Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
High spatial and temporal resolution cell manipulation techniques in microchannels
Pedro
Novo
*,
Margherita
Dell'Aica
,
Dirk
Janasek
and
René P.
Zahedi
Protein Dynamics Group, Leibniz-Institut für Analytische Wissenschaften – ISAS - e.V., Otto-Hahn-Str. 6b, 44227 Dortmund, Germany. E-mail: pedro.novo@isas.de; Web: http://www.isas.de
First published on 11th February 2016
Abstract
The advent of microfluidics has enabled thorough control of cell manipulation experiments in so called lab on chips. Lab on chips foster the integration of actuation and detection systems, and require minute sample and reagent amounts. Typically employed microfluidic structures have similar dimensions as cells, enabling precise spatial and temporal control of individual cells and their local environments. Several strategies for high spatio-temporal control of cells in microfluidics have been reported in recent years, namely methods relying on careful design of the microfluidic structures (e.g. pinched flow), by integration of actuators (e.g. electrodes or magnets for dielectro-, acousto- and magneto-phoresis), or integrations thereof. This review presents the recent developments of cell experiments in microfluidics divided into two parts: an introduction to spatial control of cells in microchannels followed by special emphasis in the high temporal control of cell-stimulus reaction and quenching. In the end, the present state of the art is discussed in line with future perspectives and challenges for translating these devices into routine applications.
1. Introduction
Purification, concentration and counting of specific cell types from complex mixtures (e.g. blood, cell culture) is often required in clinical diagnostics but also fundamental research. Well established methods include fluorescence or magnetic activated cell sorting (FACS and MACS). These techniques are readily available and widely used in laboratories as they allow effective counting and separation of cell populations. Besides routine applications, these techniques have been demonstrated to (i) detect rare cells,1–3 such as circulating tumor cells (CTC), and achieve high (ii) isolation/purification rate as well as (iii) throughput. However, FACS and MACS both lack of more sophisticated cell manipulation capabilities, e.g. to precisely control and adjust trajectories as required in elaborated cell stimulation experiments. To date, cell manipulation has been majorly performed using conventional techniques and labware such as pipettes, sample tubes, microwell plates and cell culture flasks. Notably, these strategies have only limited cell manipulation capabilities, are rather time and reagent consuming, lack sufficient precision and reproducibility and have only limited or no online monitoring capacities (e.g., integrated sensors for pH, fluorescence or magnetic field measurement).4 These limitations have motivated the development of microfluidic methods for enhanced spatial and temporal control of the neighboring cellular environment, as represented by numerous publications (>3.500 in the last two decades, according to a search report in ISI Web of Knowledge using the “cell microfluidics” keywords, October 2015).5–7Although different examples of microfluidic systems exist in nature (e.g., channel networks in paper), in literature the terms microfluidics or microchannels are generally employed for microfabricated fluidic networks that have characteristic dimensions below 100 μm. The possibility to tailor microfluidic networks enables the custom design of channels aiming at different manipulation methods. These may include a multitude of flow operations integrated in series or in parallel. As a consequence of the reduction of the characteristic dimensions in microfluidic channels, liquids flow in the laminar regime (low Reynolds numbers), allowing precise spatial and temporal control of flow patterns. Moreover, the conditions for laminar flow can be kept at high flow rate values (e.g., in the order of m s−1), opening possibilities, e.g., for high-throughput cell manipulation8,9 and fast local environment switching times.10 Channel size can be fabricated to dimensions in the order of magnitude of cells, thus allowing precise control at the single cell level, but also requiring low volumes which is highly beneficial in many biological and clinical research scenarios which deal with limited amounts (e.g., cerebrospinal fluid) or numbers (e.g., research in rare disease) of samples. Control of the flow of cells by rational structure design, such as in pinched-flow via stream thinning elements (STE)s, or deterministic lateral displacement (DLD), can be further complemented by integrated actuation methods such as those employing dielectro-, magneto- and acoustophoresis, as well as pressured valving.11 Therefore, great attention has been paid to the development of microfluidic devices for cell manipulation, reflected by a vast number of publications that may be found in literature. Importantly, the overall reduction of chip size holds a great potential for portability as well as integration of sensors and actuators, which is an important factor in current lab-on-a-chip and point-of-care applications.
Integrating sensors or detection and quantification systems in lab on chips fall outside the scope of this review and therefore will not be discussed here. Suggested literature includes.12–16
Owing to the large number of examples and broad spectrum of applications, this review will not cover the general state-of-the-art, but will rather focus on selected recent developments. We will first introduce microfluidic strategies used for fast cell manipulation, such as those for separation and concentration of CTCs. Then we will present the latest developments of microfluidic methods for fast cell stimulation and quenching reactions. Further examples such as for inertial,17 droplet,18,19 pneumatic,20,21 filter-based,22,23 electroosmotic,24 immunoaffinity25 and digital26 microfluidics, cell traps27 and optical tweezers,28 are not covered here, but we recommend corresponding in-depth literature.29–35
2. Biological relevance of cell manipulation studies
A cell is an extremely complex system comprising many different classes of biomolecules. Among those particularly proteins, lipids, and metabolites are highly dynamic and therefore key to the adaptability of cells to immediately respond to external stimuli. The human body comprises more than 100 different cell types and tissues, each evolutionary and individually tailored to fulfill specific tasks, individually or in concert with other cells and tissues. Indeed, this complexity is not limited to different cell types, but is also reflected in the heterogeneity among populations of the same cell type.Even minor dysregulation of specific cells or cell–cell interactions can have dramatic effects on the homeostasis of a tissue or even the entire organism. Consequently, the detailed and quantitative study of cellular processes and their dynamics to understand causes and consequences of (dys)regulation is imperative to combat diseases such as cancer, cardiovascular and neurodegenerative disorders. It is furthermore one of the major goals of system biology research. To elucidate these processes and to reveal their underlying principles, one needs to investigate adaptations of cells on the level of proteins, lipids and metabolites under a large variety of conditions. Indeed, microfluidic applications hold a great potential to boost systems biology research, as they offer incomparable possibilities to manipulate and perturb cells, starting from minute sample amounts, in an automated, reproducible, fast and efficient way. Therefore, in the future microfluidics might be a cornerstone of sample preparation for disciplines such as (clinical) proteomics which aim at quantifying a large number of biomolecules from complex samples, i.e. body fluids, with high reproducibility, precision and accuracy. Moreover, microfluidics offers a higher throughput, as well as novel opportunities for temporal and spatial analysis of samples, which are otherwise not achievable.
In principle, manipulation studies have four major goals: (i) separation of cells and (ii) single cell analysis, (iii) subcellular analysis, as well as (iv) temporal analysis of cellular processes and signaling.
2.1. Separation of cells
Particularly for blood samples, which are often used in clinical diagnostics, there is a considerable need to separate and analyze specific cell types. This is not only limited to the classical blood cells erythrocytes, leukocytes or platelets and their subtypes, but also applies to microparticles, CTCs or even bacterial pathogens. Conventional biochemical methods based on differential and/or density centrifugation are rather slow and laborious, and even more importantly cannot be automated and therefore lack reproducibility. More modern methods such as FACS and MACS are more powerful, however, may require specific labeling of cells, can be time and cost intensive or have limited capacity. Here, microfluidics represents a most promising alternative that may allow a fast, comparably cheap, automated and high-throughput method to separate different cell types for subsequent analysis.2.2. Single cell analysis
Since in vitro cell culture has been introduced in the early 1900's, many strategies have been developed to mimic natural cellular environments and to efficiently grow and differentiate cells. However, even in cell culture, there is a complex heterogeneity within cell populations and observed phenomena often represent average rather than individual responses.36 The main interest for individual cell analysis derives from cancer research,37,38 where precise methods are needed to define cellular phenotypes from average population data. Starting with the first single cell-like proteomic approach in Saccharomyces cerevisiae,39 many significant studies have followed and underline the importance of such focused assays.7,40 Particularly, in stem cell research, separate analyses of individual cells led to the characterization of varying levels of proteins abundance depending on time and differentiation of cells.41 The different analytical readouts used in single cell analysis comprise affinity based assays, such as flow cytometry and immune-cytochemistry, and non-affinity based assays such as mass spectrometry (MS). However, the biggest challenge in the field still remains the selective and robust isolation of single cells out of a tissue, cell culture or body fluid, often characterized by poor efficiency and reproducibility.Indeed, recent advances in single cell analysis make use of microfluidics to overcome some of the limitations. Over the past decades a large number of high quality studies on single cell analysis using microfluidic chips have been published in the literature.42–48 Typically these studies aim either at (i) studying single cell's response to stimuli49 or at (ii) detecting and concentrating specific cell types from a large population.50 The most common microfluidic methodologies for single cell analysis involve (i) mechanical trapping, such as using posts27,49 and grooves,51 (ii) spatial control during fluid flow, such as by means of deterministic lateral displacement and inertial microfluidics,50 or microdroplet encapsulation.52 For instance, Fernandes et al.27 studied the modulation of alpha-synuclein toxicity in S. cerevisiae using a microfluidic gradient generator coupled to nine parallel chambers, each containing arrays of posts where cells were individually trapped (Fig. 1 – left). Then the number of cells with alpha-synuclein inclusions was investigated by subjecting cells to different levels of FeCl3 and monitoring via fluorescence microscopy (Fig. 1 – right). Hence, this device enabled investigation of both single cell response and its magnitude upon exposure to stimuli, with applications in research on Parkinson's disease.
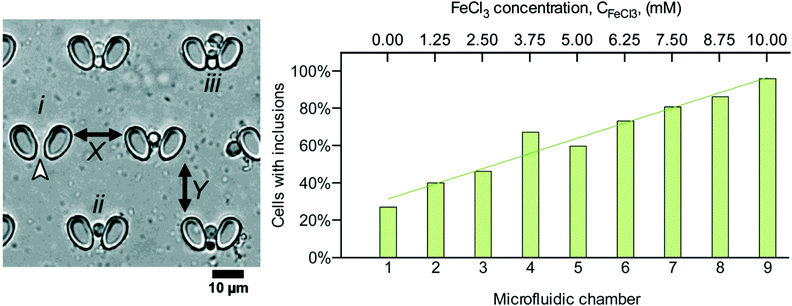 | ||
| Fig. 1 Analysis of cell's response to different stimuli concentrations using a microfluidic chip combining a gradient generator with single cell trapping chambers. (Left) each trap holds a single S. cerevisiae cell. (Right) the number of cells per trapping chamber showing alpha-synuclein inclusions is a function of the concentration of the FeCl3 stressor. Reproduced from ref. 27 with permission from the Royal Society of Chemistry. | ||
The field of single cell analysis, as can be assessed in the recommended literature, is vast and is starting to translate into successful industrial products, such as the case for the Fluidigm®C1 equipment,53 an automated solution for single-cell genomics being capable of handling up to 800 cells individually in a single run, a considerable throughput capability.
2.3. Subcellular analysis
One of the most striking limitations of -omics technologies typically used for the in-depth analysis of cells is the lack of spatial information. Nevertheless, it is well established, that e.g. (i) many proteins have multiple localizations that may lead to different functions, or (ii) may translocate either upon specific cellular stimuli54 or in disease conditions.55 Such information, however, is lost in typical large scale proteome studies. Although these can provide quantitative information on expression levels for thousands of proteins, they cannot distinguish whether the distribution of proteins between different organelles or sub-compartments has changed. Therefore, the separate analysis of isolated organelles and their content is of major interest as it allows studying the subcellular localization and distribution of proteins, representing a crucial step towards a better understanding of their individual functions and dynamics. Thus, proteomic studies analyzing isolated organelles,56–58 or sub-compartments59–61 have been mainly based on classical, laborious and non-automated methods. Typically, they encompass the mechanical rupture of the plasma membrane, homogenization of cellular matrix, and separation of its components by differential/density centrifugation. Notably, free flow electrophoresis (FFE), which allows the continuous separation of a broad variety of analytes, including cells and organelles, represents an interesting alternative to the common centrifugation steps. For instance, Lu et al.62 developed an isoelectric focusing microfluidic device for subcellular organelle sorting from crude cell lysates from NR6 murine fibroblasts. Fig. 2 shows the separation of mitochondria from nuclei under free flow electrophoresis.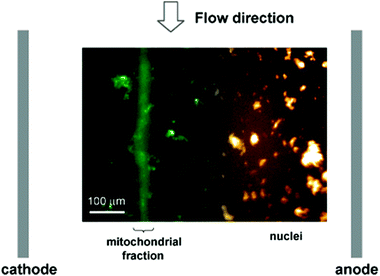 | ||
| Fig. 2 Isoelectric focusing free flow electrophoresis in microfluidics for organelle sorting from a crude cell lysate using a pH gradient between 3 and 6 and under a 20 V cm−1 electric field. Mitochondria are focused in a band closer to the cathode whereas nuclei focus closer to the anode. Reprinted with permission from ref. 62. Copyright ©2004 American Chemical Society. | ||
Alternative techniques, e.g. MACS, rely on the use of affinity tags or antibodies against specific epitopes present on the organelle's surface. Thus, microfluidics may play an important role and many recent publications have demonstrated different possibilities and prototypes for cell and organelle separations,62,63 some of which will be further described below.
2.4. Signaling dynamics
Recently, increasingly powerful mass spectrometry-based studies considerably improved our knowledge about cellular signaling, its complexity and dynamics.64–66 However, it concurrently became more and more evident, that the detailed elucidation of such dynamic processes as well as the underlying and intertwined networks requires more powerful sample preparation techniques. Notably, in cells different situations involve different timescale dimensions: processes such as regulatory protein phosphorylation upon kinase activation, or ion transport can occur within milliseconds to seconds, while others, such as changes in protein expression during cell cycle, require minutes to hours. Whereas rather slow processes can be easily followed in a time-resolved manner using conventional and mainly manual sample preparation methods, such strategies are not sufficiently reproducible and robust to study fast and short term processes. Initial signaling events are too fast to tolerate imprecise sampling and consequently measuring points; studying such fast processes demands for sub-second time resolution in combination with high accuracy and precision during sampling.3. Fast free flow cell manipulation in microfluidic devices
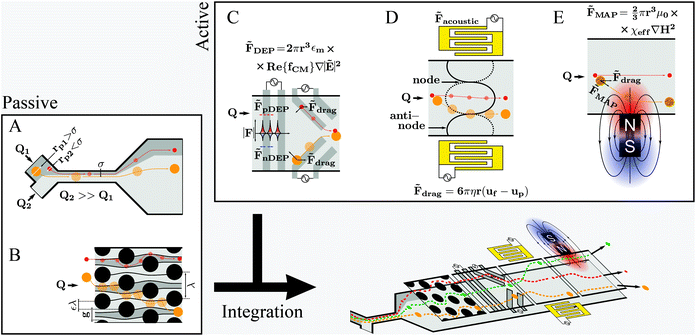
Fundamental and technological developments in science progress hand-in-hand which also holds true for the use of microfluidics to study cell properties and/or cellular processes. Consequently, new reports of novel applications, designs and capabilities of microfluidic systems for cell manipulation emerge constantly. Optimizing existing protocols and/or developing original research are the main motivation drawing researchers’ interest to the microfluidic technology. However, for many microfluidic systems handling issues have not been appropriately solved, and proper operation requires specialized professionals – often even the developers of the specific system – and well-controlled environments. This issue is relevant for future work and will be discussed later in this review.Different microfluidic technologies for cell manipulation have been described in literature. These can be divided into passive, active or integrated strategies as represented in Fig. 3. The cells’ position within microchannels can be controlled either by (i) rational design of the microfluidic structures, which results in deflection of cells’ trajectories, as for the passive strategies, or (ii) as a result of a force generated by an actuator, taking into account the cells’ electrical, magnetic and mechanical properties, as for active strategies. Often it is advantageous to combine different strategies to improve performance and functionalities within a single chip, referred to as integrated systems. However, integration is challenging, especially concerning handling simplicity, such as the need for alignment of micrometer-sized structures or fluidic and electronic connections.67–69 Firstly, we will revisit the aspects of different cell manipulation strategies and then highlight examples representing different integration designs thereof.
3.1. Geometry based manipulation techniques
Pinched flow (namely by using STEs), DLD and inertial microfluidics have been employed especially in cell sorting or controlled cell stimulation experiments. These are characterized by simplicity of fabrication and operation requiring only a flow control unit as external equipment.The magnitude of the Reynolds number (Re) (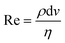 , where d is the microchannel characteristic dimension, and ρ, ν and η are the fluid's density, velocity and viscosity, respectively) indicates whether the flow is laminar (Re ≪ 1) or turbulent (Re ≫ 1). In microfluidics, flow regimes are typically laminar due to very low Re as a consequence of the small characteristic dimensions (d). Hence, a particle per cell will tend to keep its flow trajectory coincident with the streamline in which it is suspended. However, when the cell's dimension becomes comparable to the microchannel cross section width, the cell's center can be deflected into a different flow lamina. This characteristic is explored analogously in STE (Fig. 3A) and DLD (Fig. 3B).
, where d is the microchannel characteristic dimension, and ρ, ν and η are the fluid's density, velocity and viscosity, respectively) indicates whether the flow is laminar (Re ≪ 1) or turbulent (Re ≫ 1). In microfluidics, flow regimes are typically laminar due to very low Re as a consequence of the small characteristic dimensions (d). Hence, a particle per cell will tend to keep its flow trajectory coincident with the streamline in which it is suspended. However, when the cell's dimension becomes comparable to the microchannel cross section width, the cell's center can be deflected into a different flow lamina. This characteristic is explored analogously in STE (Fig. 3A) and DLD (Fig. 3B).
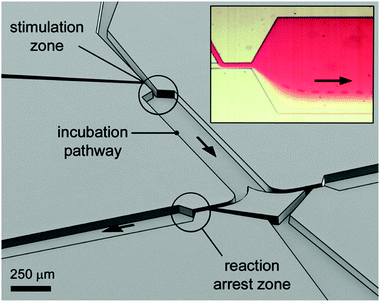 | ||
| Fig. 4 Perspective open channel representation of a two-level pinched-flow microfluidic device for high temporal resolution of cell/ligand interaction. Insert figure: optical microscope image of the “stimulation zone”. The cell, originally flowing in the transparent medium, is deflected into the red ligand containing medium at the first STE element. The time of exposure to the surface ligand is a function of fluid flow and length of the incubation path. The reaction is arrested at a second, down-stream located STE element. Reprinted with permission from ref. 10. Copyright ©2013 American Chemical Society. | ||
| Dc = 2ηgε | (1) |
Notably, although simple and attractive, both DLD and STE impose shear stress on cells which might trigger unwanted biological processes or could even disrupt cells. As there is no general way for a priori simulation of potential cell damage, assumptions are based on empirical results and, generally, shear stress is considered qualitatively rather than quantitatively. Although it is possible to simulate shear stress induced on a particle (constituted by a defined material construction) upon flow in a microchannel, calculations become very complex for cells, owing to their erratic structural constitution (e.g. cell membrane, organelles, microtubules, etc.). Therefore, assessing cell viability after flow through microfluidic channels has been the gold standard for validating design and operation. Hence, additional experiments have to be performed every time a new or modified design is employed. Thus, Varma and Voldman73 developed a cell-based shear stress sensor. They used NIH3T3 cells and created a transcriptional cell-sensor where transcription of Early Growth Factor-1 (a mechanosensitive protein) induces a fluorescent signal, achieving shear stress limits of detection down to 2 dyn cm−2 or, in SI units, 0.2 Pa, which is well below the predicted physiologic range of 0.8–3 Pa to which osteoblasts and osteocytes are exposed,74 for instance. Once biological changes induced by co-suspension of these cells with cells of interest can be excluded, it is possible to investigate simultaneously if a given shear stress value is overcome and whether this has an impact on the cells of interest.
3.2. Active manipulation techniques
Active manipulation techniques are based on the cells’ physical properties. For instance, dielectrophoresis (DEP) and acoustophoresis (ACP) rely on the dielectric and mechanical properties, respectively. The overall physical properties of cells may be influenced, for instance, by controlled biological stimuli (e.g. response to membrane shear stress in osteocytes;75 treatment of red blood cells with glutaraldehyde increases their membrane stiffness72) or by surface-labeling using antibodies attached to tags or magnetic particles. The latter is often used for specific isolation/purification of cells from a mixture by magnetophoresis (MAP) since a vast selection of antibodies for different cell surface receptors and protocols for labeling are readily available. However, such approaches depend heavily on antibody specificity as well as the availability of specific marker proteins that are uniquely expressed on one of the cell types at hand. Active manipulation techniques are not necessarily interchangeable and may actually be complementary at addressing complex cell manipulation procedures. Therefore, we will first present these techniques individually and later discuss integration developments. An advantage common to all these techniques when coupled to microfluidics is the possibility to control the position of cells in free-flow format. All following information will be related to free-flow strategies coupling DEP, ACP, MAP or combinations therefrom. For ease of reading we will not use the “free-flow” term.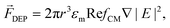 | (2) |
DEP may not be suitable for manipulating cells suspended in media of high electrical conductivity, which is typically the case for cell culture medium, and cause harm to cells by Joule heating.8 Therefore, cell medium exchange is required. Additionally, applied electric fields might be limited to avoid cell damage, concurrently limiting actuation force levels. Another concern regarding DEP is the lack of systematic studies, especially related to the use of high frequency signals (e.g. MHz) on the influence of the applied electric fields. These might cause changes observable only in the long term.
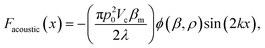 | (3) |
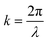 , x is the distance from a pressure node and ϕ a measure of the mechanical differences between the cell and the medium defined as:
, x is the distance from a pressure node and ϕ a measure of the mechanical differences between the cell and the medium defined as: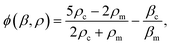 | (4) |
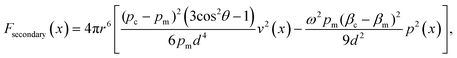 | (5) |
ACP offers an effective strategy for cell manipulation in microchannels, but contrary to DEP in which complex electrode “highways” can be fabricated for local cell manipulation, ACP techniques are only capable of focusing cells into pressure nodes or anti-nodes which are static along a channel cross section. On the other hand, the risk of affecting cell viability is much reduced as compared to DEP (is this case mainly due to Joule heating). For instance, Burguillos et al.83 demonstrated that microfluidic ACP had no impact on survival or function of Microglia, Leukocytes or tumor cells under several acoustic pressure and suspension flow conditions. Li et al.84 went further by claiming that acoustic-based separation offer excellent biocompatibility in terms of preserving both the cellular phenotype and genotype. They demonstrated the use of an ACP microfluidic device (Fig. 5) for the separation of CTCs from white blood cells (WBC) containing solutions. Recovery rates above 83% were obtained for samples with CTC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) WBC ratios of 1
WBC ratios of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100.000 and CTCs could be isolated from real clinical samples from breast cancer patients.
100.000 and CTCs could be isolated from real clinical samples from breast cancer patients.
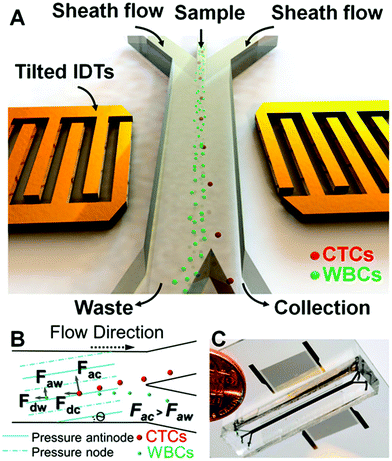 | ||
| Fig. 5 ACP microfluidic device used by Li et al.84 for isolation of CTCs from complex samples. (A) Tilted inter-digital transducers generate pressure nodes and anti-nodes within the microchannel cross-section, which force cells to separate. (B) CTCs and WBCs are separately collected into two outlets. (C) Photograph of the real microfluidic device next to a coin for size comparison. Reprinted with permission from ref. 84. | ||
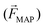 85 (see eqn (6)) only in labeled cells, is used to deplete other cells and remove the solution. Since some antibodies bind antigens, such as cell membrane receptors, with very high specificity, MAP offers a rather simplified instrumental and proceedings for cell separation, purification and concentration. According to Karabacak et al.,50 magnetophoreis has been popular because it “[...] requires simple and readily available tools”.
85 (see eqn (6)) only in labeled cells, is used to deplete other cells and remove the solution. Since some antibodies bind antigens, such as cell membrane receptors, with very high specificity, MAP offers a rather simplified instrumental and proceedings for cell separation, purification and concentration. According to Karabacak et al.,50 magnetophoreis has been popular because it “[...] requires simple and readily available tools”.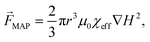 | (6) |
The above advantages, together with aforementioned aspects of microfluidics, are the motivation behind developing microfluidic MAP equivalents. In parallel to the bulk equivalent, microfluidic MAP are characterized by simplicity of fabrication and magnet integration. This strategy, is very attractive for cell separation and focusing since low intensity magnetic fields are not known to harm cells. However it is “a one way” focusing strategy that lacks the versatility of DEP, for instance, in which the cell can be guided using dedicated electrode “highways”. Moreover, it does not offer simple solutions for multiple cell sorting, in contrast to multiplexed FACS techniques available in certain flow cytometers. Recently, Giudice et al.85 reported the development of a particle sorting device employing MAP. First, a mixture of magnetic and non-magnetic beads are aligned along a streamline. Then, particles are separated into two different outlets via MAP force, yielding a deflection efficiency of 96%. See Table 1 for a summary of the above techniques' details.
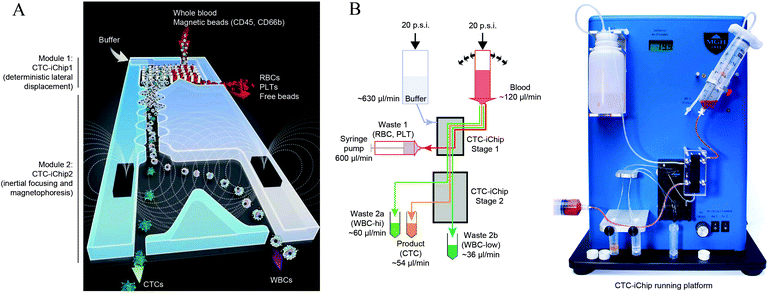
3.3. Integration
The presented techniques for cell manipulation have inherent advantages and disadvantages which do not necessarily overlap, rendering them suitable only for some applications. Thus, cells that have identical dimensions, electrical and mechanical properties, and only differ in the expression (levels) of certain membrane surface proteins can only be differentiated in microfluidic devices by using appropriate labels, such as in MAP-based separation. Most studies use proof-of-principle examples of low complexity, such as cultured cell suspensions instead of complex mixtures such as blood, for development and verification of their devices. However, often such approaches are not sufficiently flexible to be successfully translated to specific real-world applications. For instance, size-based separation of blood cells does not necessarily allow distinguishing healthy from infected red blood cells. Hence, progressively researchers invest efforts to integrate different methodologies into a single lab-on-chip (LoC) (as represented in Fig. 3-bottom) in order to either merge advantages of different strategies while canceling out handicaps, or to enhance cell manipulation control by joint forces. In the next sub-sections, we will present recent examples of integration on microchannels for improved cell manipulation.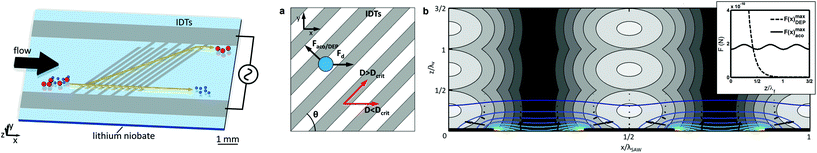 | ||
| Fig. 6 Combination of DEP and ACP by integrating electrodes in a microchannel. (Left) schematics of the integrated microfluidic constitution and operation. (Middle) the electrodes make an angle θ toward the fluid flow direction. A particle, flowing from left to right will be subjected to a combination of drag force in the direction of the fluid flow, and DEP and ACP forces both perpendicular to the electrodes. Particles/cells with dimensions above the critical diameter will be deflected along the electrodes. (Right) representation of the acoustic pressure field magnitude in gray, and the first 10 DEP force potential contours in color and the DEP force vectors. Reproduced from ref. 88 with permission from the Royal Society of Chemistry. | ||
 | ||
| Fig. 7 (A) Schematics of CTC-iChip operation. The actual device combines two separate microfluidic devices that integrate three cell manipulation techniques: DLD for removing red blood cells and platelets, an inertial focusing region, and a MAP region for separation of CTCs from WBCs. (B) CTC-iChip bench-top prototype. (Left) schematic of the microfluidic network and operation. (Right) photo of the integrated device. Reprinted by permission from Macmillan Publishers Ltd: Nature Protocols,50 Copyright ©2014. | ||
4. Fast cell stimulation and quenching
The previous sections introduced microfluidic methods that enable high spatial control of cells within microchannels. Those applications often target cell–cell separation from complex mixtures based on an invariant characteristic, or temporally static, such as whether the cell is a CTC or a white blood cell. On the other hand, cell growth, cell–cell communication and response to stimulus, for instance, are transient processes requiring not only tight control of the localization of cells but also the time lapse between events.92 This represents an additional concern and imposes design limitations. For instance, free-flow is not suitable to study processes which occur for adherent cells, or in opposition, non-adherent cells require free-flow strategies.Most recent developments on fast cell stimulation techniques make use of cell adhesion on specific parts of the microchannels. Then, cells can be subjected to a sequence of different solutions, such as growth medium exchange, stimulant containing solution and lysis or fixation buffer, simply by controlling the injection of solutions. Adsorbing, culturing and maintaining cell populations is per se not a trivial task and subject of many studies in the literature32,93,94 that will not be covered here. However, it is important to stress that, in contrast to traditional cell culture, microfluidic applications require more time and skills to successfully adhere/cultivate/maintain cells. On the other hand, microfluidics allows automation in a highly parallelized (high throughput) fashion under well-controlled conditions that are not achievable otherwise.7,9 Notably, adherent cells pose substantial limitations to the range of applications. When fast solution switching is required (e.g., in the sub-second range) two important limitations have to be taken into account: (i) cells are shear stress sensitive and will not allow the use of high flow rates, and (ii) fast solution switching while avoiding contamination is a time limiting factor, because it usually requires physical separation of solutions (e.g., using valves or air bubbles). In the next paragraphs we will present some work developed recently which is based on cell adhesion models.
4.1. Using perforated membranes
Chingozha et al.95 developed a microfluidic device using a PDMS perforated membrane to allow stimulus from one side of the membrane to reach cells located on the other side (Fig. 8A). Hence, they created a physical barrier that controls the flow rate delivered to the cells in order to control the shear stress applied. At the same time, the hydraulic resistance was tuned to ensure rapid mass transport of the ligand. Cells were seeded into the cell's compartments by gravity-driven flow. As shown in Fig. 8B, an array of single cells (Jurkat E6-1 human acute T lymphoma) were compartmentalized within the cell microfluidic compartment, separated from the active flow layer via pores. With the cell layer inlet closed, the microfluidic device was flushed sequentially with buffer and stimulant with control of the frequency of switching. The authors used FITC and fluorescence microscopy in order to characterize the device's performance in terms of (i) homogeneity of the mass delivery to the different cell locations and (ii) required time (“rise-time”) to exchange the solution at the cell location. They found a negligible difference in the periodicity of the upstream and downstream flows (<0.01%) and a rise-time of 154 ms, both values in good quantitative agreement with their computer simulations.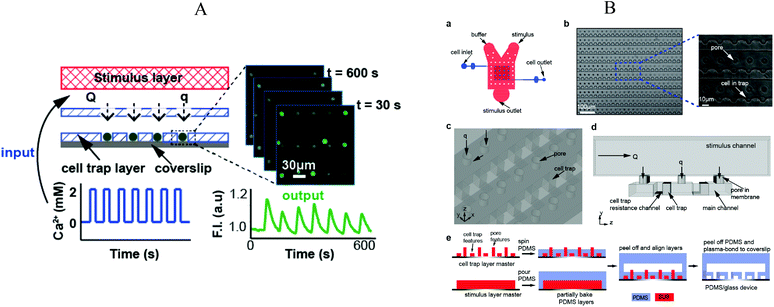 | ||
| Fig. 8 (A) Illustration of the microfluidic device divided in three layers: (top) stimulus carrying layer, (middle) perforated membrane, (bottom) high density cell trap layer. Fluorescence measurement was performed under application of an oscillating stimulus. (B) Detailed representation of the microfluidic device, structures and microfabrication process. Adapted with permission from ref. 95. Copyright ©2013 American Chemical Society. | ||
The device was used to evaluate the response of intracellular Ca2+ depleted Jurkat to alternating flows of Ca2+ and EGTA solutions, using Fluo3-AM as signal. Different switching periods between 20 s and 2 min were tested, showing the potential of this device to perform controlled stimulation experiments. Moreover, the device offers a general strategy since cell adhesion to the microfluidic walls is not mandatory. However, ensuring that cells are individually immobilized, particularly in long term experiments, is not trivial, even more when using small cells such as bacteria.
4.2. Electro wetting – digital microfluidics
Recently, Ng et al.96 described the use of digital microfluidics to perform immunocytochemistry in single cells. Digital microfluidics makes use of planar electrodes to change the contact angle of droplets in order to move them above a planar surface.97 The device comprised two parallel plates, separated in about 180 μm distance, forming an “infinite” microfluidic chamber. An array of electrodes was deposited on one of the plates, covered with a Teflon film to avoid cell adhesion. In the second plate, specific areas for cell adhesion (i.e. hydrophilic areas) were patterned into a Teflon coating (Fig. 9). Fluids where moved around by sequential actuation of the planar electrodes. Except for the cell adhesion spots all areas of the microdevice were hydrophobic, allowing precise circulation of nl volumes of solution. By flushing a solution through the hydrophilic spots a “virtual microwell” was formed by trapping a given quantity of liquid between the two parallel plates. Its lateral footprint was defined by the hydrophilic area (Fig. 10). Liquid exchange was performed by circulating a second solution through the virtual microwell area with an exchange efficiency of 93% in a single step and up to 99% in two steps.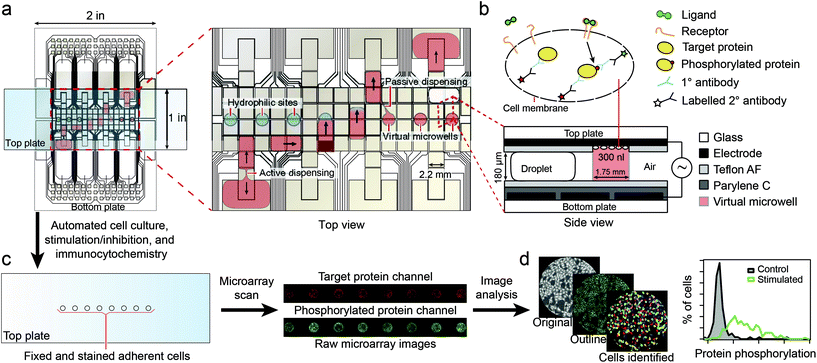 | ||
| Fig. 9 Schematics of the digital microfluidics device. Fluid droplets are circulated by actuation of integrated planar electrodes passivated with a Teflon coating. In designated areas, the absence of hydrophobic coating allows the formation of a virtual microwell where cells are trapped for further stimulation experiments. Signal inspection was performed using a microarray scanner. Reprinted with permission from ref. 96. ©2015 Macmillan Publishers Limited. | ||
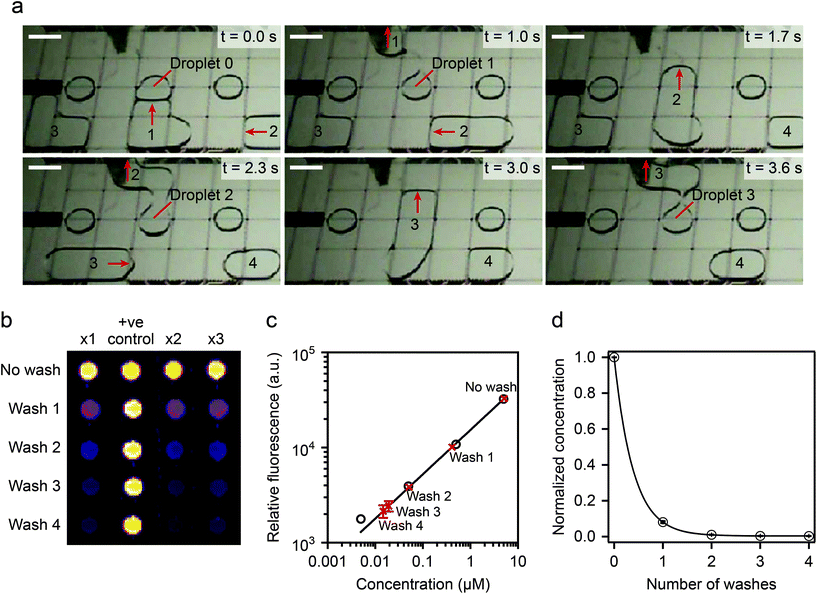 | ||
| Fig. 10 (Top) device operation details. A virtual microwell containing 5 μl of fluoresceinamine is washed sequentially and the remaining fluorescence compared to calibrated data. (Bottom) washing efficiency experimental results: two washing steps displaced >99% of the original solution. Reprinted with permission from ref. 96. ©2015 Macmillan Publishers Limited. | ||
The digital microfluidic device was used to investigate the activation dynamics of the phosphoinositide 3-kinase signaling pathway upon stimulation of fibroblasts with PDGF. Around 300–400 cells were immobilized in each virtual microwell. A series of 11 automated steps were conducted to perform cell starvation, ligand stimulation, fixation, permeabilization, blocking, staining and washing. Successive end-to-end solution exchanges were possible in about 1.3 s. Hence this device allows for fast cell stimulation assays down to the second time regime.
4.3. Chamber based microfluidics for sequential cell stimulation
Another concept to control the sequential flow of solutions to adherent cells on a chamber is by using pneumatic valving. Blazek et al.11 developed a large scale integration platform containing 16 “logic blocks”, each addressing 8 chambers for conducting high throughput studies of fast protein phosphorylation kinetics in NIH3T3 fibroblasts (Fig. 11a). The chip was supplied with all solutions required using tubbing and eventually flasks, thus avoiding any further disturbance during experiments. An automated pressure controller was used to ensure controlled flow rate delivery and timings of the different reagents used, with the full experiment taking about one and half days. The device's theoretical time resolution of 1.5 s for cell-stimulation was measured by monitoring the time required to fully exchange the cell chamber buffer at different pressure values (see Fig. 11b and c). However, the experimental resolution time was 10 s, since the authors used stimulation pulses of 5 s followed by a cell fixation pulse. Full cell activation experiments were performed on chip and signals were quantified via proximity ligand assays. Cell counts between 350 and 500 where used to ensure statistically relevant data. The characteristic phosphorylation times for different cell receptors (PDGF and IGF-1) were between 13 and 35 seconds.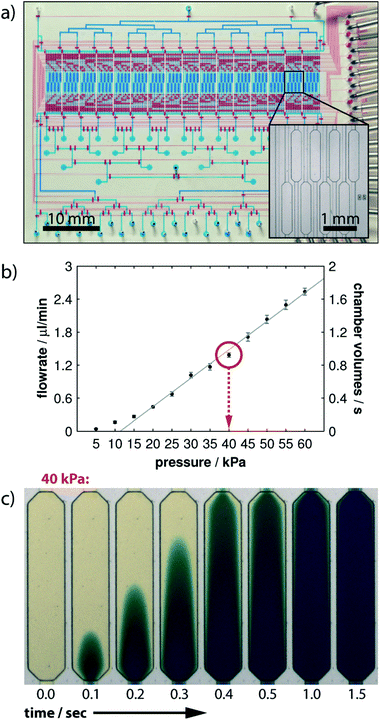 | ||
| Fig. 11 (Top) microfluidic chip overall design. Flow and control microchannel layers were filled with blue and red solutions for ease of visualization. Zoom of the cell culture chambers. (Middle) flow rate calibration results as a function of the applied pressure difference. (Bottom) time-lapsed images of buffer exchange in one cell culture chamber for an applied 40 kPa inlet pressure. Reproduced from ref. 98 with permission from the Royal Society of Chemistry. | ||
Despite reduced time resolution compared to the previous examples, this work demonstrates a highly compact and parallelized cell stimulation device that once properly interfaced with the outer world can perform complete cell stimulation experiments autonomously. However, interfacing complex tubing and pressure source connections in microfluidics is a challenging task.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7 flow ratio for instance, per ml of cell solution 7 and 49 ml of stimulation and quenching solutions are required, respectively, which may pose restrictive limitations to the concentration of the final lysate. Furthermore, for typical microfluidic device sizes and lysate collection requirements, single experiments may take hours of operation per time point experiment. Since in microfluidics liquids flow in the laminar regime, mixing of substances is controlled by diffusion. Diffusion constants are very low and especially reduced for larger molecules, hence mixing solutions in microfluidics is time inefficient. However, mixing times can be greatly reduced by increasing the number of interfaces between the liquids to mix within a microchannel cross section (i.e., diffusion distances are decreased). Microfluidics especially designed for this effect have been termed micromixers.100 Although micromixers might not offer such a high temporal resolution as STE elements, they offer the possibility to mix different solutions at comparative flow ratio values, hence avoiding dilution problems. For instance, Hirsch et al.98 used a herringbone-based micromixer to activate T cells. The microfluidic device (Fig. 12) integrated several micromixers in parallel using a common source of cells, stimulus and lysis buffers. A mixed solution of cell and stimulus buffers, obtained in only 250 ms, was split into eight different time stimulation circuits (30 s to 5 min). Cell lysis was also performed in parallel and the experiments were carried out with an equivalent of 5% of material as compared to conventional methods (Table 2).
7 flow ratio for instance, per ml of cell solution 7 and 49 ml of stimulation and quenching solutions are required, respectively, which may pose restrictive limitations to the concentration of the final lysate. Furthermore, for typical microfluidic device sizes and lysate collection requirements, single experiments may take hours of operation per time point experiment. Since in microfluidics liquids flow in the laminar regime, mixing of substances is controlled by diffusion. Diffusion constants are very low and especially reduced for larger molecules, hence mixing solutions in microfluidics is time inefficient. However, mixing times can be greatly reduced by increasing the number of interfaces between the liquids to mix within a microchannel cross section (i.e., diffusion distances are decreased). Microfluidics especially designed for this effect have been termed micromixers.100 Although micromixers might not offer such a high temporal resolution as STE elements, they offer the possibility to mix different solutions at comparative flow ratio values, hence avoiding dilution problems. For instance, Hirsch et al.98 used a herringbone-based micromixer to activate T cells. The microfluidic device (Fig. 12) integrated several micromixers in parallel using a common source of cells, stimulus and lysis buffers. A mixed solution of cell and stimulus buffers, obtained in only 250 ms, was split into eight different time stimulation circuits (30 s to 5 min). Cell lysis was also performed in parallel and the experiments were carried out with an equivalent of 5% of material as compared to conventional methods (Table 2).
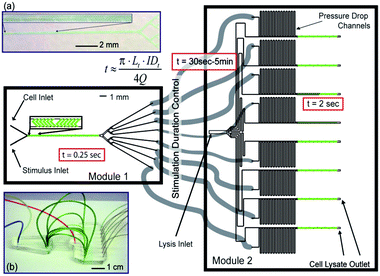 | ||
| Fig. 12 A two module microfluidic application for cell stimulation and quenching. (Top left) photograph of module 1 showing mixing of colored solutions, achieved in 0.25 s. (Middle) the mixed solution is divided through 8 outlets which are connected to module 2 via different tubbing lengths corresponding to different cell stimulation time points. (Right) the input from module 1 is mixed with lysis buffer in module 2. Reproduced from ref. 11 with permission from The Royal Society of Chemistry. | ||
| Cell handling technique | Principle for cell manipulation | Time resolution | User friendly | Generalizable |
|---|---|---|---|---|
| Perforated membrane | Controlled solution exchange | ++ | ++ | + |
| Digital microfluidics | Electro-wetting | + | ++ | + |
| Chamber and valving | Solution exchange using valving | + | — | ++ |
| Pinched flow | Deviation of cells via STE | +++ | ++ | ++ |
| Micromixers | Mixing cell and stimulus | ++ | +++ | +++ |
5. Discussion
As reviewed here, alongside high spatial resolution, high temporal control of cell manipulation in microfluidics has been performed at unprecedented levels. High temporal control is especially important when high time resolution is required and/or in characterization of fast occurring phenomena, both of which are of the interest of different fields such as research and clinics. However, key to success encompasses also technology transfer. The transfer of microfluidic technologies, such as those for cell manipulation, into real world applications faces two main obstacles, referred to in several publications in recent years.101,102 The first obstacle is to guarantee reproducibility of microfluidic operation in non-comfort zones, i.e. in the field of application rather than in the laboratory of development, without the need for specialists. Although, as demonstrated in this review, many studies have demonstrated high potential and control regarding particle per cell manipulation, issues regarding interfacing microfluidic chips to up- and downstream applications of the external world are often omitted. Importantly, in many cases this may actually compromise the practical application of otherwise elaborate devices. The above mentioned issues are well-known and in the past years researchers have put efforts into both scaling integration levels of microfluidic systems and designing practical chip-world interfaces. This applies to microfluidic devices for particle or cell manipulation, but also to other microfluidic based applications, such as biosensing, which may actually be used complementary in cell studies; for instance, early cancer diagnosis facilitated by such technologies either by detection/quantification of cancer biomarkers and CTCs may improve treatment outcomes. The second obstacle is the development of a working prototype into industrialized products. This is mainly attributed to the inability to mass-scale produce microfluidics in their typical research environment form. Thus, PDMS has been the material-of-choice for microfluidic device fabrication, but casting is incompatible with established industrial fabrication techniques such as micro-injection and compression molding, and thermoforming. Thus, whereas microfluidic devices open up novel possibilities in fundamental research, there is a still a gap to bridge for routine usage and applications, such as in clinical research.6. Perspectives
Microfluidic devices, whether in their early development stages or as integrated systems, have increasingly received attention by the research community. Among those, particularly microfluidic devices for cell manipulation have attracted attention in the field of life science and (bio)medicine. Nowadays there is a huge demand, both economically and ethically, to miniaturize procedures and develop cell-based experiments that might replace traditionally performed animal models in accordance with the 3R (Replacement, Reduction, Refinement). Along this line, there is a general trend to reduce time and costs of analyses and increase efficiency. All these concepts may greatly benefit from migration to microfluidic formats. Indeed, microfluidic technology has opened new paths to perform original studies that had been impossible previously, and many tools are available. However, what is lacking is the development and fabrication of standardized microfluidic connections and ideally of-the-shelf parts, such as in the case of electronics. Then, building a microfluidic circuit would be easily customizable, rendering it increasingly attractive to research in (bio)medicine and biology, among others. Furthermore, despite a vast research community of potential customers, the potential of microfluidics is seemingly still confined to experts. Breaking this invisible boundary by advertising its power to the general scientific public would greatly promote further interdisciplinary discussion, cooperation and development.Acknowledgements
Financial support by the Ministerium für Innovation, Wissenschaft und Forschung des Landes Nordrhein-Westfalen, the Senatsverwaltung für Wirtschaft, Technologie und Forschung des Landes Berlin, and the Bundesministerium für Bildung und Forschungis is gratefully acknowledged. Pedro Novo and Margherita Dell'Aica also thank for ISAS strategic funds.References
- Y. Hu, L. Fan, J. Zheng, R. Cui, W. Liu, Y. He, X. Li and S. Huang, Detection of circulating tumor cells in breast cancer patients utilizing multiparameter flow cytometry and assessment of the prognosis of patients in different CTCs levels, Cytometry, Part A, 2010, 77A(3), 213–219 CrossRef CAS PubMed
.
- T. Hristozova, R. Konschak, V. Budach and I. Tinhofer, A simple multicolor flow cytometry protocol for detection and molecular characterization of circulating tumor cells in epithelial cancers, Cytometry, Part A, 2012, 81A(6), 489–495 CrossRef CAS PubMed
.
- P. Li, Z. S. Stratton, M. Dao, J. Ritz and T. J. Huang, Probing circulating tumor cells in microfluidics, Lab Chip, 2013, 13(4), 602–609 RSC
.
- A. Lenshof and T. Laurell, Continuous separation of cells and particles in microfluidic systems, Chem. Soc. Rev., 2010, 39(3), 1203–1217 RSC
.
- J. El-Ali, P. K. Sorger and K. F. Jensen, Cells on chips, Nature, 2006, 442(7101), 403–411 CrossRef CAS PubMed
.
- C. Wyatt Shields IV, C. D. Reyes and G. P. López, Microfluidic cell sorting: a review of the advances in the separation of cells from debulking to rare cell isolation, Lab Chip, 2015, 15(5), 1230–1249 RSC
.
- A. J. Hughes, D. P. Spelke, Z. Xu, C.-C. Kang, D. V. Schaffer and A. E. Herr, Single-cell western blotting, Nat. Methods, 2014, 11(7), 749–755 CrossRef CAS PubMed
.
- S.-Y. Tang, W. Zhang, R. Soffe, S. Nahavandi, R. Shukla and K. Khoshmanesh, High resolution scanning electron microscopy of cells using dielectrophoresis, PLoS One, 2014, 9(8), e104109 Search PubMed
.
- N. Dénervaud, J. Becker, R. Delgado-Gonzalo, P. Damay, A. S. Rajkumar, M. Unser, D. Shore, F. Naef and S. J. Maerkl, A chemostat array enables the spatio-temporal analysis of the yeast proteome, Proc. Natl. Acad. Sci. U. S. A., 2013, 110(39), 15842–15847 CrossRef PubMed
.
- Y.-Y. Chiang, S. Haeri, C. Gizewski, J. D. Stewart, P. Ehrhard, J. Shrimpton, D. Janasek and J. West, Whole cell quenched flow analysis, Anal. Chem., 2013, 85(23), 11560–11567 CrossRef CAS PubMed
.
- M. Blazek, T. S. Santisteban, R. Zengerle and M. Meier, Analysis of fast protein phosphorylation kinetics in single cells on a microfluidic chip, Lab Chip, 2015, 15, 726–734 RSC
.
- A. van Reenen, A. M. de Jong, J. M. J. den Toonder and M. W. J. Prins, Integrated lab-on-chip biosensing systems based on magnetic particle actuation - a comprehensive review, Lab Chip, 2014, 14, 1966–1986 RSC
.
- A. L. Washburn and R. C. Bailey, Photonics-on-a-chip: recent advances in integrated waveguides as enabling detection elements for real-world, lab-on-a-chip biosensing applications, Analyst, 2011, 136, 227–236 RSC
.
- M. C. Estevez, M. Alvarez and L. M. Lechuga, Integrated optical devices for lab-on-a-chip biosensing applications, Laser Photonics Rev., 2012, 6(4), 463–487 CrossRef
.
- D. G. Rackus, M. H. Shamsi and A. R. Wheeler, Electrochemistry, biosensors and microfluidics: a convergence of fields, Chem. Soc. Rev., 2015, 44, 5320–5340 RSC
.
- D. King, M. O'Sullivan and J. Ducrée, Optical detection strategies for centrifugal microfluidic platforms, J. Mod. Opt., 2014, 61(2), 85–101 CrossRef CAS
.
- H. Amini, W. Lee and D. Di Carlo, Inertial microfluidic physics, Lab Chip, 2014, 14(15), 2739–2761 RSC
.
- R. Ramji, M. Wang, A. Asgar, S. Bhagat, D. T. S. Weng, N. V. Thakor, C. T. Lim and C.-H. Chen, Single cell kinase signaling assay using pinched flow coupled droplet microfluidics, Biomicrofluidics, 2014, 8(3), 034104 CrossRef PubMed
.
- D. H. Yoon, A. Jamshaid, J. Ito, A. Nakahara, D. Tanaka, T. Akitsu, T. Sekiguchi and S. Shoji, Active microdroplet merging by hydrodynamic flow control using a pneumatic actuator-assisted pillar structure, Lab Chip, 2014, 14(16), 3050–3055 RSC
.
- F. Volpetti, J. Garcia-Cordero and S. J. Maerkl, A microfluidic platform for high-throughput multiplexed protein quantitation, PLoS One, 2015, 10(2), e0117744 Search PubMed
.
- X Qin, S. Park, S. P. Duffy, K. Matthews, R. R. Ang, T. Todenhöfer, H. Abdi, A. Azad, J. Bazov, K. N. Chi, P. C. Black and H. Ma, Size and deformability based separation of circulating tumor cells from castrate resistant prostate cancer patients using resettable cell traps, Lab Chip, 2015, 15, 2278–2286 RSC
.
- P. Preira, V. Grandné, J.-M. Forel, S. Gabriele, M. Camara and O. Theodoly, Passive circulating cell sorting by deformability using a microfluidic gradual filter, Lab Chip, 2013, 13(1), 161–170 RSC
.
- M. G. Lee, J. H. Shin, C. Y. Bae, S. Choi and J.-K. Park, Label-free cancer cell separation from human whole blood using inertial microfluidics at low shear stress, Anal. Chem., 2013, 85(13), 6213–6218 CrossRef CAS PubMed
.
- Y. Ren, W. Liu, Y. Jia, Y. Tao, J. Shao, Y. Ding and H. Jiang, Induced-charge electroosmotic trapping of particles, Lab Chip, 2015, 15, 2181–2191 RSC
.
- L. Diéguez, M. A. Winter, K. J. Pocock, K. E. Bremmell and B. Thierry, Efficient microfluidic negative enrichment of circulating tumor cells in blood using roughened PDMS, Analyst, 2015, 140(10), 3565–3572 RSC
.
- S. C. C. Shih, H. Yang, M. J. Jebrail, R. Fobel, N. McIntosh, O. Y. Al-Dirbashi, P. Chakraborty and A. R. Wheeler, Dried blood spot analysis by digital microfluidics coupled to nanoelectrospray ionization mass spectrometry, Anal. Chem., 2012, 84(8), 3731–3738 CrossRef CAS PubMed
.
- J. Tiago, S. Fernandes, S. Tenreiro, A. Gameiro, V. Chu, T. F. Outeiro and J. P. Conde, Modulation of alpha-synuclein toxicity in yeast using a novel microfluidic-based gradient generator, Lab Chip, 2014, 14(20), 3949–3957 RSC
.
- X. Wang, S. Chen, M. Kong, Z. Wang, K. D. Costa, R. A. Li and D. Sun, Enhanced cell sorting and manipulation with combined optical tweezer and microfluidic chip technologies, Lab Chip, 2011, 11(21), 3656–3662 RSC
.
- O. O. Saeed, R. Li and Y. Deng, Microfluidic approaches for cancer cell separation: Review, J. Biomed. Sci. Eng., 2014, 07(12), 1005–1018 CrossRef
.
- Y. Chen, P. Li, P.-H. Huang, Y. Xie, J. D. Mai, L. Wang, N.-T. Nguyen and T. J. Huang, Rare cell isolation and analysis in microfluidics, Lab Chip, 2014, 14(4), 626–645 RSC
.
- H. Tsutsui and C.-M. Ho, Cell separation by non-inertial force fields in microfluidic systems, Mech. Res. Commun., 2009, 36(1), 92–103 CrossRef PubMed
.
- S. Halldorsson, E. Lucumi, R. Gómez-Sjöberg and R. M. T. Fleming, Advantages and challenges of microfluidic cell culture in polydimethylsiloxane devices, Biosens. Bioelectron., 2015, 63, 218–231 CrossRef CAS PubMed
.
- E. K. Sackmann, A. L. Fulton and D. J. Beebe, The present and future role of microfluidics in biomedical research, Nature, 2014, 507(7491), 181–189 CrossRef CAS PubMed
.
- J. Nilsson, M. Evander, B. Hammarström and T. Laurell, Review of cell and particle trapping in microfluidic systems, Anal. Chim. Acta, 2009, 649(2), 141–157 CrossRef CAS PubMed
.
- T. Laurell, F. Petersson and A. Nilsson, Chip integrated strategies for acoustic separation and manipulation of cells and particles, Chem. Soc. Rev., 2007, 36, 492–506 RSC
.
- F. S. O. Fritzsch, C. Dusny, O. Frick and A. Schmid, Single-cell analysis in biotechnology, systems biology, and biocatalysis, Annu. Rev. Chem. Biomol. Eng., 2012, 3(1), 129–155 CrossRef CAS PubMed
. PMID: 22468600.
- M. Niepel, S. L. Spencer and P. K. Sorger, Non-genetic cell-to-cell variability and the consequences for pharmacology, Curr. Opin. Chem. Biol., 2009, 13(5–6), 556–561 CrossRef CAS PubMed
.
- A. A. Cohen, N. Geva-Zatorsky, E. Eden, M. Frenkel-Morgenstern, I. Issaeva, A. Sigal, R. Milo, C. Cohen-Saidon, Y. Liron, Z. Kam, L. Cohen, T. Danon, N. Perzov and U. Alon, Dynamic proteomics of individual cancer cells in response to a drug, Science, 2008, 322(5907), 1511–1516 CrossRef CAS PubMed
.
- J. R. S. Newman, S. Ghaemmaghami, J. Ihmels, D. K. Breslow, M. Noble, J. L. DeRisi and J. S. Weissman, Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise, Nature, 2006, 441(7095), 840–846 CrossRef CAS PubMed
.
- Y. T. Chong, J. L. Y. Koh, H. Frisen, S. K. Duffy, M. J. Cox, A. Moses, J. Moffat, C. Boone and B. J. Andrews, Yeast proteome dynamics from single cell imaging and automated analysis, Cell, 2015, 161(6), 1413–1424 CrossRef CAS PubMed
.
- H. H. Chang, M. Hemberg, M. Barahona, D. E. Ingber and S. Huang, Transcriptome-wide noise controls lineage choice in mammalian progenitor cells, Nature, 2008, 453(7194), 544–547 CrossRef CAS PubMed
.
- M. Junkin and S. Tay, Microfluidic single-cell analysis for systems immunology, Lab Chip, 2014, 14, 1246–1260 RSC
.
- A. Rakszewska, J. Tel, V. Chokkalingam and W. T. S. Huck, One drop at a time: toward droplet microfluidics as a versatile tool for single-cell analysis, NPG Asia Mater, 2014, 6, e133 CrossRef CAS
.
- H. Yin and D. Marshall, Microfluidics for single cell analysis, Curr. Opin. Biotechnol., 2012, 23(1), 110–119 CrossRef CAS PubMed
. Analytical biotechnology.
- T. A. Duncombe, A. M. Tentori and A. E. Herr, Microfluidics: reframing biological enquiry, Nat. Rev. Mol. Cell Biol., 2015, 16(9), 554–567 CrossRef CAS PubMed
.
- P. K. Chattopadhyay, T. M. Gierahn, M. Roederer and J. Christopher Love, Single-cell technologies for monitoring immune systems, Nat. Immunol., 2014, 15(2), 128–135 CrossRef CAS PubMed
.
- T. P. Lagus and J. F. Edd, A review of the theory, methods and recent applications of high-throughput single-cell droplet microfluidics, J. Phys. D: Appl. Phys., 2013, 46(11), 114005 CrossRef
.
- C. Gawad, W. Koh and S. R. Quake, Single-cell genome sequencing: current state of the science, Nat. Rev. Genet., 2016 DOI:10.1038/nrg.2015.16
.
- X. J. Li, Y. Chen and P. C. H. Li, A simple and fast microfluidic approach of same-single-cell analysis (sasca) for the study of multidrug resistance modulation in cancer cells, Lab Chip, 2011, 11, 1378–1384 RSC
.
- N. M. Karabacak, P. S. Spuhler, F. Fachin, E. J. Lim, V. Pai, E. Ozkumur, J. M. Martel, N. Kojic, K. Smith, P.-i Chen, J. Yang, H. Hwang, B. Morgan, J. Trautwein, T. A. Barber, S. L. Stott, S. Maheswaran, R. Kapur, D. A. Haber and M. Toner, Microfluidic, marker-free isolation of circulating tumor cells from blood samples, Nat. Protocols, 2014, 9(3), 694–710 CAS
.
- J.-P. Frimat, M. Becker, Y.-Y. Chiang, U. Marggraf, D. Janasek, J. G. Hengstler, J. Franzke and J. West, A microfluidic array with cellular valving for single cell co-culture, Lab Chip, 2011, 11, 231–237 RSC
.
- L. Mazutis, J. Gilbert, W. Lloyd Ung, D. A. Weitz, A. D. Griffiths and J. A. Heyman, Single-cell analysis and sorting using droplet-based microfluidics, Nat. Protocols, 2013, 8(5), 870–891 CAS
.
- See https://www.fluidigm.com/products/c1-system (consulted on the 1st february 2016).
- P. E. Stenberg, R. P. McEver, M. A. Shuman, Y. V. Jacques and D. F. Bainton, A platelet alpha-granule membrane protein (GMP-140) is expressed on the plasma membrane after activation, J. Cell Biol., 1985, 101(3), 880–886 CrossRef CAS PubMed
.
- E. D. Klootwijk, M. Reichold, A. Helip-Wooley, A. Tolaymat, C. Broeker, S. L. Robinette, J. Reinders, D. Peindl, K. Renner, K. Eberhart, N. Assmann, P. J. Oefner, K. Dettmer, C. Sterner, J. Schroeder, N. Zorger, R. Witzgall, S. W. Reinhold, H. C. Stanescu, D. Bockenhauer, G. Jaureguiberry, H. Courtneidge, A. M. Hall, A. D. Wijeyesekera, E. Holmes, J. K. Nicholson, K. O'Brien, I. Bernardini, D. M. Krasnewich, M. Arcos-Burgos, Y. Izumi, H. Nonoguchi, Y. Jia, J. K. Reddy, M. Ilyas, R. J. Unwin, W. A. Gahl, R. Warth and R. Kleta, Mistargeting of peroxisomal EHHADH and inherited renal fanconi's syndrome, N. Engl. J. Med., 2014, 370(2), 129–138 CrossRef CAS PubMed
. PMID: 24401050.
- A. Sickmann, J. Reinders, Y. Wagner, C. Joppich, R. Zahedi, H. E. Meyer, B. Schönfisch, I. Perschil, A. Chacinska, B. Guiard, P. Rehling, N. Pfanner and C. Meisinger, The proteome of Saccharomyces cerevisiae mitochondria, Proc. Natl. Acad. Sci. U. S. A., 2003, 100(23), 13207–13212 CrossRef CAS PubMed
.
- S. W. Taylor, E. Fahy, B. Zhang, G. M. Glenn, D. E. Warnock, S. Wiley, A. N. Murphy, S. P. Gaucher, R. A. Capaldi, B. W. Gibson and S. S. Ghosh, Characterization of the human heart mitochondrial proteome, Nat. Biotechnol., 2003, 21(3), 281–286 CrossRef CAS PubMed
.
- J. R. Yates III, A. Gilchrist, K. E. Howell and J. J. M. Bergeron, Proteomics of organelles and large cellular structures, Nat. Rev. Mol. Cell Biol., 2005, 6(9), 702–714 CrossRef PubMed
.
- M. Niemann, S. Wiese, J. Mani, A. Chanfon, C. Jackson, C. Meisinger, B. Warscheid and A. Schneider, Mitochondrial outer membrane proteome of Trypanosoma brucei reveals novel factors required to maintain mitochondrial morphology, Mol. Cell. Proteomics, 2012, 12(2), 515–528 Search PubMed
.
- R. P. Zahedi, A. Sickmann, A. M. Boehm, C. Winkler, N. Zufall, B. Schönfisch, B. Guiard, N. Pfanner and C. Meisinger, Proteomic analysis of the yeast mitochondrial outer membrane reveals accumulation of a subclass of preproteins, Mol. Biol. Cell, 2006, 17(3), 1436–1450 CrossRef CAS PubMed
.
- F.-N. Vögtle, J. M. Burkhart, S. Rao, C. Gerbeth, J. Hinrichs, J.-C. Martinou, A. Chacinska, A. Sickmann, R. P. Zahedi and C. Meisinger, Intermembrane space proteome of yeast mitochondria, Mol. Cell. Proteomics, 2012, 11(12), 1840–1852 Search PubMed
.
- H. Lu, S. Gaudet, M. A. Schmidt and K. F. Jensen, A microfabricated device for subcellular organelle sorting, Anal. Chem., 2004, 76(19), 5705–5712 CrossRef CAS PubMed
. PMID: 15456289.
- M. Moschallski, M. Hausmann, A. Posch, A. Paulus, N. Kunz, T. T. Duong, B. Angres, K. Fuchsberger, H. Steuer, D. Stoll, S. Werner, B. Hagmeyer and M. Stelzle, MicroPrep: Chip-based dielectrophoretic purification of mitochondria, Electrophoresis, 2010, 31(15), 2655–2663 CrossRef CAS PubMed
.
- K. Sharma, R. C. J. D'Souza, S. Tyanova, C. Schaab, J. R. Wiśniewski, J. Cox and M. Mann, Ultradeep human phosphoproteome reveals a distinct regulatory nature of Tyr and Ser/Thr-based signaling, Cell Rep., 2014, 8(5), 1583–1594 CrossRef CAS PubMed
.
- Y. Zheng, C. Zhang, D. R. Croucher, M. A. Soliman, N. St-Denis, A. Pasculescu, L. Taylor, S. A. Tate, W. Rod Hardy, K. Colwill, A. Y. Dai, R. Bagshaw, J. W. Dennis, A.-C. Gingras, R. J. Daly and T. Pawson, Temporal regulation of EGF signalling networks by the scaffold protein Shc1, Nature, 2013, 499(7457), 166–171 CrossRef CAS PubMed
.
- F. Beck, J. Geiger, S. Gambaryan, J. Veit, M. Vaudel, P. Nollau, O. Kohlbacher, L. Martens, U. Walter, A. Sickmann and R. P. Zahedi, Time-resolved characterization of cAMP/PKA-dependent signaling reveals that platelet inhibition is a concerted process involving multiple signaling pathways, Blood, 2013, 123(5), e1–e10 CrossRef PubMed
.
- P. Novo, F. Volpetti, V. Chu and J. P. Conde, Control of sequential fluid delivery in a fully autonomous capillary microfluidic device, Lab Chip, 2013, 13, 641–645 RSC
.
- P. Novo, V. Chu and J. P. Conde, Integrated optical detection of autonomous capillary microfluidic immunoassays:a hand-held point-of-care prototype, Biosens. Bioelectron., 2014, 57, 284–291 CrossRef CAS PubMed
.
- E Tavares da Costa, M F. Mora, P A. Willis, C L. do Lago, H Jiao and C D. Garcia, Getting started with open-hardware: Development and control of microfluidic devices, Electrophoresis, 2014, 35(16), 2370–2377 CrossRef PubMed
.
- J. McGrath, M. Jimenez and H. Bridle, Deterministic lateral displacement for particle separation: a review, Lab Chip, 2014, 14(21), 4139–4158 RSC
.
- L. R. Huang, E. C. Cox, R. H. Austin and J. C. Sturm, Continuous particle separation through deterministic lateral displacement, Science, 2004, 304(5673), 987–990 CrossRef CAS PubMed
.
- D. Holmes, G. Whyte, J. Bailey, N. Vergara-Irigaray, A. Ekpenyong, J. Guck and T. Duke, Separation of blood cells with differing deformability using deterministic lateral displacement, Interface Focus, 2014, 4(6), 20140011–20140011 CrossRef PubMed
.
- S. Varma and J. Voldman, A cell-based sensor of fluid shear stress for microfluidics, Lab Chip, 2015, 15(6), 1563–1573 RSC
.
- S. M. Ponik, J. W. Triplett and F. M. Pavalko, Osteoblasts and osteocytes respond differently to oscillatory and unidirectional fluid flow profiles, J. Cell. Biochem., 2007, 100, 794–807 CrossRef CAS PubMed
.
- S. Weinbaum, S. C. Cowin and Y. Zeng, A model for the excitation of osteocytes by mechanical loading-induced bone fluid shear stresses, J. Biomech., 1994, 27, 339–360 CrossRef CAS PubMed
.
- H. A. Pohl and J. S. Crane, Dielectrophoresis of cells, Biophys. J., 1971, 11, 711–727 CrossRef CAS PubMed
.
- D. Das, K. Biswas and S. Das, A microfluidic device for continuous manipulation of biological cells using dielectrophoresis, Med. Eng. Phys., 2014, 36(6), 726–731 CrossRef PubMed
.
- P. Cheng, M. J. Barrett, P. M. Oliver, D. Cetin and D. Vezenov, Dielectrophoretic tweezers as a platform for molecular force spectroscopy in a highly parallel format, Lab Chip, 2011, 11(24), 4248–4259 RSC
.
- A. Salmanzadeh, L. Romero, H. Shafiee, R. C. Gallo-Villanueva, M. A. Stremler, S. D. Cramer and R. V. Davalos, Isolation of prostate tumor initiating cells (TICs) through their dielectrophoretic signature, Lab Chip, 2012, 12, 182–189 RSC
.
- X. Ding, Z. Peng, S.-C. S. Lin, M. Geri, S. Li, P. Li, Y. Chen, M. Dao, S. Suresh and T. J. Huang, Cell separation using tilted-angle standing surface acoustic waves, Proc. Natl. Acad. Sci. U. S. A., 2014, 111(36), 12992–12997 CrossRef CAS PubMed
.
- J. Nam, H. Lim and S. Shin, Manipulation of microparticles using surface acoustic wave in microfluidic systems: a brief review, Korea-Aust. Rheol. J., 2011, 23(4), 255–267 CrossRef
.
- H. Bruus, Acoustofluidics 10: Scaling laws in acoustophoresis, Lab Chip, 2012, 12(9), 1578–1586 RSC
.
- M. A. Burguillos, C. Magnusson, M. Nordin, A. Lenshof, P. Augustsson, M. J. Hansson, E. Elmér, H. Lilja, P. Brundin, T. Laurell and T. Deierborg, Microchannel acoustophoresis does not impact survival or function of microglia, leukocytes or tumor cells, PLoS One, 2013, 8(5), e64233 CAS
.
- P. Li, Z. Mao, Z. Peng, L. Zhou, Y. Chen, P.-H. Huang, C. I. Truica, J. J. Drabick, W. S. El-Deiry, M. Dao, S. Sureshe and T. J. Huang, Acoustic separation of circulating tumor cells, Proc. Natl. Acad. Sci. U. S. A., 2015, 112(16), 4970–4975 CrossRef CAS PubMed
.
- F. D. Giudice, H. Madadi, M. M. Villone, G. D'Avino, A. M. Cusano, R. Vecchione, M. Ventre, P. L. Maffettone and P. A. Netti, Magnetophoresis ‘meets’ viscoelasticity: deterministic separation of magnetic particles in a modular microfluidic device, Lab Chip, 2015, 15, 1912–1922 RSC
.
- T. P. Hunt, D. Issadore and R. M. Westervelt, Integrated circuit/microfluidic chip to programmably trap and move cells and droplets with dielectrophoresis, Lab Chip, 2008, 8(1), 81–87 RSC
.
- D. W. Inglis, R. Riehn, R. H. Austin and J. C. Sturm, Continuous microfluidic immunomagnetic cell separation, Appl. Phys. Lett., 2004, 85(21), 5093–5095 CrossRef CAS
.
- D. J. Collins, T. Alan and A. Neild, Particle separation using virtual deterministic lateral displacement (vDLD), Lab Chip, 2014, 14(9), 1595 RSC
.
- J. P. Beech, P. Jonsson and J. O. Tegenfeldt, Tipping the balance of deterministic lateral displacement devices using dielectrophoresis, Lab Chip, 2009, 9, 2698–2706 RSC
.
- H. Shafiee, J. L. Caldwell and R. V. Davalos, Selective isolation of live/dead cells using contactless dielectrophoresis (cDEP), Lab Chip, 2010, 10, 438–445 RSC
.
- J. D. Adams, P. Thévoz, H. Bruus and H. Tom Soh, Integrated acoustic and magnetic separation in microfluidic channels, Appl. Phys. Lett., 2009, 95(25), 254103 CrossRef PubMed
.
- See Fig. 1 of “chapter 4: Rates and duration” from “cell biology by the numbers” by ron milo and rob phillips, also available online on http://book.bionumbers.org/rates-and-duration-introduction/ (consulted on the 3rd of december 2015).
- M.-H. Wu, S.-B. Huang and G.-B. Lee, Microfluidic cell culture systems for drug research, Lab Chip, 2010, 10, 939–956 RSC
.
- I. Meyvantsson and D. J. Beebe, Cell culture models in microfluidic systems, Annu. Rev. Anal. Chem., 2008, 1(1), 423–449 CrossRef CAS PubMed
. PMID: 20636085.
- L. Chingozha, M. Zhan, C. Zhu and H. Lu, A generalizable, tunable microfluidic platform for delivering fast temporally varying chemical signals to probe single-cell response dynamics, Anal. Chem., 2014, 86(20), 10138–10147 CrossRef CAS PubMed
. PMID: 25254360.
- A. H. C. Ng, M. Dean Chamberlain, H. Situ, V. Lee and A. R. Wheeler, Digital microfluidic immunocytochemistry in single cells, Nat. Commun., 2015, 6, 1–12 Search PubMed
.
- R. B. Fair, Digital microfluidics: is a true lab-on-a-chip possible?, Microfluid. Nanofluid., 2007, 3(3), 245–281 CrossRef CAS
.
- A. M. Hirsch, C. A. Rivet, B. Zhang, M. L. Kemp and H. Lu, Parallel multi-time point cell stimulation and lysis on-chip for studying early signaling events in T cell activation, Lab Chip, 2009, 9, 536–544 RSC
.
- F. A. Solari, M. Dell'Aica, A. Sickmann and R. P. Zahedi, Why phosphoproteomics is still a challenge, Mol. BioSyst., 2015, 11, 1487–1493 RSC
.
- N.-T. Nguyen and Z. Wu, Micromixers - a review, J. Micromech. Microeng., 2005, 15(2), R1–R16 CrossRef
.
- B. D. Plouffe and S. K. Murthy, Perspective on microfluidic cell separation: A solved problem?, Anal. Chem., 2014, 86(23), 11481–11488 CrossRef CAS PubMed
.
- G. M. Whitesides, Cool, or simple and cheap? Why not both?, Lab Chip, 2013, 13, 11–13 RSC
.
| This journal is © The Royal Society of Chemistry 2016 |