Open Access Article
Open Access ArticleTwo-channel image analysis method for the screening of OBOC libraries†
Dorothea
Helmer‡
a,
Kevin
Brahm‡
b,
Christian
Helmer
c,
Julia Susanne
Wack
b,
Gerald
Brenner-Weiss
d and
Katja
Schmitz
 *b
*b
aInstitute of Microstructure Technology (IMT), Karlsruhe Institute of Technology, Hermann-von-Helmholtz-Platz 1, 76344 Eggenstein-Leopoldshafen, Germany
bClemens-Schöpf Institute, Technische Universität Darmstadt (TU Darmstadt), Alarich-Weiss-Straße 8, 64287 Darmstadt, Germany. E-mail: schmitz@biochemie.tu-darmstadt.de
cInstitute for Program Structures and Data Organisation (IPD), Karlsruhe Institute of Technology, Am Fasanengarten 5, 76131 Karlsruhe, Germany
dInstitute of Functional Interfaces (IFG), Karlsruhe Institute of Technology, Hermann-von-Helmholtz-Platz 1, 76344 Eggenstein-Leopoldshafen, Germany
First published on 29th April 2016
Abstract
The split-synthesis approach offers a quick and easy method for producing a large diversity of substances for the discovery of novel protein ligands. Screening of the resulting one-bead-one-compound (OBOC) libraries with fluorescently labelled proteins is not trivial, however, since the resin beads can display significant autofluorescence. Here we present a simple two-channel microscopy image-based screening method for OBOC libraries on TentaGel MB-HMBA resin using tracers labelled with fluorophores. Bead images are taken at short exposure times in the RHO and FITC channels to keep photobleaching of the fluorophores at a minimum. Simple RGB colour vector addition ensures the identification of beads with high fluorescence intensities in the RHO channel. Pre-sorting of the library to exclude highly fluorescent beads is not necessary. The presented method is especially suited for small laboratories without automation equipment. By screening a library with a maximum of 117![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 649 peptoid hexamers we discovered 18 sequences that bind the human chemokine interleukin-8 (CXCL8) with affinities between 11 μM and 112 μM.
649 peptoid hexamers we discovered 18 sequences that bind the human chemokine interleukin-8 (CXCL8) with affinities between 11 μM and 112 μM.
Introduction
The screening of large compound libraries is a promising method for the discovery of novel protein ligands. Combinatorial chemistry provides the tools for the effective synthesis of large compound collections. A simple method for the generation of large combinatorial libraries is the mix-and-split method developed by Furka1 that relies on the statistical splitting and recombination of resin-bound substances. This method was further developed by Lam2 to give one-bead-one-compound (OBOC) libraries, where in the final bead-mixture ideally every resin bead carries a different compound.The most convenient method for OBOC library screening is the on-bead screen, where a target-molecule or a cell that is coupled to a marker is incubated with the library beads and binds to the beads that carry potential target-ligands. The beads carrying ligands of interest are then identified by the marker. In their work Lam et al. used alkaline phosphatase coupled to the target protein that yielded a blue stain of positive beads when reacted with BCIP (5-bromo-4-chloro-3-indolyl phosphate)3. In the following years, more screening methods were reported: radioligand binding assays on bead,4 screening with magnetic bead-labelling,5,6 and detection of whole-cells bound to beads.7,8 One of the most common methods is to employ a fluorescently labelled target that may be identified by fluorescence-activated cell sorting (FACS)9 or fluorescence microscopy10,11 or more recently in an automated fashion by a Complex Object Parametric Analyser and Sorter (COPAS bead sorter).11–13 The process of OBOC screening is followed by the separation of hits and sequence analysis. While in most methods, beads of interest have to be isolated by micropipette manipulators or manually, techniques like FACS and COPAS combine screening with bead isolation. The final step, identification of the compound on the isolated bead, can be achieved by sequencing (peptides, DNA/RNA) or matrix-assisted laser desorption ionization (MALDI) time-of-flight (TOF)/TOF mass spectrometry (peptides, peptidomimetics). For this purpose, a unique DNA or isotope label may be attached to each bead during combinatorial synthesis.
The combination of on-bead synthesis with consecutive on-bead screening demands that the solid support needs good swelling abilities in organic solvent during synthesis as well as provide good ligand accessibility in aqueous environment for screening. TentaGel resin, a hydrophilic tentacle-polymer based on polyethylene glycol (PEG),14 unites both properties and is therefore widely used for on-bead screening of OBOC libraries. However, two major issues complicate efficient fluorescence-based screening of TentaGel OBOC libraries: first, the autofluorescence of TentaGel beads leads to low signal-to-noise ratios and complicates the identification of hits, and second, unspecific interactions lead to false positive screening results. One of the simplest approaches to overcome problems due to bead autofluorescence is a pre-sorting routine to exclude the beads with the highest fluorescence from the screening process, which can be performed manually or with a COPAS bead sorter.11 In general, screening with a red fluorophore is favourable since TentaGel autofluorescence is highly significant in the FITC (fluorescein isothiocyanate) channel.15,16 Further improvement can be achieved by the use of quantum dots that offer high quantum yields and photostability.15,17 Moreover, internal quenchers can be incorporated into the beads to unambiguously identify fluorescence of labelled target molecules interacting with ligands at the bead surface in confocal scanning.18
The detection of false positives during the screening process usually requires a second screening step (re-screening) of the potential hits. One example is the dual colour screening method where positive beads are identified by a labelled primary antibody in the first step and re-screened with a primary antibody plus labelled secondary antibody in the second step.19 Non-specific interactions are kept at a minimum with the image subtraction method where beads are incubated with a protein mixture and then immobilised and incubated with the second mixture that contains the protein of interest.20 Comparison of the images of the beads before and after the second incubation step identifies the beads that exclusively bind to the target protein. As an advanced screening method, CONA (confocal nanoscanning and bead picking)21 identifies positive beads by the fluorescence “halo” on the outer rim in contrast to a dark interior of the bead. Likewise, two-channel COPAS screening22 enables the reliable identification and isolation of hits. Chen et al. showed that the decrease of ligand concentration on the beads led to a significantly reduced amount of false positives due to the reduction of nonspecific binding caused by avidity effects.23 While many of the traditional methods are labour-intensive and prone to observation bias, most advanced methods rely on expensive equipment that may not be available to smaller labs.
We report here a simple, microscopy-based two-channel screening method for TentaGel-bound peptoid OBOC libraries with DyLight 550-labelled interleukin 8 (CXCL8). Our method is applicable to other peptide mimetics and target proteins and it is well-suited for small laboratories. Screening is facilitated by a fluorescence microscope with an automated xy-table. We show that overlay images of FITC and rhodamine (RHO) channel images in the RGB colour space facilitate the identification of screening hits and eliminate the need for bead pre-sorting. Image-based screening ensures a minimal exposure time of fluorophores to light which minimizes photobleaching. With our method, we identified 77 positive beads from a library of 117![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 649 theoretical members, which is a number easily manageable in a re-screen. Validation yielded 18 peptoid hexamers that bind CXCL8 (interleukin-8) with affinities between Kd = 11 μM and 112 μM.
649 theoretical members, which is a number easily manageable in a re-screen. Validation yielded 18 peptoid hexamers that bind CXCL8 (interleukin-8) with affinities between Kd = 11 μM and 112 μM.
Results and discussion
TentaGel autofluorescence
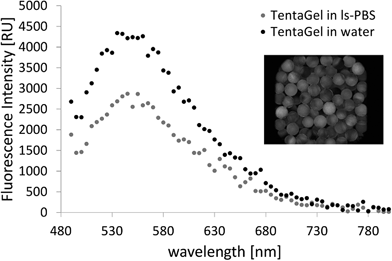
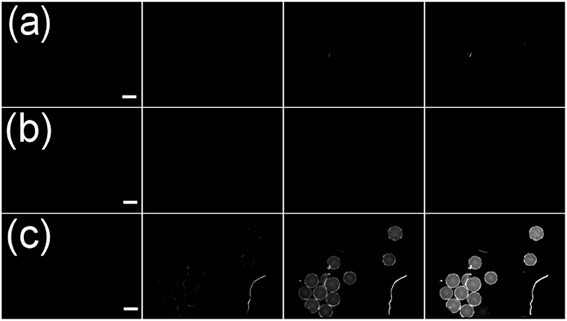
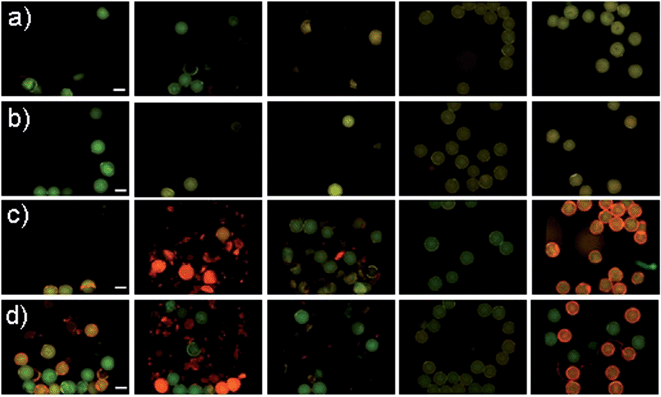
TentaGel MB HMBA resin (300 μm) was used throughout this work as it provides good solubility in organic solvents and aqueous media and low levels of non-specific binding. The untreated beads showed a broad emission spectrum with a maximum of 550 nm upon excitation at 480 nm at high amplification in the plate reader (see Fig. 1). The fluorescence intensity in water and PBS buffer was low and differed for individual beads.TentaGel MB (macrobeads) HMBA resin autofluorescence was not detectable in the FITC or RHO channel under a fluorescence microscope at exposure times below 100 ms, the resin autofluorescence was only observed in the DAPI channel (see Fig. 2c). However, during the synthesis of peptides and peptoids, the resin acquired considerable autofluorescence in the FITC channel (see Fig. 3). This concept of “acquired fluorescence” has previously been described by Hintersteiner et al.22 as opposed to the concept of “autofluorescence” of the TentaGel resin itself.15,16 The effect of enhanced fluorescence of functionalised beads makes the screening of TentaGel OBOC libraries with fluorophore-labelled target proteins difficult.
Overlay images in the RGB colour space
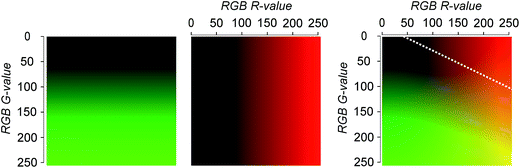
To overcome problems related to bead autofluorescence and screening, we investigated overlay microscopy images of beads in the FITC and RHO channel. A FITC (“green”) channel image is depicted by pixels with RGB values 0/x/0 (R/G/B), while the values y/0/0 (R/G/B) are employed for the RHO image (“red”) even though the real emission (here: 570–640 nm) rather corresponds to yellow to orange. The overlay image of the two channels then has pixels with the colour values y/x/0 in the RGB space as expected by vector addition (see Fig. 4). Colour perception in human trichromats is subjective and depends on the number of photoreceptor cells (cones) on the retina. Three types of cones are known with sensitivity maxima of 420 nm (S-cones), 534 nm (M-cones) and 564 nm (L-cones).24 Due to the overlap of M- and L-cones, the human eye is more sensitive to green-yellow colour than to red colour. This is why in the R/G RGB overlap space (Fig. 4 on the right) the area perceived as “red” is limited to the upper right corner of high R values and G-values below approximately 110 (indicated by the white dashed line). This way, pixels with high values in the green channel (>110) will not be perceived red in the overlay image. Therefore, the false-colour representation assigned to emission filter sets of different wavelengths increases the sensitivity by which positives can be identified. Pre-sorting to exclude beads with high green fluorescence is therefore not necessary when overlay images are analysed, since the green colour will always supersede the red colour unless a bead possesses very high R-values and G-values below 110.Evaluation as screening method
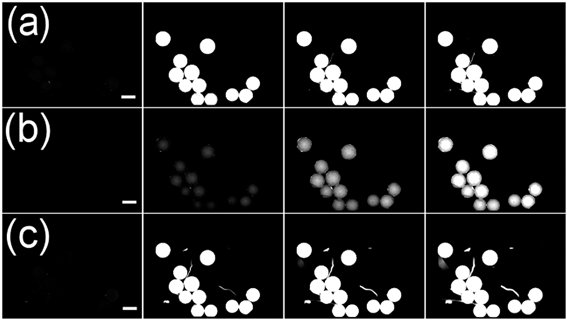
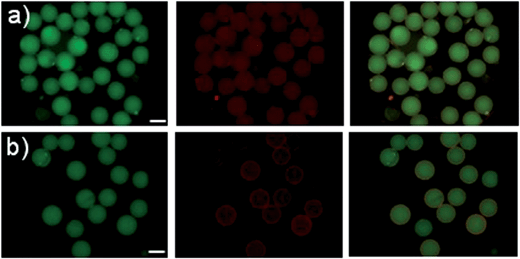
To test whether this effect would be useful for screening we selected three different peptides with reported affinity to streptavidin and known dissociation constants based on their different physicochemical properties to estimate the impact of peptide length, calculated pI and dissociation constant on performance in our assay (see Table 1).25–27These peptides were synthesized on TentaGel MB HMBA resin with 6-aminohexanoic acid as a linker. After attachment of the last amino acid and cleavage of the protecting groups, we incubated the resin-bound peptides with a streptavidin-DyLight 550 conjugate (yellow fluorophore). As negative control, the corresponding peptide-loaded beads as well as unfunctionalised TentaGel MB HMBA beads were incubated with ls-PBS and unlabelled streptavidin, respectively. As a positive control, beads functionalised with biotin were incubated with streptavidin-DL550. The two-colour overlay images of FITC and RHO channel were displayed and analysed by eye. As expected positives were easily spotted by their red colour in the false-colour image.
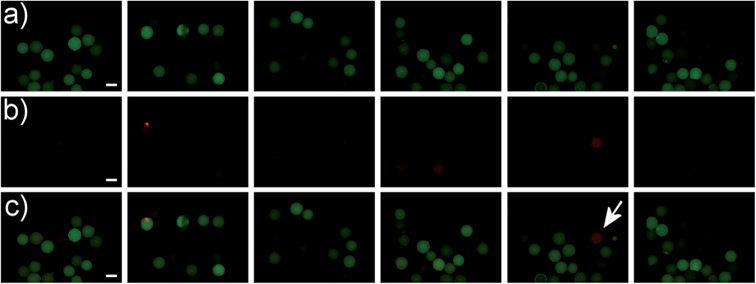
As shown in Fig. 5 positive hits with Kd values in the ten micromolar range could be easily detected at exposure times of 40 ms. The low-affinity peptide S3 showed no significant fluorescence increase in the RHO channel after incubation with streptavidin-DL550. We assume that an avidity effect in binding of the smaller peptide S2 yields an enhanced signal for this particular sequence in comparison to S1. In further experiments we could show that electrostatic attraction of the negatively charged streptavidin-dye conjugate and positively charged beads leads to no false positive results (see Fig. 6). This shows that this assay is in principle suitable for screening short peptide or peptoid sequences.
Library synthesis and screening
We incubated a peptoid library with a protein labelled by DyLight 550 and analysed the FITC/RHO channel overlay images without pre-sorting of the beads. Peptoids are an attractive class of peptide mimetics as protein ligands due to their structural similarity to peptides and comparably simple synthesis combined with their superior features like stability towards proteases and higher oral availability.28,29,30 The library with a maximum of 117![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 649 = 76 peptoid hexamers was synthesised from seven different amines by the submonomer method reported by Zuckermann et al.31 For peptoid nomenclature rules see Table 2. The amines used for submonomer synthesis were: isobutylamine (Nleu+0), isopropylamine (Nval+0), 2-phenylethylamine (Nphe+1), glycine (Nasp+0) protected as tert-butyl ester acetate (tBu-Ac-Nasp+0), ethanolamine (Nser+1) protected as triisopropylsilyl ether (TIPS-Nser+1), 1,4-diaminobutane (Nlys+0) with a Boc protecting group (Boc-Nlys+0) and tryptamine (Ntrp+1) with a Boc protecting group (Boc-Ntrp+1) (see ESI Fig. S1†). As a linker, 6-aminohexanoic acid was attached to TentaGel MB HMBA resin. After attachment of the last amine and cleavage of the protecting groups, the resin was thoroughly washed and dried.
649 = 76 peptoid hexamers was synthesised from seven different amines by the submonomer method reported by Zuckermann et al.31 For peptoid nomenclature rules see Table 2. The amines used for submonomer synthesis were: isobutylamine (Nleu+0), isopropylamine (Nval+0), 2-phenylethylamine (Nphe+1), glycine (Nasp+0) protected as tert-butyl ester acetate (tBu-Ac-Nasp+0), ethanolamine (Nser+1) protected as triisopropylsilyl ether (TIPS-Nser+1), 1,4-diaminobutane (Nlys+0) with a Boc protecting group (Boc-Nlys+0) and tryptamine (Ntrp+1) with a Boc protecting group (Boc-Ntrp+1) (see ESI Fig. S1†). As a linker, 6-aminohexanoic acid was attached to TentaGel MB HMBA resin. After attachment of the last amine and cleavage of the protecting groups, the resin was thoroughly washed and dried.
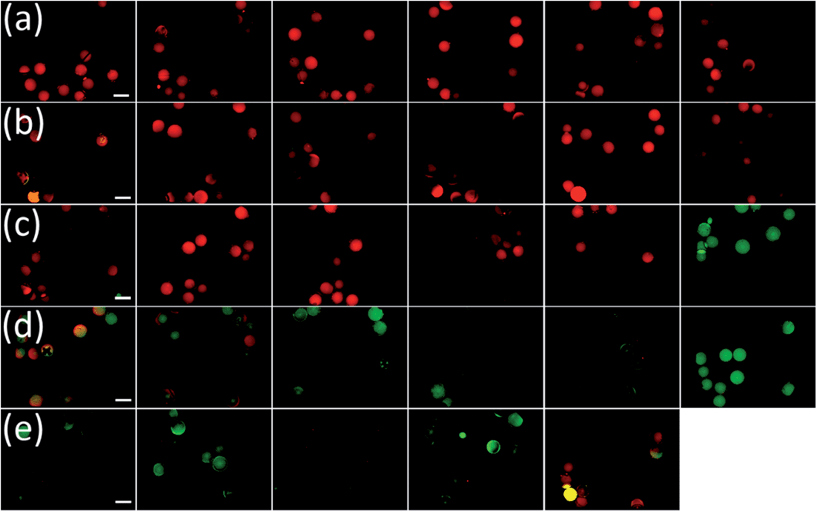
Resin beads of the seven individual synthesis vessels were washed with low-salt PBS (ls-PBS) buffer (40 mM Na2HPO4, 35 mM NaCl, pH 7.4), consecutively incubated with 10 μM CXCL8-DyLight 550 conjugate (CXCL8-DL550) and washed with ls-PBS buffer. When analysing the beads in a larger vessel like a Petri dish we found that the beads would roll when the sample table was moved, which complicated bead identification and made it impossible to re-identify beads from pictures taken of several areas of the dish. We therefore distributed the beads manually into 384-well transparent-bottom microtiter plates (MTP). Images of the beads were taken in the FITC and RHO channel at 40 ms exposure time. The short exposure times kept bleaching effects at a minimum, as opposed to real-time observation of the beads under the microscope. The two-coloured overlay images of FITC and RHO channel were displayed and analysed by eye. As expected from the consideration above positives were easily spotted by their red-colour in the false-colour image. The peptoid library included a large number of beads that displayed fluorescence in the green channel (see Fig. 7a). Removing these beads in a pre-screening procedure would have been laborious, but could be omitted due to the new method.
Potential hits were isolated and analysed by MALDI TOF MS/MS mass spectrometry and sequences were identified by using an Excel macro with a fragment-search option. 44 peptoid sequences were identified, one sequence was found twice. 14 of these sequences were highly hydrophobic (see ESI Table T1†). These peptoids were likely to have a low bioavailability and were not investigated any further. The 29 remaining sequences (see Table 2) were re-synthesised on TentaGel MB HMBA resin and re-screened by incubation with CXCL8-DL550. The re-screening (same procedure as the initial screening experiment) revealed 11 false positive sequences that failed to bind CXCL8-DL550 (see Fig. 8). Peptoid hits were synthesised on 2-chlorotritylchloride resin with an N-terminal 5(6)-carboxyfluorescein label without 6-aminohexanoic acid linker. Dissociation constants, Kd, were determined by fluorescence polarization in ls-PBS buffer (see ESI Fig. S2–S7† for anisotropy data). For the 18 peptoids that bound to CXCL8, dissociation constants between 12 μM and 112 μM were discovered (see Table 2). This is well in the usual range of screening positives in the first generation, especially for hexamers. A large number of approaches exists to mature such sequences towards high affinity-binders.32 Efforts to modify these lead sequences to increase affinity are ongoing.
| Peptoid | Sequence of the peptoid hits discovered in the library screena, C-terminus on the left | K d of the corresponding N-terminally fluorescein-labelled peptoid [μM] |
|---|---|---|
| a Peptoid nomenclature: Ahx: 6-aminohexanoic acid; N indicates the N-substitution of the backbone; the three letter code is analogous to the corresponding amino acids of the side chain. The number in superscript indicates that the main chain of the sidechain is equivalent (+0) to the peptide side chain or one carbon atom longer (+1). Cleavage from TentaGel HMBA-AM resin with ammonia vapour yielded amides. | ||
| 1 | H2N–Ahx–Nlys+0Nlys+0Nlys+0Nval+0Nlys+0Nleu+0–H | 11.6 ± 1.8 |
| 2 | H2N–Ahx–Nlys+0Nlys+0Nlys+0Nleu+0Nval+0Nasp+0–H | 20.0 ± 2.9 |
| 3 | H2N–Ahx–Nlys+0Nlys+0Nser+1Nlys+0Nlys+0Nleu+0–H | 42.9 ± 9.2 |
| 4 | H2N–Ahx–Nlys+0Nlys+0Nleu+0Nlys+0Nval+0Nleu+0–H | 65.3 ± 1.3 |
| 5 | H2N–Ahx–Nlys+0Nlys+0Nval+0Nlys+0Nphe+1Nleu+0–H | 41.4 ± 17.6 |
| 6 | H2N–Ahx–Nlys+0Nlys+0Nval+0Nleu+0Nleu+0Nleu+0–H | 57.2 ± 19.7 |
| 7 | H2N–Ahx–Nlys+0Nlys+0Nval+0Nlys+0Nphe+1Nasp+0–H | 19.0 ± 2.4 |
| 8 | H2N–Ahx–Nlys+0Nlys+0Nphe+1Nlys+0Nphe+1Nleu+0–H | 16.7 ± 4.8 |
| 9 | H2N–Ahx–Nlys+0Nleu+0Nlys+0Nlys+0Nphe+1Nasp+0–H | 25.0 ± 7.4 |
| 10 | H2N–Ahx–Nlys+0Nleu+0Nlys+0Nphe+1Nlys+0Nasp+0–H | 43.3 ± 5.1 |
| 11 | H2N–Ahx–Nleu+0Nlys+0Nlys+0Nleu+0Nlys+0Nleu+0–NH2 | 47.6 ± 12.8 |
| 12 | H2N–Ahx–Nleu+0Nlys+0Nlys+0Nval+0Nphe+1Nasp+0–NH2 | 112.3 ± 36.1 |
| 13 | H2N–Ahx–Nleu+0Nleu+0Nlys+0Nlys+0Nphe+1Nasp+0–NH2 | 71.4 ± 35.3 |
| 14 | H2N–Ahx–Nval+0Nlys+0Nlys+0Nphe+1Nlys+0Nleu+0–NH2 | 51.1 ± 15.4 |
| 15 | H2N–Ahx–Nphe+1Nlys+0Nlys+0Nleu+0Nlys+0Nleu+0–NH2 | 39.8 ± 13.9 |
| 16 | H2N–Ahx–Nphe+1Nlys+0Nlys+0Nval+0Nphe+1Nasp+0–NH2 | 35.4 ± 10.9 |
| 17 | H2N–Ahx–Nphe+1Nlys+0Nser+0Nlys+0Nval+0Nleu+0–NH2 | 21.4 ± 6.1 |
| 18 | H2N–Ahx–Nphe+1Ntrp+1Nphe+1Ntrp+1Nphe+1Nlys+0–NH2 | n.d. |
| 19 | H2N–Ahx–Ntrp+1Ntrp+1Nphe+1Nlys+0Nlys+0Nleu+0–NH2 | n.d. |
| 20 | H2N–Ahx–Ntrp+1Ntrp+1Nlys+0Nlys+0Ntrp+1Nlys+0–NH2 | n.d. |
| 21 | H2N–Ahx–Ntrp+1Ntrp+1Nser+1Nphe+1Ntrp+1Nlys+0–NH2 | n.d. |
| 22 | H2N–Ahx–Ntrp+1Nphe+1Nval+0Ntrp+1Nser+1Nlys+0–NH2 | n.d. |
| 23 | H2N–Ahx–Ntrp+1Nphe+1Ntrp+1Nphe+1Nser+1Nser+1–NH2 | n.d. |
| 24 | H2N–Ahx–Ntrp+1Nphe+1Ntrp+1Nval+0Nleu+0Nser+1–NH2 | n.d. |
| 25 | H2N–Ahx–Ntrp+1Nleu+0Ntrp+1Nval+0Nlys+0Nlys+0–NH2 | n.d. |
| 26 | H2N–Ahx–Ntrp+1Nleu+0Ntrp+1Nleu+0Nser+1Nser+1–NH2 | n.d. |
| 27 | H2N–Ahx–Ntrp+1Nleu+0Ntrp+1Nval+0Nval+0Nser+1–NH2 | n.d. |
| 28 | H2N–Ahx–Ntrp+1Nleu+0Ntrp+1Nphe+1Nser+1Nser+1–NH2 | n.d. |
| 29 | H2N–Ahx–Nphe+1Ntrp+1Nphe+1Ntrp+1Nphe+1Nlys+0–NH2 | 27.6 ± 6.4 |
Conclusion
It was shown, that with a simple screening method exploiting the false-colour visualization in two-channel fluorescence microscopy an OBOC peptoid library could be screened for new ligands for CXCL8. This method can be employed for the screening of other types of libraries and different fluorophores can be used as long as the fluorescence of the fluorophore chosen for protein labelling is distinct from the autofluorescence and acquired fluorescence of the beads. Overlay image analysis using false colour representation in green (background fluorescence) and red (labelled protein) requires no pre-sorting of beads and simplifies the identification of hits by only displaying beads with high intensities in the red channel in the overlay images due to RGB colour vector addition. Furthermore, low exposure times in the automated acquisition of the microscopy images leads to minimal photobleaching of the fluorophores. Starting from a combinatorial library of a maximum of 117![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 649 members we were able to identify 18 peptoid hexamers that bind CXCL8 with affinities between 12 μM and 112 μM. We believe this simple technique is a useful tool to efficiently screen OBOC libraries in a timely manner with minimal automation equipment and without any advanced labelling techniques.
649 members we were able to identify 18 peptoid hexamers that bind CXCL8 with affinities between 12 μM and 112 μM. We believe this simple technique is a useful tool to efficiently screen OBOC libraries in a timely manner with minimal automation equipment and without any advanced labelling techniques.
Experimental
Isobutylamine (Nleu+0), isopropylamine (Nval+0) and 2-phenylethyl amine (Nphe+1) were purchased at Sigma Aldrich, glycine tert-butyl ester acetate (tBu-Ac-Nasp+0) was purchased at Merck. Streptavidin DyLight 550 conjugate and streptavidin were purchased at VWR. Fmoc-amino acids and coupling reagents were purchased at Carbolution. Ethanolamine (Nser+1), 1,4-diaminobutane (Nlys+0) and tryptamine (Ntrp+1) were purchased at Sigma-Aldrich and equipped with the respective protecting groups as described below.2-[(Triisopropylsilyl)oxy]ethylamine (TIPS-Nser+1)33
6.30 mL (92.7 mmol, 4.00 eq.) ethanolamine were mixed with 25 mL of dichloromethane (DCM) in a round-bottom flask. 5.00 mL (23.5 mmol, 1.00 eq.) triisopropylsilyl chloride were added to the mixture under vigorous stirring. The solution was stirred overnight at rt. 50.0 mL DCM were added and the solution was washed three times with saturated sodium bicarbonate, three times with water and three times with saturated sodium chloride solution. The organic phase was dried over magnesium sulphate. Removal of the solvent in vacuo yielded 4.85 g of a colourless oil (95%). 1H-NMR (300 MHz, CDCl3), δ/ppm: 1.07–1.03 (m, 18H, CH3); 2.79 (t, 3J = 5.26 Hz, 2H, CH2); 3.63 (bs, 2H, NH2); 3.70 (t, 3J = 5.31 Hz, 2H, CH2).N-(4-Aminobutyl)-carbamic acid tert-butyl ester (Boc-Nlys+0)34
57.0 g (647 mmol, 1.00 eq.) 1,4-diaminobutane were dissolved in DCM and cooled on an ice bath. 14.1 g (64.7 mmol, 0.13 eq.) di-tert-butyl dicarbonate were dissolved in 200 mL DCM and added dropwise to the 1,4-diaminobutane under vigorous stirring. The white precipitate was filtered off and the solvent and residual 1,4-diaminobutane were removed in vacuo. The reaction yielded 9.9 g (81%) of a slightly yellow oil. 1H-NMR (300 MHz, CDCl3), δ/ppm: 1.41 (s, 9H, Boc); 1.43–1.51 (m, 4H, CH2); 2.69 (t, 3J = 6.70 Hz, 2H, CH2); 3.09–3.10 (m, 2H, CH2); 4.73 (bs, 1H, NH).N-(2-(1H-Indol-3-yl)ethyl)-2,2,2-trifluoroacetamide (TFA-Ntrp+1)35
5.40 g (34.0 mmol, 1.00 eq.) tryptamine were dissolved in 50.0 mL DCM. 25.0 mL (310 mmol, 9.11 eq.) pyridine were added to give a brown solution. 5.00 mL (34.0 mmol, 1.05 eq.) trifluoroacetic acid were added dropwise under vigorous stirring. The solution was stirred for 48 h at rt. 100 mL DCM were added and the organic phase was washed three times with saturated sodium bicarbonate solution, three times with a saturated ammonium chloride solution and three times with water. The organic phase was dried over magnesium sulphate. The solvent was removed in vacuo. Residual pyridine was removed by adding small amounts of toluene to the residue and removing the liquid in vacuo. The reaction yielded 6.80 g (78%) of a brown solid. 1H-NMR (300 MHz, CDCl3), δ/ppm: 2.94 (t, 3J = 6.70 Hz, 2H, CH2); 3.60 (dd, 3J = 6.50 Hz, 2H, CH2); 6.00 (bs, 1H, NH); 6.93 (d, 3J = 2.30 Hz, 1H, Ar–H); 7.04 (t, 3J = 7.60 Hz, 1H, Ar–H); 7.13 (t, 3J = 7.60 Hz, 1H, Ar–H); 7.28 (d, 3J = 8.10 Hz, 1H, Ar–H); 7.48 (d, 3J = 7.90 Hz, 1H, Ar–H); 8.03 (bs, 1H, NH).tert-Butyl-3-(2-(2,2,2-trifluoroacetamido)ethyl)-1H-indole-1-carboxylate (TFA-Boc-Ntrp+1)35
3.40 g (13.3 mmol, 1.00 eq.) of TFA-Ntrp+1 were dissolved in 25.0 mL 1,4-dioxane. 3.48 g (16 mmol, 1.2 eq.) di-tert-butyl dicarbonate and 0.08 g (0.67 mmol, 0.05 eq.) 4-dimethylaminopyridine were added to the solution. The reaction mixture was heated to 40 °C for 3 h. The solution was diluted with DCM and the organic phase was washed four times with water and once with saturated sodium chloride solution. The organic phase was dried over magnesium sulphate. The solvent was removed in vacuo and the residue was purified by chromatography on silica gel (5% (v/v) ethyl acetate in cyclohexane). Purification yielded 2.86 g (60%) of a yellow oil that freezes to a white solid upon storage at −8 °C. 1H-NMR (300 MHz, CDCl3), δ/ppm: 1.68 (s, 9H, Boc); 3.00 (t, 3J = 6.90 Hz, 2H, CH2); 3.70 (dd, 3J = 6.60 Hz, 2H, CH2); 7.27 (t, 3J = 7.60 Hz, 1H, Ar–H); 7.36 (t, 3J = 7.70 Hz, 1H, Ar–H); 7.45 (bs, 1H, Ar–H); 7.53 (d, 3J = 7.70 Hz, 1H, Ar–H); 8.16 (d, 3J = 7.70 Hz, 1H, Ar–H).1-Boc-tryptamine (Boc-Ntrp+1)35
1.70 g (4.78 mmol, 1.00 eq.) of TFA-Boc-Ntrp+1 were dissolved in 20.0 mL water![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol mixture (1
methanol mixture (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4). 1.73 g (12.5 mmol, 2.62 eq.) potassium carbonate were added and the reaction mixture was stirred for 4 d at rt. The reaction mixture was poured into 140 mL of water and the product was extracted with DCM. The organic phase was washed with saturated sodium chloride solution and dried over magnesium sulphate. The solvent was removed in vacuo. The reaction yielded 1.13 g (91%) of a colourless oil. 1H-NMR (300 MHz, CDCl3), δ/ppm: 1.67 (s, 9H, Boc); 2.84 (dt, 3J = 6.60 Hz, 4J = 0.90 Hz, 2H, CH2); 3.05 (t, 3J = 6.50 Hz, 2H, CH2); 7.14–7.27 (m, 1H, Ar–H); 7.29–7.35 (m, 1H, Ar–H); 7.42 (bs, 1H, Ar–H); 7.54 (dq, 3J = 7.70 Hz, 4J = 0.70 Hz, 1H, Ar–H); 8.14 (d, 3J = 8.11 Hz, 1H, Ar–H).
4). 1.73 g (12.5 mmol, 2.62 eq.) potassium carbonate were added and the reaction mixture was stirred for 4 d at rt. The reaction mixture was poured into 140 mL of water and the product was extracted with DCM. The organic phase was washed with saturated sodium chloride solution and dried over magnesium sulphate. The solvent was removed in vacuo. The reaction yielded 1.13 g (91%) of a colourless oil. 1H-NMR (300 MHz, CDCl3), δ/ppm: 1.67 (s, 9H, Boc); 2.84 (dt, 3J = 6.60 Hz, 4J = 0.90 Hz, 2H, CH2); 3.05 (t, 3J = 6.50 Hz, 2H, CH2); 7.14–7.27 (m, 1H, Ar–H); 7.29–7.35 (m, 1H, Ar–H); 7.42 (bs, 1H, Ar–H); 7.54 (dq, 3J = 7.70 Hz, 4J = 0.70 Hz, 1H, Ar–H); 8.14 (d, 3J = 8.11 Hz, 1H, Ar–H).
Synthesis of peptoid library (with 6-aminohexanoic acid linker)
2.40 g TentaGel MB HMBA resin (0.576 mmol, 1.00 eq.) was swollen in DCM (1 mL per 50.0 mg resin) and drained. 610 mg (1.728 mmol, 3.00 eq.) Fmoc-6-aminohexanoic acid (Fmoc-Ahx) were dissolved in DCM and cooled to 0 °C in an ice bath. 469 μL of a 15 mg mL−1 solution of 4-dimethylaminopyridine in DMF (0.058 mmol, 0.1 eq.) and 357 μL N,N′-diisopropylcarbodiimide (2.30 mmol, 4.00 eq.) were added and the mixture was stirred for 5 min. The mixture was then added to the resin. The suspension was shaken overnight at rt at 600 rpm and the resin was washed five times with DMF. The Fmoc protecting group was removed by treatment with 20% piperidine in DMF for 20 min. The resin was evenly distributed into 7 frits. Peptoids were synthesised by the submonomer method:31 bromoacetic acid was dissolved in DMF to give a 1.20 M solution. 10.0 eq. bromoacetic acid were added to the resin and consecutively 9.00 eq. N,N′-diisopropylcarbodiimide were added and the suspension was shaken for 1 h at 600 rpm. The resin was washed five times with DMF. 10.0 eq. of the respective amine were added in DMF and the mixture was shaken for 1.5 h at rt. The resin was washed and the content of the 7 frits was pooled and re-distributed into 7 frits for the next synthesis cycle. After 6 cycles protecting groups were removed by 4 h treatment of the resin with 95% trifluoroacetic acid, 2.5% water and 2.5% triisopropyl silane. The resin was washed with DCM, DMF and low-salt PBS (40 mM Na2HPO4, 35 mM NaCl in deionised water, pH 7.41) until the supernatant was not acidic anymore. The procedure yielded seven synthesis frits with deprotected hexameric peptoid sequences.Library screening
Each of the seven filter frits from the library synthesis (see above) was incubated with 1 mL of 10 μM CXCL8-DL550 (CXCL8 labelled with DyLight 550 fluorescent dye) for 5 h at rt. The supernatant was removed and the resin was washed three times with 2 mL low-salt-PBS per frit and consecutively suspended in 2 mL ls-PBS. The wells of a 384-well black, transparent-bottom MTP (Corning #3675) were pre-filled with 20 μL ls-PBS and droplets of the bead suspension were placed into the upper corners of the MTP cavities. The MTP was centrifuged and images were taken in the FITC and RHO channel at 40 ms exposure time. One synthesis frit was distributed into at least two 384-well plates; imaging took about 7 min per plate. Images were analysed by eye and potential hits were pipetted out of the respective MTP well. These beads were distributed into a new MTP pre-filled with 20 μL ls-PBS and imaged. Single beads that appeared red in the two-colour overlay were placed into microcentrifuge tubes and dried.MALDI analysis
Isolated beads were placed in a desiccator with 25% aqueous ammonia solution. Vacuum was applied to produce an ammonia atmosphere. Beads were left in the ammonia vapour for 4 h to cleave the peptoids from the TentaGel MB-HMBA beads. 5 μL of a water![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) acetonitrile (1
acetonitrile (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, (v/v)) mixture with 0.1% trifluoroacetic acid (TFA) were added to the individual beads. A matrix-solution of 10 mg mL−1 α-cyano-4-hydroxy cinnamic acid (CHCA) in water
1, (v/v)) mixture with 0.1% trifluoroacetic acid (TFA) were added to the individual beads. A matrix-solution of 10 mg mL−1 α-cyano-4-hydroxy cinnamic acid (CHCA) in water![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) acetonitrile (1
acetonitrile (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) with 0.1% TFA was prepared. 1 μL matrix-solution was mixed with 1 μL bead-supernatant and two droplets of 1 μL each were spotted onto a 384-spot MALDI plate. MALDI-TOF-MS and TOF/TOF-MS/MS was performed using a 4800plus instrument from AB SCIEX in reflector mode (see ESI† for peptoid MS data).
1) with 0.1% TFA was prepared. 1 μL matrix-solution was mixed with 1 μL bead-supernatant and two droplets of 1 μL each were spotted onto a 384-spot MALDI plate. MALDI-TOF-MS and TOF/TOF-MS/MS was performed using a 4800plus instrument from AB SCIEX in reflector mode (see ESI† for peptoid MS data).
Synthesis of peptides
600 mg TentaGel MB HMBA resin (0.144 mmol, 1.00 eq.) was swollen in DCM (1 mL per 50.0 mg resin) for 30 min and drained. 153 mg (0.432 mmol, 3.00 eq.) Fmoc-6-aminohexanoic acid were dissolved in DCM and cooled to 0 °C in an ice bath. 118 μL of a 15 mg mL−1 solution of 4-dimethylaminopyridine in DMF (0.015 mmol, 0.1 eq.) and 89 μL of N,N′-diisopropylcarbodiimide (0.575 mmol, 4.00 eq.) were added and the mixture was stirred for 5 min. The mixture was then added to the resin. The suspension was shaken overnight at rt at 600 rpm and the resin was washed five times with DMF. The Fmoc protecting group was removed by treatment with 20% piperidine in DMF for 20 min. The resin was evenly distributed into three frits. Amino acids were coupled in a double-coupling procedure. 3 eq. Fmoc-protected amino acid, 3 eq. HBTU and 6 eq. DIPEA were dissolved in 1 mL DMF and shaken for 5 min at rt and 600 rpm. The mixture was then added to the resin and stirred for 40 min. After washing five times with DMF and DCM the Fmoc protecting group was removed by treatment with 20% piperidine in DMF for 20 min. After deprotection of the final N-terminal amino acid protecting groups were removed by 2 h treatment of the resin with 88% trifluoroacetic acid, 5% water, 5% 1,4-dithiothreitol and 2% triisopropyl silane. The resin was washed with DCM, DMF and low-salt PBS until the supernatant was not acidic anymore, and finally dried. The procedure yielded three deprotected peptide sequences attached to TentaGel resin. For mass analysis approximately 2 mg of deprotected beads were placed separately in a desiccator with 25% aqueous ammonia solution. Vacuum was applied to produce an ammonia atmosphere. Beads were left in the ammonia vapour overnight to cleave the peptides from the TentaGel MB HMBA resin. 50 μL of a 50% acetonitrile solution in water were added and the cleaved peptides were analysed with ESI-MS.Test of model peptides for streptavidin binding
A few resin beads of each of the three synthesis vessels were washed with low-salt PBS, incubated with 2 μM streptavidin-DyLight 550 conjugate (Strep-DL550) and washed with ls-PBS buffer. As negative control some of the synthesis resin as well as unfunctionalised TentaGel MB HMBA beads were separately incubated in ls-PBS and streptavidin. As a positive control beads functionalised with biotin were used. We distributed the beads manually into 384-well transparent-bottom MTPs. The most efficient way of manual distribution was to pre-fill each cavity of a 384-well MTP with 20 μL of ls-PBS and to place small droplets of the bead suspension into the upper corners of the MTP wells with a 200 μL pipette tip with a cut-off end. The MTP was centrifuged to evenly distribute the beads at the bottom of each well and images of the beads were taken in the FITC channel at 40 ms exposure time and RHO channel at exposure times from 1 to 180 ms. The two-colour overlay images of FITC and RHO channel were displayed and analysed by eye.Synthesis of fluorescently labelled peptoids (without 6-aminohexanoic acid linker)
310 mg 2-chlorotritylchloride resin (0.465 mmol, 1.00 eq.) was swollen in DCM and drained. 1136 mg (4.65 mmol, 3.00 eq.) bromoacetic acid (1.2 M solution in N-methyl-2-pyrrolidone) were mixed with 635 μL (4.65 mmol, 3.00 eq.) diisopropylethylamine and the mixture was added to the resin. The mixture was shaken at 600 rpm overnight. The resin was split into aliquots of 10 mg and reacted with 10 equivalents of the first submonomer amine. Bromoacetic acid was attached by adding 10 eq. of bromoacetic acid and 9 eq. of N,N′-diisopropylcarbodiimide under shaking for 1 h at room temperature. Further amines were attached by adding 10 eq. of amine to the bromoacetylated sequence and shaking for 1.5 h at room temperature. The fluorescence label was attached by adding 3 eq. of 5(6)-carboxyfluorescein (dissolved in a minimum amount of DMF), 3 eq. of 1-hydroxybenzotriazole (300 mg mL−1 in DMF) and 3 eq. of N,N′-diisopropylcarbodiimide to the resin. The mixture was shaken for 4 h at rt. The resin was washed five times with DMF, five times with 20% piperidine in DMF and five times with DCM and dried. The resin was treated with 95% trifluoroacetic acid, 2.5% water and 2.5% triisopropyl silane for 4 h at rt to remove protecting groups and to cleave the fluorescently labelled peptoids. Peptoids were purified by HPLC (C18 RP column). Mass spectra were recorded as described above (see ESI† for peptoid MS data).Protein expression
E. coli BL21 DE3 RIL were transformed with a pET-22b vector containing the sequence of a 72 amino acid form of CXCL8 with a C-terminal cysteine residue (CXCL8S72C). The proteins were expressed and purified by a procedure modified from Wiese and Schmitz.36 Briefly, the transformed cells were grown in LB-medium containing ampicillin (60 μg mL−1). At an OD600 of 0.6–0.8 expression was induced by addition of IPTG to a final concentration of 0.1 mM. The expression was continued for 3 h at 30 °C and cells were harvested by centrifugation (45 min at 5000 × g) and resuspended in lysis buffer (40 mM Na2HPO4, 90 mM NaCl, pH 7.4) supplemented with 1 mM EDTA, 0.2 mg mL−1 lysozyme, 0.1 mg mL−1 DNase I and incubated on ice for 1.5 h. After addition of Triton X-100, the suspension was sonified three times for 30 s each at 50% intensity (Sonopuls, Bandelin Electronics). After incubation with DNase I the sample was heated to 70 °C for 10 min to precipitate host cell proteins. The lysate was centrifuged at 4 °C and 4500 × g for 45 min. The protein was purified from the supernatant by HPLC with a 5 mL HiTrap SP FF column (GE Healthcare).Protein labelling
150 μL (1.20 μmol, 4.80 eq.) tris(2-carboxyethyl)-phosphine reducing gel (TCEP gel, Thermo Fisher Scientific) was washed three times with 400 μL of PBS with 20.0 mM EDTA (pH 7.00) and once with 400 μL low-salt PBS. 2.10 mg (0.250 μmol, 1.00 eq.) CXCL8S72C dissolved in 500 μL low-salt PBS was added to the TCEP gel and incubated for 30 min at rt. The supernatant was transferred into another microcentrifuge tube and the TCEP gel was washed twice with 200 μL low-salt PBS. The washing solutions were combined with the supernatant. 0.50 mg (0.469 μmol, 1.88 eq.) of DyLight 550 maleimide (Thermo Fisher Scientific, 10 mg mL−1 in DMF, 1065 g mol−1) were added to the supernatant and incubated for 2.5 h at rt. The product was isolated by HPLC (C8-RP column). The procedure yielded 1.00 mg (0.106 μmol) CXCL8S72C-DL550 (42%). MALDI-TOF (matrix: CHCA, linear mode), m/z (%): 9396.98 (100) [M + H]+, 4688.07 (25) [M + H]2+, 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 786.84 (6) [2M + H]+.
786.84 (6) [2M + H]+.
Microscopy
All images were taken on a Zeiss Axio Observer. Z1 microscope equipped with a Hal 100 halogen lamp for brightfield images and a HXP 120 C lamp for fluorescence imaging. The microscope was equipped with an automated x/y-table and was operated by the AxioVision (Rel. 4.8) software. Images were taken with the AxioCam MR 12 bit black and white camera. The following filter sets were used: filter set 49 (DAPI, excitation 300–400 nm, emission 420–470 nm), filter set 38HE (FITC, excitation 450–490 nm, emission 500–550 nm), filter set 43HE (RHO, excitation 538–562 nm, emission 570–640 nm).Fluorescence anisotropy
Measurements were conducted in 384-well black, low-volume, transparent-bottom MTPs (Corning #3540) on a Tecan Infinite M1000 plate reader according to the method of Moerke.37 16 consecutive 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 dilutions of CXCL8 in ls-PBS were prepared to give a final volume of 31.5 μL each. 3.5 μL of fluorescently labelled peptoid (20 nM or 50 nM solution in ls-PBS) was added to the dilutions. Additionally, a free ligand control of 31.5 μL ls-PBS with 3.5 μL of fluorescently labelled peptoid was prepared. As a blank, ls-PBS was measured. Each mixture was measured in triplicate (10 μL for each measurement). Excitation: 470 nm, emission: 520 nm. Non-linear regression of the anisotropy data was performed in SigmaPlot using the one-site saturation model A = Amin + Amax × c(CXCL8)/(Kd + c(CXCL8)).
2 dilutions of CXCL8 in ls-PBS were prepared to give a final volume of 31.5 μL each. 3.5 μL of fluorescently labelled peptoid (20 nM or 50 nM solution in ls-PBS) was added to the dilutions. Additionally, a free ligand control of 31.5 μL ls-PBS with 3.5 μL of fluorescently labelled peptoid was prepared. As a blank, ls-PBS was measured. Each mixture was measured in triplicate (10 μL for each measurement). Excitation: 470 nm, emission: 520 nm. Non-linear regression of the anisotropy data was performed in SigmaPlot using the one-site saturation model A = Amin + Amax × c(CXCL8)/(Kd + c(CXCL8)).
Acknowledgements
The authors wish to thank Ina Rink and Anke Imrich for providing purified CXCL8 protein, Stefanie Eckes for helping with microscopy imaging and Bernhard Valldorf for conducting ESI-MS measurements. We thank the Fonds der Chemischen Industrie for financial support. This work received financial support by the ‘Concept for the Future’ of the Karlsruhe Institute of Technology (KIT) within the framework of the German Excellence Initiative (Research Group 26-2).References
- A. Furka, F. Sebestyen, M. Asgedom and G. Dibo, Int. J. Pept. Protein Res., 1991, 37, 487–493 CrossRef CAS PubMed.
- K. S. Lam, S. E. Salmon, E. M. Hersh, V. J. Hruby, W. M. Kazmierski and R. J. Knapp, Nature, 1991, 354, 82–84 CrossRef CAS PubMed.
- S. Miyamoto, R. W. Liu, S. S. Hung, X. B. Wang and K. S. Lam, Anal. Biochem., 2008, 374, 112–120 CrossRef CAS PubMed.
- J. Z. Wu, Q. Y. N. Ma and K. S. Lam, Biochemistry, 1994, 33, 14825–14833 CrossRef CAS PubMed.
- G. Fassina, M. R. Bellitti and G. Cassani, Protein Pept. Lett., 1994, 1, 15–18 CAS.
- J. M. Astle, L. S. Simpson, Y. Huang, M. M. Reddy, R. Wilson, S. Connell, J. Wilson and T. Kodadek, Chem. Biol., 2010, 17, 38–45 CrossRef CAS PubMed.
- K. S. Lam, Q. Lou, Z. G. Zhao, J. Smith, M. L. Chen, E. Pleshko and S. E. Salmon, Biomed. Pept., Proteins Nucleic Acids, 1995, 1, 205–210 CAS.
- L. Peng, R. W. Liu, J. Marik, X. B. Wang, Y. Takada and K. S. Lam, Nat. Chem. Biol., 2006, 2, 381–389 CrossRef CAS PubMed.
- M. C. Needels, D. G. Jones, E. H. Tate, G. L. Heinkel, L. M. Kochersperger, W. J. Dower, R. W. Barrett and M. A. Gallop, Proc. Natl. Acad. Sci. U. S. A., 1993, 90, 10700–10704 CrossRef CAS.
- J. K. Chen, W. S. Lane, A. W. Brauer, A. Tanaka and S. L. Schreiber, J. Am. Chem. Soc., 1993, 115, 12591–12592 CrossRef CAS.
- T. Kodadek and K. Bachhawat-Sikder, Mol. BioSyst., 2006, 2, 25–35 RSC.
- M. M. Marani, M. C. M. Ceron, S. L. Giudicessi, E. de Oliveira, S. Cote, R. Erra-Balsells, F. Albericio, O. Cascone and S. A. Camperi, J. Comb. Chem., 2009, 11, 146–150 CrossRef CAS PubMed.
- C. F. Cho, B. B. Azad, L. G. Luyt and J. D. Lewis, ACS Comb. Sci., 2013, 15, 393–400 CrossRef CAS PubMed.
- E. Bayer, M. Dengler and B. Hemmasi, Int. J. Pept. Protein Res., 1985, 25, 178–186 CrossRef CAS.
- H. I. Olivos, K. Bachhawat-Sikder and T. Kodadek, ChemBioChem, 2003, 4, 1242–1245 CrossRef CAS PubMed.
- P. G. Alluri, M. M. Reddy, K. Bachhawat-Sikder, H. J. Olivos and T. Kodadek, J. Am. Chem. Soc., 2003, 125, 13995–14004 CrossRef CAS PubMed.
- D. G. Udugamasooriya, S. P. Dineen, R. A. Brekken and T. Kodadek, J. Am. Chem. Soc., 2008, 130, 5744–5752 CrossRef CAS PubMed.
- J. Townsend, A. Do, A. Lehman, S. Dixon, B. Sanii and K. S. Lam, Comb. Chem. High Throughput Screening, 2010, 13, 422–429 CrossRef CAS PubMed.
- K. S. Lam, S. Wade, F. Abdullatif and M. Lebl, J. Immunol. Methods, 1995, 180, 219–223 CrossRef CAS PubMed.
- A. Lehman, S. Gholami, M. Hahn and K. S. Lam, J. Comb. Chem., 2006, 8, 562–570 CrossRef CAS PubMed.
- M. Hintersteiner, C. Buehler, V. Uhl, M. Schmied, J. Muller, K. Kottig and M. Auer, J. Comb. Chem., 2009, 11, 886–894 CrossRef CAS PubMed.
- B. Hintersteiner and M. Auer, Methods Appl. Fluoresc., 2013, 1, 1–7 Search PubMed.
- X. W. Chen, P. H. Tan, Y. Y. Zhang and D. H. Pei, J. Comb. Chem., 2009, 11, 604–611 CrossRef CAS PubMed.
- J. K. Bowmaker and H. J. A. Dartnall, J. Physiol., 1980, 298, 501–511 CrossRef CAS.
- T. Lamla and V. A. Erdmann, J. Mol. Biol., 2003, 329(2), 381–388 CrossRef CAS PubMed.
- T. G. Schmidt, J. Koepke, R. Frank and A. Skerra, J. Mol. Biol., 1996, 255(5), 753–766 CrossRef CAS PubMed.
- P. C. Weber, M. W. Pantoliano and L. T. Thompson, Biochemistry, 1992, 31(39), 9350–9354 CrossRef CAS PubMed.
- S. M. Miller, R. J. Simon, S. Ng, R. N. Zuckermann, J. M. Kerr and W. H. Moos, Drug Dev. Res., 1995, 35, 20–32 CrossRef CAS.
- E. W. Taylor, J. A. Gibbons and R. A. Braeckman, Pharm. Res., 1997, 14, 572–577 CrossRef.
- R. J. Simon, E. J. Martin, S. M. Miller, R. N. Zuckermann, J. M. Blaney and W. H. Moos, Tech. Protein Chem. V, [Pap. Symp. Protein Soc.], 7th, 1994, 533–539 CAS.
- R. N. Zuckermann, J. M. Kerr, S. B. H. Kent and W. H. Moos, J. Am. Chem. Soc., 1992, 114, 10646–10647 CrossRef CAS.
- K. V. Reddy, R. D. Yedery and C. Aranha, Int. J. Antimicrob. Agents, 2004, 24(6), 536–547 CrossRef PubMed.
- R. N. Zuckermann, Abstr. Pap. Am. Chem. Soc., 1994, 208, 376–377 Search PubMed.
- I. Peretto, R. M. Sanchez-Martin, X. H. Wang, J. Ellard, S. Mittoo and M. Bradley, Chem. Commun., 2003, 2312–2313 RSC.
- J. D. White, Y. Li and D. C. Ihle, J. Org. Chem., 2010, 75, 3569–3577 CrossRef CAS PubMed.
- D. Wiese and K. Schmitz, J. Immunol. Methods, 2011, 364, 77–82 CrossRef CAS PubMed.
- N. Moerke, Curr. Protoc. Chem. Biol., 2009, 1, 1–15 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5ay02981c |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2016 |