Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Rapid detection and identification of bacterial meningitis pathogens in ex vivo clinical samples by SERS method and principal component analysis†
Agnieszka
Kamińska
*a,
Evelin
Witkowska
a,
Aneta
Kowalska
a,
Anna
Skoczyńska
b,
Patrycja
Ronkiewicz
b,
Tomasz
Szymborski
a and
Jacek
Waluk
ac
aInstitute of Physical Chemistry, Polish Academy of Sciences, Kasprzaka 44/52, 01-224 Warsaw, Poland. E-mail: akamin@ichf.edu.pl
bNational Medicines Institute, Chełmska 30/34, 00-725 Warsaw, Poland
cFaculty of Mathematics and Natural Sciences, College of Science, Cardinal Stefan Wyszyński University, Dewajtis 5, 01-815 Warsaw, Poland
First published on 9th May 2016
Abstract
Three of the most common meningitis pathogens, Neisseria meningitidis, Haemophilus influenzae, and Streptococcus pneumoniae, have been successfully detected and identified in clinical cerebrospinal fluid (CSF) samples using a new class of a surface-enhanced Raman scattering (SERS) assay. Bacterial meningitis is a disease of the nervous system that is extremely serious and often fatal (an inflammation encompasses the lining around the brain and spinal cord). The approach presented in this study challenges the current SERS-based method of microorganism detection in terms of sensitivity and, more importantly, reveals a simple, quick (on a timescale of seconds), label-free detection of multiple components from very small volumes of clinical samples. This new SERS class of assay, based on the combination of two types of Au/Ag-coated, nuclepore track-etched polycarbonate membranes, allow simultaneous filtration of CSF and immobilization of CSF components, enhancing their Raman signals and enabling detection of the spectra of a single bacteria cell present in the analyzed CSF samples. The multivariate statistical method, principal component analysis (PCA), was applied (i) to extract the biochemical information from the recorded bacterial spectra, (ii) to perform the statistical classification of analyzed microorganisms, and, finally, (iii) to identify the spectrum of an unknown sample by comparing it to the library of known bacterial spectra. The three meningitis pathogens, namely, N. meningitidis, H. influenzae, and S. pneumoniae, were detected and identified simultaneously using a label-free SERS method. This method of detection produces consistent results faster and cheaper than traditional laboratory techniques and demonstrates the powerful potential of SERS technique in medical applications. Additionally, the present study was undertaken to evaluate the CSF neopterin level in patients with diagnosed meningococcal meningitis. The results of this study confirmed that bacterial meningitis caused by N. meningitidis, H. influenzae, and S. pneumoniae is associated with elevated cerebrospinal fluid neopterin levels compared with control CSF samples. The neopterin concentration can be used to predict meningitis, but cannot be applied to qualify the species of bacteria inducing the meningitis infection.
1. Introduction
Surface-enhanced Raman scattering (SERS) spectroscopy has recently received increasing attention due to its huge potential in the highly selective and sensitive detection of various types of molecules, even at a single-molecule detection level.1,2 SERS, in which the scattering cross-section is considerably enhanced for molecules upon their adsorption onto metallic nanostructures, is a promising technique for various applications in biochemical and analytical studies,3,4 clinical diagnosis,5,6 environmental monitoring,7 forensics,8 and other areas. Especially, SERS holds great possibility for investigating biological materials, from single macromolecules to whole tissues.9–11 SERS analysis of biological molecules can provide rich structural information as well as quantitative and qualitative analysis, with the ability to simultaneously detect multiple analytes. Moreover, it can be applied to samples under physiological conditions in a non-destructive way.12 Despite the great advantages of this technique, development of practical applications in biomedical and analytical tests is still hampered by difficulties in the generation of SERS-active nanostructures. Classical SERS substrates are based on three metals, Au, Ag, and Cu, which have localized surface plasmon resonances in the visible and near-infrared spectral regions. An ideal SERS substrate should exhibit a uniform and high enhancement factor (EF), chemical stability, and a cheap and reproducible production method. Researchers have explored numerous techniques that could be used to prepare efficient SERS substrates. These include electrochemical methods,13 nanosphere lithography,14 electron-beam lithography,15 nanoimprinting lithography,16 vapor layer deposition,17 and colloidal suspension.18 In the last few years, our research group has developed several types of novel SERS substrates that satisfy all the requirements mentioned above and show great potential for biological and medical applications.19–22 In this study, we present a new SERS assay format that uses a combination of different SERS-active surfaces based on Au–Ag coated polycarbonate membranes for the label-free and rapid detection of Neisseria meningitidis, Streptococcus pneumoniae, and Haemophilus influenzae,23 the three main bacteria that cause acute bacterial meningitis. Meningitis is an inflammation of the protective membranes covering the brain and spinal cord, known as the meninges, and may be caused by infection with bacteria, viruses, or other microorganisms, and less commonly by certain drugs.24 Meningitis caused by above-mentioned bacteria is usually very serious (5% to 10% of patients die, typically within 24 to 48 hours after the onset of symptoms) and requires a rapid detection method and urgent medical attention with appropriate antibiotic therapy. Taking into account the high mortality rates, the rapid detection of these bacteria in body fluids and subsequent effective treatment is essential. Recently, Gracie et al.25 presented a disease-specific DNA-based SERS assay for the detection of these three bacteria. This identification assay was based on the detection of labeled DNA sequences coding for N. meningitidis, S. pneumoniae, and H. influenzae. Detection was carried out in a multi-step process by monitoring the characteristic SERS peaks of the fluorescent dyes (Raman probes) attached at the 5′ end of phosphate-modified oligonucleotides. Therefore, the approach presented in this paper is a promising alternative to that described above and to the conventional biological methods; it enables rapid and accurate detection of pathogenic bacteria in biological fluids.Additionally, we broaden our research attempt to investigate the presence of neopterin, a new diagnostics marker used in the determination of bacterial meningitis infections. Neopterin is produced by activated macrophages, monocytes and dendritic cells upon stimulation by interferon gamma produced by T-lymphocytes.26 The presence of neopterin indicates the state of activation of the cellular immune system during subsequent stages of various diseases, such as rheumatoid arthritis (RA),27 neuropsychiatric abnormalities,28 cardiovascular disease,29 insulin resistance,30 allograft rejection, some tumors,31 viral infections32–34 (hepatitis A, B, and C; cytomegalovirus; measles; rubella; influenzae) and bacterial infections.35
The measurement of neopterin levels can provide reliable information regarding the disease diagnosis, disease stage, prognosis, and the evaluation of therapy. Bacterial meningitis might be associated with both elevated serum and cerebrospinal fluid (CSF) neopterin levels compared with control samples.36 In this study, we have determined the neopterin level in CSF clinical samples of patients with meningitis in relation to the analyzed bacteria that caused the disease.
Moreover, in this work, principal component analysis (PCA) was performed over the pre-processed SERS spectra in order (i) to evaluate the spectral differences among the clinical samples infected by N. meningitidis, S. pneumoniae, and H. influenzae along with the normal (control) type and (ii) to develop models allowing the simultaneous discrimination and classification of these three meningitis pathogens in clinical samples.
2. Experimental section
2.1. Chemicals and materials
Neopterin was purchased from Tocris Bioscience (Bristol, UK). Phosphate-buffered saline (PBS) packs (10 mM, pH = 7.2) from Sigma-Aldrich (Dorset, UK) were used without further purification. Water (resistivity over 18 MΩ), purified using a Milli-Q plus 185 system, was used throughout the process. In our experiments, we used CSF samples with N. meningitidis and S. pneumoniae obtained from the National Reference Centre for Bacterial Meningitis (NRCBM) in the National Medicines Institute (NMI) in Warsaw. H. influenzae strain was obtained from the Institute of Microbiology, University of Warsaw. CSF samples were evaluated for the presence of neopterin by using the commercial enzyme-linked immunoassay test (ELISA, IBL International GmbH, Hamburg).2.2. Instrumentation
SERS spectroscopy measurements were carried out with a Renishaw inVia Raman system equipped with a 785 nm diode laser. The light from the laser passed through a line filter and was focused on a sample mounted on an X–Y–Z translation stage with a 20× microscope objective, NA = 0.25. The beam diameter was approximately 5 μm. The laser power at the sample was 0.5 mW or less. The microscope was equipped with 1200 grooves per mm grating, cutoff optical filters, and a 1024 × 256 pixel Peltier-cooled RenCam CCD detector, which allowed the registration of the Stokes part of the Raman spectra with 5–6 cm−1 spectral resolution and 2 cm−1 wavenumber accuracy. The experiments were performed at ambient conditions using a back-scattering geometry.The obtained spectra were processed with the Opus software provided by Bruker. Some of the spectra required post-processing involving spike removal, smoothing and six-point baseline correction.
SEM measurements were performed under high vacuum using the FEI Nova NanoSEM 450 with an accelerating voltage of 2 kV under high vacuum.
2.3. Specimen collection
Clinical samples were obtained as part of a routine activity of the NRCBM. All experiments were performed in compliance with the European Parliament and Council decision for the epidemiological surveillance and control of communicable disease in the European Community.37,38 During the study, we used N. meningitidis of serogroup B (603/2011) and S. pneumoniae ATCC 49619 obtained from the NRCBM, and H. influenzae type b (Hib) obtained from the University of Warsaw, which served as reference strains in PCR reactions.2.4. Chemometrics
The SERS spectra were prepared for principal component analysis (PCA) using a two-step approach. First, using OPUS software (Bruker Optic GmbH 2012 version), the spectra were smoothed with Savitzky–Golay filter, the background was removed using baseline correction, and then the spectra were normalized using the so-called min–max normalization (the area of band around 963 cm−1). All the data were transferred to the Unscrambler software (CAMO software AS, version 10.3, Norway), where the PCA analysis was performed. PCA is a multivariate technique that reduces the dimensionality of complex spectroscopic data from many wavenumber assignments to a few principal components (PCs), making it easier to identify the majority of variations within the spectra.43PCA is based on a linear transformation of spectra regarded as N-dimensional vectors, where N is the number of data points in a single spectrum, into a new coordinate system in which the new vectors are orthogonal to each other. All the spectra can then be expressed in a much simpler manner through a small number of principal components (PCs) that account for the maximum variance in the data. In a PCA model, the matrix containing the set of spectra (X) is decomposed into two smaller matrices (the scores (T) and the loadings (P): X = TPT + E), where E is the residual containing information that is not described by the multiplication of the scores and the loadings for each spectrum. Typically, systems of orthogonal axes (score plots) of two or three components (PCs) are used to represent the point-spectra, where the first PC maximizes the spectral variance, the second is orthogonal to the first and maximizes the residual variance, etc. By plotting the principal component scores, similarities between the samples are revealed. Additionally, plotting loadings as a function of the wavenumber reveals the most important diagnostic vibrations in the spectra.
In the present work, PCA was carried out on five different data sets consisting of spectra obtained for normal CSF (without microbiological confirmation of N. meningitidis, S. pneumonia, and H. influenzae infections), for three CSF samples infected by these pathogens separately, and for one CSF sample containing all three bacteria.
3. Results and discussion
3.1. Detection of N. meningitidis, S. pneumoniae, and H. influenzae in clinical CSF samples
The most common causes of bacterial meningitis are meningococci (N. meningitidis), pneumococci (S. pneumoniae), group B streptococci (Streptococcus agalactiae), and rod-like H. influenzae. Conventional detection of the meningitis pathogens is based on phenotypic and biochemical characterization. However, biochemical methods require time-consuming bacteria cultivation and have relatively low sensitivity, especially when antibiotics have been given to the patient prior to sampling.44,45 It has been reported that identification of the above-mentioned bacteria can take up to 36 hours46 using the traditional method. The use of nucleic acid amplification tests, such as quantitative polymerase chain reaction (qPCR), has enabled more sensitive and rapid detection of pathogens in respiratory secretions and cerebrospinal fluid. Several qPCR assays for the detection of S. pneumoniae, H. influenzae, and N. meningitidis have been developed,47–49 and multiplex detection of several target DNAs in a single tube is achievable.50 Still, the specificity of methods used is an underestimated problem, and commonly used targets have been shown to be unspecific and cause misleading results.In this study, we present a new class of SERS assay based on two types of polycarbonate membranes with different pore sizes coated with Au–Ag for the label-free detection of N. meningitides, S. pneumoniae, and H. influenzae in clinical CSF samples. The morphology of these SERS substrates is presented in ESI, Fig. S1.†
Fig. 1 shows the experimental setup used for the detection of bacterial meningitis pathogens. The applied technique is simple and allows filtration and/or concentration of CSF composites (leucocytes, albumins, globulins, and bacteria) within a small area of the platform. The setup for the proposed method consists of (1) a high-precision syringe pump system (NE-1000, New Era Pump Systems Inc., USA) used for automated control of flow, (2) syringe, and (3) two syringe holders built-in with two types of SERS substrates based on Au/Ag-coated, nuclepore track-etched polycarbonate membranes with different pore sizes (0.3 μm and 3 μm).
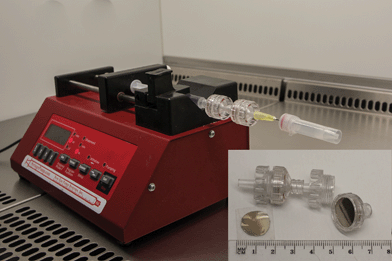 | ||
| Fig. 1 Experimental setup of the SERS-based detection method of bacteria from CSF samples. This setup consists of syringe pump, syringe, filter holders with Au/Ag polycarbonate membranes (working as SERS-active platforms) and polymer vial (working as a waste collector). Please see ESI† for SEM pictures of SERS-active platforms based on nuclepore track-etched polycarbonate membranes. | ||
In the first step (Fig. 1), 1 mL of saline solution was flushed through the SERS assay (coupled syringe filters with SERS substrates) at the rate of 3.5 μL min−1 in order to remove air bubbles and fill the dead space volume in syringe filters. In the next step, the appropriate CSF samples were injected into the SERS assay at the rate of 1.5 μL min−1.
Taking into account the size of pores in the first and second membrane, 3 and 0.3 μm, respectively, the size of CSF components (leucocytes from 10 μm up to 15 μm,51 albumins from 3 up to 10 nm,52 and the sizes of the analyzed pathogens (N. meningitidis ranges from 0.6 to 1.0 μm;53S. pneumoniae—from 0.5 to 1.25 μm;54 and H. influenzae—is about 1 × 0.3 μm (ref. 55)), we were able to separate these bacterial cells from CSF components. The largest leukocytes (the main component of CSF) remain on the first membrane, while the smaller proteins and bacteria pass through and finally fall on the second membrane with about 0.3 μm pore size.
A total of 350 spectra were collected, including 100 spectra from 10 control patients without meningitis and 250 spectra from 38 meningococcal meningitis patients. The SERS spectra of control and patients with meningitis are depicted in Fig. 2.
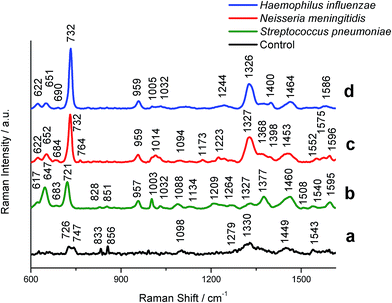 | ||
| Fig. 2 SERS spectra of control CSF (a) and CSF infected by (b) S. pneumoniae; (c) N. meningitidis, and (d) H. influenzae bacteria recorded on the polycarbonate membrane (0.3 μm pore size) according to the procedure illustrated in Fig. 1. Experimental conditions: 0.5 mW, 785 nm excitation, 30 seconds acquisition time (mapping mode). The SERS spectra have been baseline-corrected, normalized and shifted vertically for better visualization. Each SERS spectrum was averaged from 20 measurements in different places of the SERS platform. | ||
Fig. 2 shows typical SERS spectra (after averaging, baseline correction, smoothing and normalization) of the bacteria filtrated and detected from clinical samples according to the procedure described above (Fig. 1). The SERS spectra were analyzed to obtain general biochemical information and to perform the band assignment for each data group: (i) control group and groups infected by experimental (ii) S. pneumoniae, (iii) H. influenzae, and (iv) N. meningitidis samples.
We have performed the SERS measurements and band assignments of these bacteria, S. pneumoniae, H. influenzae, and N. meningitidis, inoculated onto BHI agar plates directly from pre-cultures in order to obtain the reference spectra of these pathogens. Fig. S2† presents the SERS spectra of bacteria multiplied and then deposited on the Au/Ag polycarbonate membranes. These results show no differences (except for the recorded SERS signal intensities) between the SERS spectra of particular bacterial species trapped via SERS-based assay (Fig. 2) and the SERS spectra of the reference bacteria (Fig. S2†). As can be seen in Fig. 2b–d, the SERS spectra exhibit common spectral features for all bacteria with some differences in the band position, their relative intensity ratios and/or the appearance of new peaks. The spectra depicted in Fig. 2b–d reveal a combination of SERS bands typical for bacteria and arising from the molecular vibrations of nucleic acid, amides, n-alkanes, flavins, and other constituent compounds of bacterial cell.56,57 All tested bacterial species show a common intense spectral feature at about 732 cm−1 (H. influenzae and N. meningitidis) and 721 cm−1 (S. pneumoniae). This band is observed also in many types of bacterial species, such as Escherichia coli, Salmonella enterica, Staphylococcus epidermidis, Staphylococcus aureus and Bacillus megaterium. It is assigned to the C–N stretching mode of the adenine part of the lipid layer in the cell wall or to the purine ring breathing mode.58,59 Jarvis et al.,60 on the other hand, attributed the same peak to the glycosidic ring mode from cell wall peptidoglycan building blocks, N-acetyl-D-glucosamine (NAG) and N-acetylmuramic acid (NAM). All spectra of S. pneumoniae, H. influenzae, and N. meningitidis show also a common peak at 957 cm−1 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C deformation or C–N stretching), 1003 cm−1 (phenylalanine, C–C aromatic ring stretching), 1327 cm−1 (amide III (protein), C–H deformation), 1338 cm−1 (phospholipids), and 1460 cm−1 (CH2 deformation).61 Apart from these common bands, all bacteria species reveal their own individual spectral characteristics, which aid in the whole-organism fingerprint analysis. For example, the band at 647 cm−1 can be seen in the S. pneumoniae spectrum, but not in N. meningitidis and H. influenzae spectra. To distinguish between S. pneumoniae and N. meningitidis, the ratio of intensities of the peaks at 647 cm−1 and 731 cm−1 can be used. S. pneumoniae can be distinguished from other bacteria using bands at 1209 cm−1 for H. influenzae and 1173 cm−1, 1223 cm−1, and 1540 cm−1 for N. meningitidis. Table 1 shows the main bands observed in S. pneumoniae, H. influenzae, and N. meningitidis spectra and the corresponding band assignments.
C deformation or C–N stretching), 1003 cm−1 (phenylalanine, C–C aromatic ring stretching), 1327 cm−1 (amide III (protein), C–H deformation), 1338 cm−1 (phospholipids), and 1460 cm−1 (CH2 deformation).61 Apart from these common bands, all bacteria species reveal their own individual spectral characteristics, which aid in the whole-organism fingerprint analysis. For example, the band at 647 cm−1 can be seen in the S. pneumoniae spectrum, but not in N. meningitidis and H. influenzae spectra. To distinguish between S. pneumoniae and N. meningitidis, the ratio of intensities of the peaks at 647 cm−1 and 731 cm−1 can be used. S. pneumoniae can be distinguished from other bacteria using bands at 1209 cm−1 for H. influenzae and 1173 cm−1, 1223 cm−1, and 1540 cm−1 for N. meningitidis. Table 1 shows the main bands observed in S. pneumoniae, H. influenzae, and N. meningitidis spectra and the corresponding band assignments.
| Assignment | Range | Neisseria meningitidis | Streptococcus pneumoniae | Haemophilus influenzae |
|---|---|---|---|---|
| a Assignments: − absent; + present; ++ strong. | ||||
| Guanine, tyrosine | 640–675 | ++ | + | + |
| Adenine, glycoside | 713–740 | ++ | ++ | ++ |
| Cytosine, uracil | 745–790 | + | − | − |
| O–P–O (RNA) | 800–815 | + | − | + |
C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C deformation, C–N stretching C deformation, C–N stretching |
930–990 | + | + | + |
| Phenylalanine, C–C aromatic ring stretching | 1000–1010 | + | + | + |
| C–C stretching (phospholipids, carbohydrates), C–N stretching | 1025–1060 | + | + | + |
| O–P–O (DNA), C–C or C–O–C stretching (carbohydrates) | 1080–1105 | + | + | − |
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–O–C C–O–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) (unsaturated fatty acids in lipids) (unsaturated fatty acids in lipids) |
1130–1145 | − | + | − |
| C–O ring, aromatic amino acids in proteins | 1150–1185 | + | − | − |
| Amide III (random), thymine | 1215–1295 | + | + | + |
| Amide III (protein), C–H deformation | 1315–1325 | − | ||
| Adenine, guanine, C–H deformation | 1330–1345 | + | ++ | ++ |
| COO– symmetric stretching | 1390–1415 | + | − | + |
| CH2 deformation | 1440–1475 | + | + | + |
| Amide II | 1510–1560 | + | − | + |
| Adenine, guanine (ring stretching) | 1570–1595 | + | + | + |
The SERS spectrum of control CSF samples, analyzed in the same way as bacterial meningitis samples using the proposed SERS assay, reveals very weak bands at 726, 747, 833, 856, 1098, 1330, 1449 and 1543 cm−1 (Fig. 2a). These bands correspond to vibrations of small proteins62 that can pass through pores of SERS substrates. For example, the band at 1330 cm−1 is assigned to amide III vibrations of proteins, and the band at 1543 cm−1 corresponds to δ(NH3+) and/or amide II vibrations of proteins. Most of these bands, but with higher intensity, we can find in both control and meningitis pathogens infected CSF spectra recorded before filtration via SERS-assay (see Fig. 3Sa†). For example, in H. influenzae-infected CSF samples (Fig. 3Sd†), the aromatic amino acid residues, tyrosine, phenylalanine and tryptophan have bands at 720, 852, 1000 and 1330 cm−1. The intensive bands in the CSF spectra at 728, 1096, 1135, 1337, 1466 and 1596 cm−1 (see Fig. 3Sa†) were assigned to vibrations of the nitrogenous bases of DNA and lipids.63
Additionally, the reproducibility of S. pneumoniae, N. meningitides, and H. influenzae recorded SERS spectra were calculated with a Savitzky–Golay second derivative method (see Fig. 4S and Table 1S in ESI†). The lowest value of the calculated average correlation coefficient is 0.92, which is sufficient for identification purposes.
All these results clearly indicate that the proposed new class of SERS assay, based on the combination of two types of Au/Ag polycarbonate membranes with different pore sizes, enables simultaneous filtration of CSF components, thus enhancing their Raman signals, and detection of spectra from single bacteria cell present in the analyzed CSF samples.
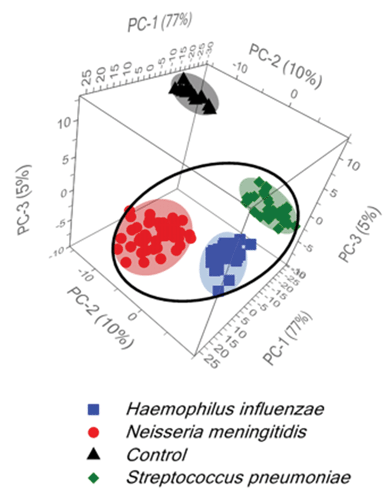
Additionally, for further development of diagnostic algorithms for improving: (i) the discrimination among tested bacteria and (ii) the differentiation between normal and the bacterial meningitis samples, principal component analysis (PCA) has been applied. The main information obtained from the PCA is explained by the three first principal components PC-1, PC-2, and PC-3, which explain 92% of the total variance in the data. Fig. 3 shows two large clusters corresponding to the control and meningitis CSF samples and three clusters inside the meningitis group associated with three types of bacterial pathogens that cause acute bacterial meningitis. Fig. 3 displays the score plot of the first three principal components, PC-1 (77% of the total variance), PC-2 (10% of the total variance) and PC-3 (5% of the total variance), of the control CSF and CSF infected with bacteria.
The statistical data demonstrate the significant differences between normal and meningitis groups and also reveal a huge difference among meningitis samples for the classification of S. pneumoniae, H. influenzae, and N. meningitidis bacterial infections.
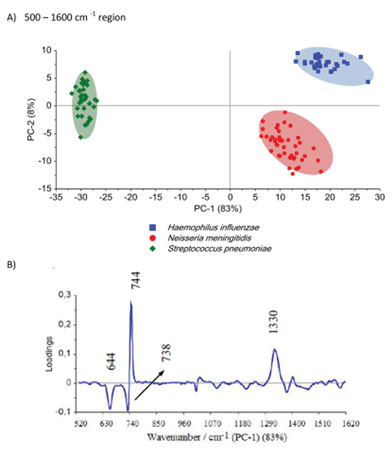
The differences among the S. pneumoniae, H. influenzae and N. meningitidis are due to the peaks analysed above and are clearly illustrated by loadings plots presented in Fig. 4B and Fig. S5B.† The loadings of the PCs provide information on the variables (wavenumbers of the spectra) that are important for group separation. Fig. 4B displays the loadings plot of PC-1 for the entire wavenumber region, 500–1600 cm−1, for all studied meningococcal meningitis samples. By analyzing this plot, one can recognize the most important diagnostic variables in the analyzed data set. Variables with high loading values are the most important for diagnostic purposes. Unlike the SERS spectrum, the loading spectrum contains positive and negative bands, and their corresponding frequencies can be correlated with some of the major variations in the molecular structure among analyzed samples. On the basis of SERS spectra analysis and loadings plots from PC-1, the most important variations among these three meningitis pathogens, N. meningitidis, H. influenzae, and S. pneumoniae, were found to be associated with the peaks at 644, 738, 744, and 1330 cm−1 corresponding to the main compounds of bacterial cells (see Fig. 2 and Table 1). The PCA analysis performed for these three pathogens in the entire fingerprint area gives the sum of PC-1 and PC-2 equal to 91% of total variance. Moreover, taking into account the loadings plot versus variables, calculations of PCA in the area of the most pronounced marker bands at 644, 738, and 744 cm−1 were performed. The PCA scores calculated for regions of the chosen markers give a value of 97% (PC-1 and PC-2) of the total variance in studied samples (see Fig. S5, ESI†).
Importantly, the differences between samples belonging to the same group of classification (one type of pathogen) are diagnostically significant. These results clearly demonstrate that PCA-based SERS technique has potential for the detection and discrimination of the meningitis pathogen in clinical CSF samples.
3.2. Detection of neopterin level in CSF samples as a marker of bacterial meningitis
Neopterin appears in cerebrospinal human fluid, blood, and urine, and its increased levels indicate activation of the immune system involved in the pathogenesis and/or affected by malignant diseases. An earlier literature report68 reveals that neopterin characterization can serve as a useful method of the diagnosis and monitoring of many diseases, such as rheumatoid arthritis (RA),27 neuropsychiatric abnormalities,28 cardiovascular disease,29 allograft rejection, some tumors,31 and viral and bacterial infections.32,33Our previous reports19 showed that neopterin as a biomarker is biologically and chemically stable in human biological fluids, gives very strong SERS signals, and can be easily quantified using surface-enhanced Raman spectroscopy.
The present study was undertaken to evaluate the CSF neopterin levels in patients with diagnosed meningitis caused by three meningitis pathogens: S. pneumoniae, H. influenzae, and N. meningitidis. In the first step, the SERS spectroscopic characterization of CSF samples from both healthy subjects and patients with confirmed bacterial meningitis caused by the three analyzed pathogens was performed. Fig. S2† presents the normalized SERS spectra of CSF samples infected by N. meningitidis (b), S. pneumoniae (c), H. influenzae, (d) and the control CSF samples (a) deposited onto the SERS substrate. Detailed SERS analysis of both control and infected CSF samples is presented in ESI.† There are distinctive SERS features and intensity differences for normal and infected CSF samples in the spectral ranges of 1200–1260 cm−1, 1270–1380 cm−1 and 1500–1650 cm−1. These differences in the SERS spectral pattern could reflect changes in the quantity and structure of proteins and peptides of CSF associated with the abnormal metabolism of patients with bacterial infections. It should be highlighted that the major difference between control and infected CSF samples concerns the band at 695 cm−1, which corresponds to the C–C vibration and ring modes of neopterin. It can be observed only in the infected samples, but not in control subjects. Fig. S6† presents the SERS spectrum of neopterin adsorbed onto the polycarbonate membrane-based SERS platform from 35.0 nmol L−1 neopterin solution in PBS buffer.
In order to test the performance of the SERS method and to calculate the neopterin concentration in the analyzed clinical samples, the calibration curve, i.e. plot of SERS intensity of the marker band at 695 cm−1versus the concentration of neopterin in CSF, was constructed. Fig. S7 (ESI†) shows a calibration curve obtained by plotting the intensity of this marker band versus the concentration of neopterin in the range from 0.0 to 90.0 nmol L−1 (neopterin was artificially added to normal CSF solution). Based on this calibration line, the concentration of neopterin in clinical samples of CSF infected by the three meningitis pathogens and control CSF samples was estimated. The same samples were also evaluated for neopterin concentration by using a commercial enzyme-linked immunoassay (ELISA, IBL International) (Table 2).
| SERS (new method) | ELISA (reference method) | |
|---|---|---|
| Control CSF; nmol L−1 | 3.8 ± 0.7 | 4.0 ± 1.3 |
| Infected CSF; nmol L−1 (with N. meningitidis) | 30.0 ± 4.1 | 36.0 ± 5.2 |
| Infected CSF; nmol L−1 (with S. pneumoniae) | 38.0 ± 4.8 | 45.0 ± 6.6 |
| Infected CSF; nmol L−1 (with H. influenzae) | 32 ± 4.5 | 40.0 ± 5.8 |
The level of neopterin was significantly higher in CSF samples infected by N. meningitidis, (30 nmol L−1), S. pneumoniae (38 nmol L−1), H. influenzae (32 nmol L−1) compared to normal (control) CSF samples (3.8 nmol L−1). It should be highlighted that, taking into account the error bars of performed calculations, the value of neopterin concentration can be used to predict meningococcal meningitis, but cannot be applied to qualify the type of bacteria inducing the meningitis infection.
To summarize, the results of our study clearly demonstrate that: (i) neopterin may be used as a novel marker in the screening tests to detect bacterial meningitis and (ii) in the positive response, the simultaneous identification of three of the most common meningitis pathogens, e.g., Neisseria meningitidis, Haemophilus influenzae, and Streptococcus pneumoniae in clinical cerebrospinal fluid sample is possible using our new class of SERS substrates. Therefore, the approaches presented in this study challenge current methods of meningitis microorganism detection from body fluids in terms of using a very small volume of clinical materials, which are additionally selected by the neopterin test from all bacterial meningitis-suspected CSF specimens.
4. Conclusions
We have demonstrated that SERS spectroscopy combined with multivariate analysis (PCA) can be applied with high accuracy to label-free, simultaneous and rapid identification of all three meningitis pathogens: Neisseria meningitidis, Haemophilus influenzae, and Streptococcus pneumoniae in clinical CSF samples. PCA (i) permits the selection of significant variables from the spectra among three analyzed meningitis pathogens: N. meningitidis, H. influenzae, and S. pneumoniae present in CSF samples; (ii) shows the possibility of their simultaneous differentiation in a mixture within the CSF clinical samples; and (iii) allows differentiation of healthy and meningitis patients using neopterin test.In this study, we also confirmed that neopterin is an important inflammatory marker and a strong predictor of central nervous system disease caused by meningitis pathogens. Cerebrospinal fluid neopterin concentrations from 38 meningitis patients have been measured using our newly developed label-free SERS technique. The level of neopterin was significantly higher in CSF samples infected by N. meningitidis, (30 nmol L−1), S. pneumoniae (38 nmol L−1), and H. influenzae (32 nmol L−1) compared to normal (control) samples (3.8 nmol L−1).
The results of this study illustrate the huge potential of SERS spectroscopy for the rapid detection of a variety of bacterial diseases. The developed method is of practical interest, as it is fast, it requires very simple sample preparation without the need for sample amplification by a bacterial culture and by polymerase chain reaction (PCR), and no chemical reagents are necessary to specifically mark cell components.
Future studies should be designed to investigate the correlation between elevated neopterin level and the stage of infection, length of hospitalization, age and gender of patients. Such detailed cerebrospinal fluid neopterin characterization can serve in monitoring the central nervous system inflammatory effects and can give valuable information regarding the cause of ongoing brain pathology. Future studies should be designed also to examine additional meningitis markers such as C-reactive protein (CRP) or interleukins (IL) from both CSF and clinical blood samples to search for other clinical characteristics of this illness.
Acknowledgements
We would like to thank Dorota Korsak from University of Warsaw (Applied Microbiology, Institute of Microbiology, Faculty of Biology) for preparation of sBHI agar medium and cultivation of H. influenzae. The authors would like to acknowledge the support from NCN under grant UMO-2015/17/B/ST4/04128.References
- E. C. Le Ru and P. G. Etchegoin, in Annual Review of Physical Chemistry, ed. M. A. Johnson and T. J. Martinez, Annual Reviews, Palo Alto, 2012, vol. 63, pp. 65–87 Search PubMed.
- S. M. Nie and S. R. Emery, Science, 1997, 275, 1102–1106 CrossRef CAS PubMed.
- T. G. Spiro, Biological Applications of Raman Spectroscopy, John Wiley & Sons Canada, Limited, 1988 Search PubMed.
- E. Koglin and J. M. Sequaris, Top. Curr. Chem., 1986, 134, 1–57 CrossRef CAS.
- M. A. Woo, S. M. Lee, G. Kim, J. Baek, M. S. Noh, J. E. Kim, S. J. Park, A. Minai-Tehrani, S. C. Park, Y. T. Seo, Y. K. Kim, Y. S. Lee, D. H. Jeong and M. H. Cho, Anal. Chem., 2009, 81, 1008–1015 CrossRef CAS PubMed.
- H. Chon, S. Lee, S. W. Son, C. H. Oh and J. Choo, Anal. Chem., 2009, 81, 3029–3034 CrossRef CAS PubMed.
- Y. W. C. Cao, R. C. Jin and C. A. Mirkin, Science, 2002, 297, 1536–1540 CrossRef CAS PubMed.
- C. Rodger, G. Dent, J. Watkinson and W. E. Smith, Appl. Spectrosc., 2000, 54, 1567–1576 CrossRef CAS.
- A. Bonifacio, D. Millo, C. Gooijer, R. Boegschoten and G. van der Zwan, Anal. Chem., 2004, 76, 1529–1531 CrossRef CAS PubMed.
- I. R. Nabiev, H. Morjani and M. Manfait, Eur. Biophys. J., 1991, 19, 311–316 CrossRef CAS PubMed.
- H. Morjani, J. F. Riou, I. Nabiev, F. Lavelle and M. Manfait, Cancer Res., 1993, 53, 4784–4790 CAS.
- M. Jackson and H. Mantsh, Pathology by infrared and Raman spectroscopy, J. Wiley, 2002 Search PubMed.
- H. H. Wang, C. Y. Liu, S. B. Wu, N. W. Liu, C. Y. Peng, T. H. Chan, C. F. Hsu, J. K. Wang and Y. L. Wang, Adv. Mater., 2006, 18, 491–495 CrossRef CAS.
- L. A. Dick, A. D. McFarland, C. L. Haynes and R. P. Van Duyne, J. Phys. Chem. B, 2002, 106, 853–860 CrossRef CAS.
- D. P. Fromm, A. Sundaramurthy, A. Kinkhabwala, P. J. Schuck, G. S. Kino and W. E. Moerner, J. Chem. Phys., 2006, 124, 4 CrossRef PubMed.
- R. Alvarez-Puebla, B. Cui, J. P. Bravo-Vasquez, T. Veres and H. Fenniri, J. Phys. Chem. C, 2007, 111, 6720–6723 CAS.
- D. J. Semin and K. L. Rowlen, Anal. Chem., 1994, 66, 4324–4331 CrossRef CAS.
- K. Faulds, W. E. Smith and D. Graham, Anal. Chem., 2004, 76, 412–417 CrossRef CAS PubMed.
- A. A. Kowalska, A. Kaminska, W. Adamkiewicz, E. Witkowska and M. Tkacz, J. Raman Spectrosc., 2015, 46, 428–433 CrossRef CAS.
- A. Kaminska, A. A. Kowalska, D. Snigurenko, E. Guziewicz, J. Lewinski and J. Waluk, Analyst, 2015, 140, 5090–5098 RSC.
- A. Kaminska, I. Dziecielewski, J. L. Weyher, J. Waluk, S. Gawinkowski, V. Sashuk, M. Fialkowski, M. Sawicka, T. Suski, S. Porowski and R. Holyst, J. Mater. Chem., 2011, 21, 8662–8669 RSC.
- T. Szymborski, E. Witkowska, W. Adamkiewicz, J. Waluk and A. Kaminska, Analyst, 2014, 139, 5061–5064 RSC.
- P. B. McIntyre, K. L. O'Brien, B. Greenwood and D. van de Beek, Lancet, 2012, 380, 1703–1711 CrossRef.
- S. A. E. Logan and E. MacMahon, Br. Med. J., 2008, 336, 36–40 CrossRef PubMed.
- K. Gracie, E. Correa, S. Mabbott, J. A. Dougan, D. Graham, R. Goodacre and K. Faulds, Chem. Sci., 2014, 5, 1030–1040 RSC.
- C. Huber, J. R. Batchelor, D. Fuchs, A. Hausen, A. Lang, D. Niederwieser, G. Reibnegger, P. Swetly, J. Troppmair and H. Wachter, J. Exp. Med., 1984, 160, 310–316 CrossRef CAS PubMed.
- W. Kullich, Clin. Rheumatol., 1993, 12, 387–391 CrossRef CAS PubMed.
- S. Zeuzem, S. V. Feinman, J. Rasenack, E. J. Heathcote, M. Y. Lai, E. Gane, J. O'Grady, J. Reichen, M. Diago, A. Lin, J. Hoffman and M. J. Brunda, N. Engl. J. Med., 2000, 343, 1666–1672 CrossRef CAS PubMed.
- X. Garcia-Moll, D. Cole, E. Zouridakis and J. C. Kaski, Heart, 2000, 83, 346–350 CrossRef CAS PubMed.
- M. Ledochowski, C. Murr, B. Widner and D. Fuchs, Clin. Chim. Acta, 1999, 282, 115–123 CrossRef CAS.
- C. Murr, A. Bergant, M. Widschwendter, K. Heim, H. Schrocksnadel and D. Fuchs, Clin. Chem., 1999, 45, 1998–2004 CAS.
- G. Wernerfelmayer, E. R. Werner, D. Fuchs, A. Hausen, G. Reibnegger and H. Wachter, Cancer Res., 1990, 50, 2863–2867 CAS.
- D. Fuchs, A. Hausen, G. Reibnegger, E. R. Werner, M. P. Dierich and H. Wachter, Immunol. Today, 1988, 9, 150–155 CrossRef CAS PubMed.
- D. Fuchs, G. Weiss, G. Reibnegger and H. Wachter, Crit. Rev. Clin. Lab. Sci., 1992, 29, 307–341 CrossRef CAS PubMed.
- D. Fuchs, A. Hausen, M. Kofler, H. Kosanowski, G. Reibnegger and H. Wachter, Lung, 1984, 162, 337–346 CrossRef CAS PubMed.
- L. Hagberg, L. Dotevall, G. Norkrans, M. Larsson, H. Wachter and D. Fuchs, J. Infect. Dis., 1993, 168, 1285–1288 CrossRef CAS PubMed.
- The European Parliament, the Council of the EU, European Union website. Decision number 2119/98/EC of the European Parliament and of the Council of 24 September 1998: setting up a network for the epidemiological surveillance and control of communicable diseases in the community: Official J Eur Communities L268/1, http://eur-lex.europa.eu/LexUriServ/LexUriServ.douri=CELEX:31998D2119:EN:NOT, 1998.
- The European Commission of the European Communities, European Union website, Commission decision of 22 December 1999 on the communicable diseases to be progressively covered by the community network under decision number 2119/98/EC of the Parliament and of the Council, Official J Eur Communities L28/50, 2000.
- M. Guiver, R. Borrow, J. Marsh, S. J. Gray, E. B. Kaczmarski, D. Howells, P. Boseley and A. J. Fox, FEMS Immunol. Med. Microbiol., 2000, 28, 173–179 CrossRef CAS PubMed.
- M. K. Taha, J. Clin. Microbiol., 2000, 38, 855–857 CAS.
- P. Toikka, S. Nikkari, O. Ruuskanen, M. Leinonen and J. Mertsola, J. Clin. Microbiol., 1999, 37, 633–637 CAS.
- J. C. McAvin, P. A. Reilly, R. M. Roudabush, W. J. Barnes, A. Salmen, G. W. Jackson, K. K. Beninga, A. Astorga, F. K. McCleskey, W. B. Huff, D. Niemeyer and K. L. Lohman, J. Clin. Microbiol., 2001, 39, 3446–3451 CrossRef CAS PubMed.
- S. Mabbott, A. Eckmann, C. Casiraghi and R. Goodacre, Analyst, 2013, 138, 118–122 RSC.
- G. Tzanakaki, M. Tsopanomichalou, K. Kesanopoulos, R. Matzourani, M. Sioumala, A. Tabaki and J. Kremastinou, Clin. Microbiol. Infect., 2005, 11, 386–390 CrossRef CAS PubMed.
- K. Dalhoff, J. Braun, H. Hollandt, R. Lipp, K. J. Wiessmann and R. Marre, Infection, 1993, 21, 291–296 CrossRef CAS PubMed.
- X. Wang, M. J. Theodore, R. Mair, E. Trujillo-Lopez, M. du Plessis, N. Wolter, A. L. Baughman, C. Hatcher, J. Vuong, L. Lott, A. von Gottberg, C. Sacchi, J. M. McDonald, N. E. Messonnier and L. W. Mayer, J. Clin. Microbiol., 2012, 50, 702–708 CrossRef CAS PubMed.
- C. E. Corless, M. Guiver, R. Borrow, V. Edwards-Jones, A. J. Fox and E. B. Kaczmarski, J. Clin. Microbiol., 2001, 39, 1553–1558 CrossRef CAS PubMed.
- S. Deutch, J. K. Moller and L. Ostergaard, Scand. J. Infect. Dis., 2008, 40, 607–614 CrossRef CAS PubMed.
- S. T. Hedberg, P. Olcen, H. Fredlund and P. Molling, Apmis, 2009, 117, 856–860 CrossRef CAS PubMed.
- G. M. K. Abdeldaim, K. Stralin, P. Olcen, J. Blomberg and B. Herrmann, Diagn. Microbiol. Infect. Dis., 2008, 60, 143–150 CrossRef CAS PubMed.
- R. I. Handin, S. E. Lux and T. P. Stossel, Blood: Principles and Practice of Hematology, Lippincott Williams & Wilkins, 2003 Search PubMed.
- L. Noorani, M. Stenzel, R. Liang, M. H. Pourgholami and D. L. Morris, J. Nanobiotechnol., 2015, 13, 12 CrossRef PubMed.
- J. S. Swartley, A. A. Marfin, S. Edupuganti, L. J. Liu, P. Cieslak, B. Perkins, J. D. Wenger and D. S. Stephens, Proc. Natl. Acad. Sci. U. S. A., 1997, 94, 271–276 CrossRef CAS.
- K. K. Todar, Streptococcus pneumoniae: Pneumococcal pneumonia, Todar's Online Textbook of Bacteriology, 2003 Search PubMed.
- H. Gmuender, K. Kuratli, K. Di Padova, C. P. Gray, W. Keck and S. Evers, Genome Res., 2001, 11, 28–42 CrossRef CAS PubMed.
- A. E. Grow, L. L. Wood, J. L. Claycomb and P. A. Thompson, J. Microbiol. Methods, 2003, 53, 221–233 CrossRef CAS PubMed.
- A. A. Guzelian, J. M. Sylvia, J. A. Janni, S. L. Clauson and K. M. Spencer, in Vibrational Spectroscopy-Based Sensor Systems, ed. S. D. Christesen and A. J. Sedlacek, Spie-Int Soc Optical Engineering, Bellingham, 2002, vol. 4577, pp. 182–192 Search PubMed.
- T. Luna-Pineda, K. Soto-Feliciano, E. De la Cruz-Montoya, L. C. P. Londono, C. Rios-Velazquez and S. P. Hernandez-Rivera, in Chemical and Biological Sensing VIII, ed. A. W. Fountain, Spie-Int Soc Optical Engineering, Bellingham, 2007, vol. 6554, p. K5540 Search PubMed.
- A. Walter, A. Marz, W. Schumacher, P. Rosch and J. Popp, Lab Chip, 2011, 11, 1013–1021 RSC.
- R. M. Jarvis, A. Brooker and R. Goodacre, Faraday Discuss., 2006, 132, 281–292 RSC.
- C. Fan, Z. Q. Hu, A. Mustapha and M. S. Lin, Appl. Microbiol. Biotechnol., 2011, 92, 1053–1061 CrossRef CAS PubMed.
- E. Podstawka, Y. Ozaki and L. M. Proniewicz, Appl. Spectrosc., 2005, 59, 1516–1526 CrossRef CAS PubMed.
- J. W. Chan, D. S. Taylor, S. M. Lane, T. Zwerdling, J. Tuscano and T. Huser, Anal. Chem., 2008, 80, 2180–2187 CrossRef CAS PubMed.
- M. Kahraman, A. I. Zamaleeva, R. F. Fakhrullin and M. Culha, Anal. Bioanal. Chem., 2009, 395, 2559–2567 CrossRef CAS PubMed.
- X. Yang, C. Gu, F. Qian, Y. Li and J. Z. Zhang, Anal. Chem., 2011, 83, 5888–5894 CrossRef CAS PubMed.
- M. C. Demirel, P. Kao, N. Malvadkar, H. Wang, X. Gong, M. Poss and D. L. Allara, Biointerphases, 2009, 4, 35–41 CrossRef CAS PubMed.
- J. Guicheteau and S. D. Christesen, in Chemical and Biological Sensing VII, ed. P. J. Gardner and A. W. Fountain, Spie-Int Soc Optical Engineering, Bellingham, 2006, vol. 6218, p. G2180 Search PubMed.
- L. Hagberg, P. Cinque, M. Gisslen, B. J. Brew, S. Spudich, A. Bestetti, R. W. Price and D. Fuchs, AIDS Res. Ther., 2010, 7, 12 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6ay01018k |
| This journal is © The Royal Society of Chemistry 2016 |