differ significantly from the signals of empty gel elements. Good discriminatory results were obtained for pairs of monospecific C5 and A11 immobilized antibodies and oligospecific SCC107 and H31 developing antibodies. Although the signal-to-noise ratio was high enough in several antibody pairs, they were not equivalent. If SCC107 and H31 were immobilized and C5 or A11 were the developing antibodies, two developing antibodies with two different dyes would be required for the simultaneous detection of SCCA1 and SCCA2 in one analysis. This would complicate the process, especially when the list of analytes is expanded, and the sensitivity of the assay may suffer. Therefore, we decided to use monospecific C5 and A11 antibodies for the capture.
| Protein | Immobilized antibodies | Developing antibodies | |||
|---|---|---|---|---|---|
| SCC107-Cy5 | H31-Cy5 | C5-Cy5 | A11-Cy5 | ||
| a Signals from gel elements without any immobilized antibodies, which are used as controls to determine the fluorescence resulting from the non-specific adsorption of reagents. | |||||
| SCCA1 | SCC107 | 2.4 | 13.2 | 63.6 | 0.8 |
| C5 | 94.0 | 62.7 | 1.6 | 1.0 | |
| A11 | 1.7 | 1.3 | 0.8 | 0.9 | |
| H31 | 6.3 | 2.7 | 16.2 | 1.1 | |
| SCCA2 | SCC107 | 2.6 | 10.0 | 2.4 | 92.2 |
| C5 | 4.1 | 1.7 | 1.0 | 1.2 | |
| A11 | 45.8 | 57.4 | 1.1 | 1.0 | |
| H31 | 6.5 | 2.4 | 1.2 | 34.1 | |
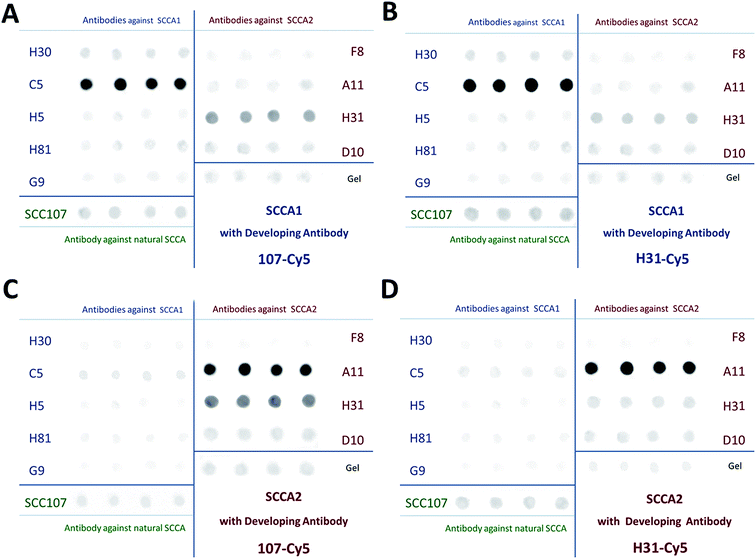
The specificity of the differential determination of SCCA1 and SCCA2 on a biochip with immobilized C5 and A11 antibodies was confirmed by comparative individual sandwich analysis using two developing antibodies, SCC107-Cy5 (A and C) and H31-Cy5 (B and D) (Fig. 4).
The specificity of the SCCA1 and SCCA2 antibodies selected for differential determination was estimated as follows: in the case of SCCA1 analysis, we determined the ratio of fluorescence signals from the gel elements containing C5 antibody (IFC5) to the fluorescence signals from the elements containing A11 antibody (IFA11). In the case of SCCA2, the corresponding ratio (IFA11/IFC5) was determined.
As a result of the sandwich analysis, specific pairs of antibodies to each of the proteins were determined. These were C5/SCC107-Cy5 and C5/H31-Cy5 for SCCA1 and A11/SCC107-Cy5 and A11/H31-Cy5 for SCCA2. It should be noted that the H31-Cy5 (IFA11/IFC5 = 44.9) developing antibody allows SCCA2 determination with a higher specificity than SCC107-Cy5 (IFA11/IFC5 = 13.3), as the IFA11/IFC5 value for them is considerably higher. On the other hand, for the purpose of SCCA1 measurement, both developing antibodies are comparably specific, as the ratio of fluorescence signals from the elements with immobilized C5 to the signals from the elements with immobilized A11 is rather high for both selected developing antibodies (using SCC107-Cy5 IFC5/IFA11 = 67.0, using H31-Cy5 IFC5/IFA11 = 53.5). Therefore, for the simultaneous independent determination of SCCA1 and SCCA2, it is preferable to use the H31-Cy5 developing antibody.
3.3. Analytical characteristics of the method developed
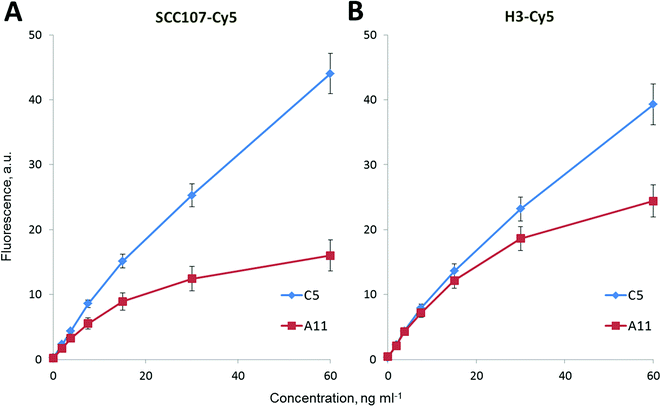
To provide a quantitative determination of SCCA1 and SCCA2 in a sample, we obtained fluorescence signal – antigen concentration calibration curves for individual sandwich analyses (data not shown) and when both compounds were present, calibration curves were obtained for two developing antibodies, SCC107-Cy5 and H31-Cy5 (Fig. 5).Using the calibration curves obtained, the sensitivity (LOD) and reproducibility of the SCCA1 and SCCA2 analyses on a biochip were determined. The CV% for various SCCA1 and SCCA2 concentrations determined both separately and simultaneously did not exceed 10% in our experiments (Fig. 5). The analytical sensitivity was calculated as the concentration corresponding to the fluorescence value that exceeds the mean fluorescence signal from the zero calibration sample measured ten times by no less than two standard deviations.
The analytical sensitivity of the simultaneous analysis of SCCA1 and SCCA2 on a biochip was 0.006 ng ml−1 for SCCA1 and 0.011 ng ml−1 for SCCA2 with SCC107-Cy5 as the developing antibody, or 0.014 ng ml−1 for SCCA1 and 0.01 ng ml−1 for SCCA2 with H31-Cy5 as the developing antibody. This finding suggests some sort of steric hindrance of H31-Cy5 binding to SCCA1, as its performance is negatively impacted while SCCA2 detection is unchanged. This result is superior to the analytical sensitivity values of available commercial test systems (0.3 ng ml−1 – CanAg SCC EIA, Sweden).
The stability of the biochips was measured by periodically checking the fluorescence signals after the sandwich immunoassay. The biochips were stored at +4 °C. The value of the fluorescence signals in the same experiment on the biochips after one month and three months of storage exhibited no significant difference.
The selectivity of the biochip with anti-SCCA antibodies was tested for its role in analysing biological samples. The effect of the cross-reactivity of antibodies C5, A11, SCC107 and H31 with other tumour markers had been investigated. An SCCA1 and SCCA2 (10 ng ml−1) solution containing 100 ng ml−1 (or 100 U ml−1 for the last five markers) of each of the tumour markers (AFP, CEA, NSE, PSA total, PSA free, HCG, CA 19-9, CA 72-4, CA 242 and CA 125) was measured on a biochip. As a result, the current variation due to the other tumour markers was less than 5% of that without interference. These results indicated that the biochip selectivity was acceptable.
3.4. Application to the detection of serum SCCA1 and SCCA2
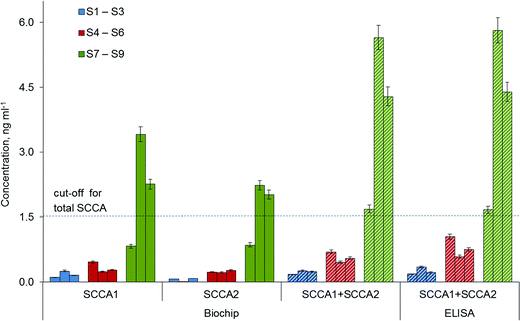
The feasibility of the immunoassay system for clinical applications was investigated by analysing nine blood samples of patients with adenocarcinoma (S1–S3), healthy donors (S4–S6) and patients with SCC (S7– S9). The blood samples were centrifuged for 10 min at 2000 rpm, and the serum supernatant was stored at −80 °C for later use. Each sample was analysed in replicates on three biochips (each containing 4 gel elements per antibody). The final concentration was calculated as the average of the three median values, measured on the same device. To demonstrate the validity of the proposed immunoassay protocol, the tumour marker concentrations were also measured by ELISA for the same serum samples, and the concentrations are shown in Fig. 6. There was no significant difference in SCCA1 and SCCA2 values between the two methods, based on the sandwich immunoassay technique. Thus, we have shown that simultaneous immunoassay on a biochip containing immobilized antibodies C5 and A11 with H31-Cy5 developing antibodies allows a quantitative determination of each of the two homologous proteins, SCCA1 and SCCA2, in one analysis using human serum.4. Conclusions
The modern approach to the complex diagnosis of diseases is provided by multiplex diagnostic tests that can detect different analytes in a single assay. However, in the case of an immunoassay, the simultaneous analysis of highly homologous compounds is limited, primarily due to the antibody specificity and to the steric and conformational difficulties associated with protein immobilization. In this paper, we have presented the complete development cycle of simultaneous analysis of SCCA1 and SCCA2, from specific antibody production to the final system tests, which allows for the differential quantification of proteins with 92% homology in amino acid composition in human serum using a single assay.Acknowledgements
This work was financially supported by the Ministry of Education and Science, Agreement No. 14.604.21.0103 from August 5, 2014, applied research project ID RFMEFI60414X0103. The project is part of the Federal Target Programme “Research and Development in Priority Areas of the Science and Technology Complex of Russia in 2014–2020”, Decree No. 1096 of the Government of the Russian Federation from November 28, 2013.References
- H. Kato and T. Torigoe, Cancer, 1977, 40, 1621–1628 CrossRef CAS PubMed.
- S. S. Schneider, C. Schick, K. E. Fish, E. Miller, J. C. Pena, S. D. Treter, S. M. Hui and G. A. Silverman, Proc. Natl. Acad. Sci. U. S. A., 1995, 92, 3147–3151 CrossRef CAS.
- M. Gatto, L. Iaccarino, A. Ghirardello, N. Bassi, P. Pontisso, L. Punzi, Y. Shoenfeld and A. Doria, Clin. Rev. Allergy Immunol., 2013, 45, 267–280 CrossRef CAS PubMed.
- S. Cataltepe, E. R. Gornstein, C. Schick, Y. Kamachi, K. Chatson, J. Fries, G. A. Silverman and M. P. Upton, J. Histochem. Cytochem., 2000, 48, 113–122 CrossRef CAS PubMed.
- J. M. Catanzaro, J. L. Guerriero, J. Liu, E. Ullman, N. Sheshadri, J. J. Chen and W.-X. Zong, PLoS One, 2011, 6, e19096 CAS.
- M. Guido, T. Roskams, P. Pontisso, M. Fassan, S. N. Thung, L. Giacomelli, A. Sergio, F. Farinati, U. Cillo and M. Rugge, J. Clin. Pathol., 2008, 61, 445–447 CrossRef CAS PubMed.
- T. Vassilakopoulos, T. Troupis, C. Sotiropoulou, P. Zacharatos, P. Katsaounou, D. Parthenis, O. Noussia, G. Troupis, S. Papiris, C. Kittas, C. Roussos, S. Zakynthinos and V. Gorgoulis, Lung Cancer, 2001, 32, 137–144 CrossRef CAS PubMed.
- S. Badola, H. Spurling, K. Robison, E. R. Fedyk, G. A. Silverman, J. Strayle, R. Kapeller and C. A. Tsu, Genomics, 2006, 88, 173–184 CrossRef CAS PubMed.
- P. A. Brioschi, P. Bischof, C. Delafosse and F. Krauer, Int. J. Cancer, 1991, 47, 376–379 CrossRef CAS PubMed.
- G. Scambia, P. B. Panici, G. Baiocchi, M. Amoroso, E. Foti, S. Greggi and S. Mancuso, Am. J. Obstet. Gynecol., 1991, 164, 631–636 CrossRef CAS PubMed.
- H. Y. S. Ngan, S. Y. W. Chan, L. C. Wong, D. T. K. Choy and H. K. Ma, Gynecol. Oncol., 1990, 37, 260–263 CrossRef CAS PubMed.
- M. Takeda, N. Sakuragi, K. Okamoto, Y. Todo, S.-I. Minobe, E. Nomura, H. Negishi, M. Oikawa, R. Yamamoto and S. Fujimoto, Acta Obstet. Gynecol. Scand., 2002, 81, 451–457 CrossRef PubMed.
- Y. Sun, N. Sheshadri and W.-X. Zong, Semin. Cell Dev. Biol., 2016 DOI:10.1016/j.semcdb.2016.09.005.
- C. A. Burtis, E. R. Ashwood and D. E. Bruns, Tietz Textbook of Clinical Chemistry and Molecular Diagnostics, 2012 Search PubMed.
- M. Crescenzi, A. Tessari, A. Biasiolo, A. Padoan, A. Gallotta, G. Fassina, C. Panciatichi, O. Rossetto, P. Pontisso, D. Basso and M. Plebani, Anal. Methods, 2015, 7, 629–637 RSC.
- F. Calabrese, F. Lunardi, E. Balestro, G. Marulli, E. Perissinotto, M. Loy, N. Nannini, M. Valente, M. Saetta, C. Agostini and F. Rea, Pathology, 2012, 44, 192–198 CrossRef CAS PubMed.
- C. Turato, M. A. Buendia, M. Fabre, M. J. Redon, S. Branchereau, S. Quarta, M. Ruvoletto, G. Perilongo, M. A. Grotzer, A. Gatta and P. Pontisso, Eur. J. Cancer, 2012, 48, 1219–1226 CrossRef CAS PubMed.
- K.-Y. Ho, H. H. Huang, K.-F. Hung, J.-C. Chen, C.-Y. Chai, W.-T. Chen, S.-M. Tsai, C.-Y. Chien, H.-M. Wang and Y.-J. Wu, Laryngoscope, 2012, 122, 2818–2823 CrossRef CAS PubMed.
- R. D. Petty, K. M. Kerr, G. I. Murray, M. C. Nicolson, P. H. Rooney, D. Bissett and E. S. R. Collie-Duguid, J. Clin. Oncol., 2006, 24, 1729–1744 CrossRef CAS PubMed.
- E. S. R. Collie-Duguid, K. Sweeney, K. N. Stewart, I. D. Miller, E. Smyth and S. D. Heys, Breast Cancer Res. Treat., 2012, 132, 807–818 CrossRef CAS PubMed.
- J. Stenman, J. Hedström, R. Grénman, I. Leivo, P. Finne, A. Palotie and A. Orpana, Int. J. Cancer, 2001, 95, 39–43 CrossRef CAS PubMed.
- Z. Deng, M. Hasegawa, Y. Yamashita, S. Matayoshi, A. Kiyuna, S. Agena, T. Uehara, H. Maeda and M. Suzuki, Cancer Sci., 2012, 103, 2127–2134 CrossRef CAS PubMed.
- K.-F. Hsu, S.-C. Huang, A.-L. Shiau, Y.-M. Cheng, M.-R. Shen, Y.-F. Chen, C.-Y. Lin, B.-H. Lee and C.-Y. Chou, Int. J. Gynecol. Cancer, 2007, 17, 174–181 CrossRef PubMed.
- S. Ohta, R. Shibata, Y. Nakao, Y. Azuma, K. Taniguchi, K. Arima, S. Suzuki, H. Shiraishi, T. Iwasaka and K. Izuhara, Ann. Clin. Biochem., 2012, 49, 277–284 CrossRef CAS PubMed.
- E. V. Konovalova, E. N. Savvateeva, E. I. Dementieva, M. A. Filippova, A. Y. Turygin, T. V. Osipova, T. P. Ryabykh, A. Y. Rubina and A. S. Zasedatelev, Mol. Biol., 2007, 41, 665–669 CrossRef CAS.
- V. I. Butvilovskaya, M. V. Tsybulskaya, A. A. Tikhonov, V. O. Talibov, P. V. Belousov, A. Y. Sazykin, A. M. Schwartz, S. A. Surzhikov, A. A. Stomakhin, O. N. Solopova and A. Y. Rubina, Mol. Biol., 2015, 49, 705–713 CrossRef CAS.
- G. Köhler and C. Milstein, Nature, 1975, 256, 495–497 CrossRef.
- E. Engvall, K. Jonsson and P. Perlmann, Biochim. Biophys. Acta, Protein Struct., 1971, 251, 427–434 CrossRef CAS.
- A. Y. Rubina, A. Kolchinsky, A. A. Makarov and A. S. Zasedatelev, Proteomics, 2008, 8, 817–831 CrossRef CAS PubMed.
- K. Nustad, B. L. Dowell, G. J. Davis, K. Stewart, O. Nilsson, E. Röijer, Y. Suminami, S. Nawata, S. Cataltepe, G. A. Silverman, H. Kato and H. W. A. De Bruijn, Tumor Biol., 2004, 25, 69–90 CrossRef CAS PubMed.
- A. Y. Rubina, M. A. Filippova, G. U. Feizkhanova, A. O. Shepeliakovskaya, E. I. Sidina, K. M. Boziev, A. G. Laman, F. A. Brovko, Y. V. Vertiev, A. S. Zasedatelev and E. V. Grishin, Anal. Chem., 2010, 82, 8881–8889 CrossRef CAS PubMed.
- A. Y. Rubina, E. I. Dementieva, A. A. Stomakhin, E. L. Darii, S. V. Pan'kov, V. E. Barsky, S. M. Ivanov, E. V. Konovalova and A. D. Mirzabekov, BioTechniques, 2003, 34, 1008–1014 CAS.
- A. Y. Rubina, S. Pan'kov, E. Dementieva, D. Pen'kov, A. Butygin, V. Vasiliskov, A. Chudinov, A. Mikheikin, V. Mikhailovich and A. Mirzabekov, Anal. Biochem., 2004, 325, 92–106 CrossRef CAS PubMed.
- D. A. Zubtsov, E. N. Savvateeva, A. Y. Rubina, S. V. Pan'kov, E. V. Konovalova, O. V. Moiseeva, V. R. Chechetkin and A. S. Zasedatelev, Anal. Biochem., 2007, 368, 205–213 CrossRef CAS PubMed.
- V. Barsky, A. Perov, S. Tokalov, A. Chudinov, E. Kreindlin, A. Sharonov, E. Kotova and A. Mirzabekov, J. Biomol. Screening, 2002, 7, 247–257 CrossRef CAS PubMed.
- E. Roijer, H. W. A. de Bruijn, U. Dahlén, K. ten Hoor, M. Lundin, K. Nilsson, K. Soderstrom and O. Nilsson, Tumour Immunobiol., 2006, 27, 142–152 CAS.
- S. Saffarian and E. L. Elson, Biophys. J., 2003, 84, 2030–2042 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2016 |