Pseudo isobaric peptide termini labelling for relative proteome quantification by SWATH MS acquisition†
Abstract
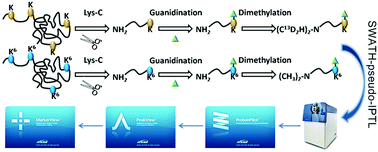
The pseudo isobaric peptide termini labeling (IPTL) method is a remarkable strategy in quantitative proteomics, and has been efficiently applied in biological studies due to its high quantitative accuracy. However, irreproducible precursor ion selection caused by data dependent acquisition and the chromatographic shift caused by isotope effects limit the wide application of this method. Herein, we expand the use of pseudo IPTL to SWATH MS application and develop a novel quantitative strategy, termed SWATH-pseudo-IPTL, by which the relative quantification could be achieved by comparing the “complete” extracted ion chromatogram (XIC) intensity of MS/MS scan instead of a single intensity measurement in DDA-pseudo-IPTL which only reflected the peptide abundances at that given time. The quantitative analysis of various proportions of mixed HeLa samples revealed the strong accuracy and precision of our SWATH-pseudo-IPTL method, both of which were better than that of the DDA-pseudo-IPTL strategy. SWATH-pseudo-IPTL was also applied to the quantitative profiling of the proteome from human hepatocellular carcinoma cell lines with high and low metastatic potential, and most of the differentially expressed proteins were related to tumorigenesis and tumor metastasis, demonstrating the feasibility of this methodology for biological applications.

 Please wait while we load your content...
Please wait while we load your content...