A comparison of two classic Pb2+-dependent RNA-cleaving DNAzymes†
Runjhun
Saran
and
Juewen
Liu
*
Department of Chemistry, Waterloo Institute for Nanotechnology, Waterloo, Ontario, Canada N2L 3G1. E-mail: liujw@uwaterloo.ca
First published on 11th January 2016
Abstract
Pb2+ is a very important metal cofactor in DNAzyme catalysis. GR5 is the first reported DNAzyme, and 17E is the most thoroughly studied. Both have the highest activity with Pb2+ and are by far the fastest RNA-cleaving DNAzymes. GR5 reacts only with Pb2+ while 17E is also active with a number of other divalent metal ions. It is also interesting to note that Pb2+ shows activity with most RNA-cleaving DNAzymes. To understand these Pb2+-dependent DNAzymes and the occurrence of DNAzyme sequences, herein systematic mutation studies are performed on GR5. A comparison with 17E is also made. The A6, G7, C13, and G14 positions in 17E have been previously established to be crucial and we report A6, G7, C14, and G15 in GR5 to have the same role. The guanine at the cleavage site dinucleotide junction of the substrate strand is also mutated to hypoxanthine, 2-aminopurine, and adenine. Again, both enzymes show the same trend of activity change. Our results suggest that both DNAzymes have a similar binding pocket for Pb2+. The reason for Pb2+ being active in many DNAzymes is attributed to its simple binding motif requirement. Finally, we propose that 17E is a special form of GR5. They both have the simple sequence requirements needed for Pb2+-dependent activity, but 17E has additional motifs making it active also with other divalent metal ions.
Introduction
DNAzymes are catalysts made of DNA. They are attractive molecules due to excellent stability, high catalytic efficiency, programmability, and ease of modification.1–5 DNAzymes have found a diverse range of applications in anti-viral research,6 nanotechnology,7 organic synthesis,8 and biosensor development.9,10 With limited chemical functionality and a highly negatively charged backbone, DNAzymes often need to recruit metal ions for catalysis and different metal ions may prefer different DNA sequences.3,11,12 To date, a number of RNA-cleaving DNAzymes have been reported to be specific for Pb2+,13–15 Zn2+,16,17 UO22+,18 Hg2+,19,20 trivalent lanthanides,21–23 Na+,24 and other metal ions.25,26This field has grown tremendously by studying the Pb2+-specific DNAzymes. The first DNAzyme (named GR5) was selected in the presence of Pb2+ in 1994.13 GR5 had not attracted much attention then, since the initial DNAzyme research was focused on RNA cleavage (e.g. anti-viral applications). GR5 cannot cleave all-RNA substrates; it only cleaves DNA/RNA chimera.13 Three years later, the landmark paper by Santoro and Joyce reported two general-purpose RNA-cleaving DNAzymes.6 One of them is called the 8–17 DNAzyme. A variant of it, 17E, is highly active with Pb2+.14,16 Since then, 17E has been coupled to many signaling mechanisms to develop biosensors.14,27–30 The GR5 DNAzyme recently revived as a biosensor,31,32 since the Lu group reported its exceptionally high selectivity for Pb2+ (much better than 17E).33 It is also interesting to note that Pb2+ is active with many other DNAzymes,21,23,34,35 especially at above neutral pH.
We reason that GR5 and 17E form a good pair for comparison, since both have the highest activity with Pb2+. Both are small DNAzymes with only ∼15 nucleotides in the catalytic core and share the same substrate sequence. Extensive biochemical,6,36–39 and spectroscopic studies have been carried out on 17E.40–44 In particular, many active 17E mutants have been identified.38,45,46 However, relatively little is known regarding GR5.13,15,32,33 A closer examination of these two DNAzymes indicates that they contain similar conserved nucleotides. Therefore, an interesting question is whether they work with the same mechanism (e.g. they are just mutants). Another intriguing observation is that 17E occurred from at least six independent selections,6,16,45,47–50 but GR5 was reported only once.13 In this work, we aim to answer these fundamental questions based on a side-by-side comparison.
Materials and methods
Chemicals
All the enzyme strands and their mutants were purchased from Eurofins (Huntsville, AL). The FAM-labeled substrate was from Integrated DNA Technologies Inc. and purified by HPLC (Coralville, IA). The sequences of the DNA used in this work are listed in Table S1.† All the metals and buffers were purchased from Sigma-Aldrich and Amresco respectively.Biochemical assays
To prepare the wild-type and mutant DNAzymes, the respective enzyme (5 μM) and FAM-labeled substrate strands (3.3 μM) were mixed in buffer and the tubes were incubated in a water bath at 85 °C for 1 min and then slowly cooled to room temperature. Then the DNAzyme complex was further diluted with buffer and the final concentration of the enzyme strand is 0.7 μM for the assay. The assays for the DNAzymes bearing the substrate strand mutations (with wild-type enzyme strand) were carried out in 50 mM MES (pH 6.0), 25 mM NaCl, while the assays with the enzyme strand mutations (with wild type substrate strand) were carried out in 50 mM HEPES (pH 7.6), 25 mM NaCl. To determine the DNAzyme activity, metal ion solution (final concentration of 10 mM Mg2+ or 10 μM Pb2+) was added to initiate the reaction. At designated time points (from 15 s to 2 h), a small aliquot was taken out and the reaction was quenched by mixing with gel loading buffer (5 mM EDTA, 8 M urea). The samples were analyzed using denaturing polyacrylamide gel electrophoresis (15%). Gel images were taken with a Bio-Rad ChemiDoc MP imaging system and DNA bands were quantified using software Image Lab 4.1. Two independent experiments were performed with each DNAzyme construct, and the data obtained were fitted according to the first-order rate equation Yt = Y0 + a(1 − e−bx) using SigmaPlot12.5 where Yt and Y0 is cleavage fraction at a given reaction time t and 0 min respectively and b is the observed rate constant.Results and discussion
Activity of the 17E mutants
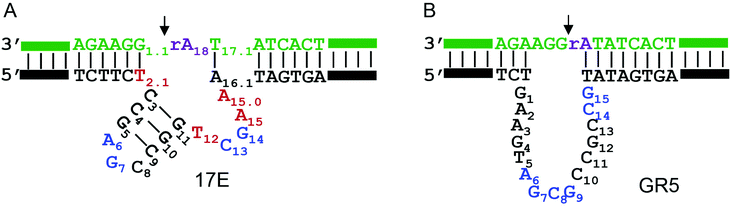
The secondary structures of the 17E and GR5 DNAzymes are shown in Fig. 1A and B, respectively. They share the same substrate sequence (in green). The cleavage junction is 5′rAdG3′ (see the arrowhead). Here, rA means ribo-adenosine, which is the only RNA linkage in the substrate. Each enzyme binds the substrate via two base paired arms, and the catalytically important nucleotides are located in the bulged loops. In the presence of Pb2+, the phosphodiester bond linking, the rA·G is cleaved. With extensive biochemical studies, four highly conserved nucleotides have been identified in 17E (A6, G7, C13, and G14, highlighted in blue, Fig. 1A). Mutation to any of them leads to over 100-fold decrease in activity.38,45,46The 17E DNAzyme has the highest activity with Pb2+, but it is also active with high concentrations of many other metals such as Mg2+, Ca2+, Mn2+, Cd2+ and Zn2+. For 17E assays, Mg2+ has been the most common metal cofactor.6,36,51 However, GR5 has no activity with 50 mM to 1 M Mg2+.15,33 For a meaningful comparison, we employed Pb2+ as the common metal here. Pb2+ has a much higher apparent binding constant and can be used at much lower concentrations.37 This would confer less perturbation to the properties of DNA. Pb2+ also has a well-established nucleotide coordination chemistry,52–54 making it easier to probe metal binding sites.
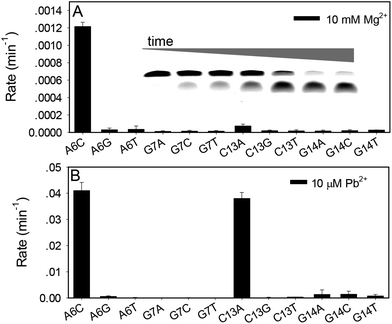
Although very rich information is available for 17E with Mg2+, we need to first confirm whether these can be applied to Pb2+. For this purpose, a mutation study was performed with 17E in the presence of Mg2+ and Pb2+. Each of the four highly conserved nucleotides (highlighted in blue in Fig. 1A) was mutated and the cleavage rate was measured. The wild-type 17E has a rate of 0.44 min−1 with 10 mM Mg2+ at pH 7.6 and a gel image is shown in the inset of Fig. 2A. The fraction of cleavage was calculated at each time point to extract the kinetic information. All the mutations significantly reduced activity (Fig. 2A). Even the most active A6C mutant, (e.g. the adenine at position 6 mutated to cytosine) has a rate of only 0.0012 min−1, an ∼370-fold drop compared to the wild type. Other mutants barely had any activity, which is consistent with the literature on Mg2+.36,37,45 The activity of the wild-type 17E is much higher with 10 μM Pb2+, which is estimated to be greater than >11.9 min−1 at pH 7.6. Note that the rate is too fast to be accurately measured by manual pipetting. With such high activity in the presence of Pb2+, two mutations, A6C and C13A showed moderate activity (Fig. 2B). Again, even for the most active mutant, the rate is still >300-fold less than the wild-type. Therefore, the nucleotides important for Mg2+ are also important for Pb2+. It is also interesting to note that the C13A mutant is relatively more active with Pb2+, which hints that Pb2+ might have better tolerance to mutations.
GR5 mutation studies
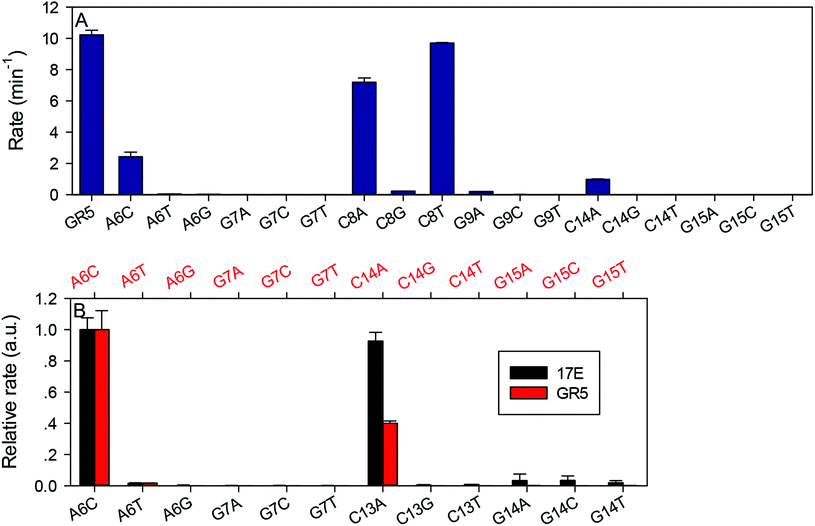
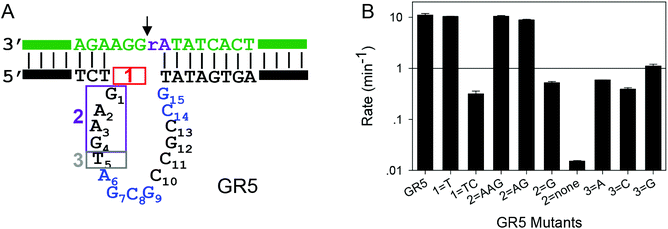
After confirming the important nucleotides for Pb2+ in 17E, we next studied GR5. So far, no systematic mutation studies have been carried out on GR5. From its sequence alignment,13 six highly conserved nucleotides were identified in the original paper by Breaker and Joyce (Fig. 1B, in blue). These nucleotides segregate into two groups: four (A6G7C8G9) in the middle and two (C14G15) towards the end. The other nucleotides appear to be less important since their sequence and length can vary while still retaining activity.13 Therefore, we first focused on these six conserved nucleotides. It is interesting to note that 17E has the conserved AG followed by CG. In GR5, the conserved nucleotides can be considered to be AG followed by CG and another CG. Therefore, it is possible that 17E and GR5 have similar Pb2+ binding pockets and even similar mechanisms.We systematically mutated these six conserved nucleotides in GR5 (Fig. 3A). The wild-type GR5 has a rate greater than 10.2 min−1 at pH 7.6 with 10 μM Pb2+. The A6C mutant retained a high activity (∼2 min−1), but any other mutations to A6 abolished activity. Mutations made to G7 abolished the activity as well. This trend is exactly the same as that in 17E for its A6 and G7. Therefore, these two nucleotides are likely to play the same role in these two DNAzymes. For GR5, C8 can be mutated to A or T (activity almost fully retained) but not G. Any mutations made to G9 abolished the activity. This pattern is however different from that in 17E for its C13G14. On the other hand, the activities of C14G15 mutants in GR5 are comparable with that of the C13G14 in 17E, where only the C14A mutant retained partial activity. We plotted their relative rates (normalized to the A6C mutant of both DNAzymes) in Fig. 3B. Using the 17E mutants as a benchmark, we reason that A6G7 and C14G15 in GR5 serve similar catalytic roles to those corresponding nucleotides in 17E. It is interesting to note that the GR5 mutants have higher activity: the rates of the three most active GR5 mutants are greater than 1 min−1, while the 17E mutants under the same conditions are less than 0.05 min−1. We compared the wild-type DNAzymes at pH 5.5, and GR5 is ∼4-fold more active, which partially accounts for the 20-fold difference in rate for their mutants. To test whether the mutations can change the folding of the GR5 core, we analyzed all the mutants using Mfold.55 Under our experimental conditions (25 mM NaCl), the GR5 DNAzyme folds into a structure with a two base-pair hairpin (Fig. S1†). However, these two base pairs involve the highly conserved C14G15, and therefore, are unlikely to be real. For this reason, we herein followed the simple loop GR5 secondary structure as shown in the original paper.13 According to Mfold predictions, most mutants follow the same structure as the original GR5, and the few that showed alternative folding are listed in Fig. S1.† None of these listed mutants are active. For them, we cannot rule out misfolding to be a reason for their inactivity.
Cleavage junction mutations
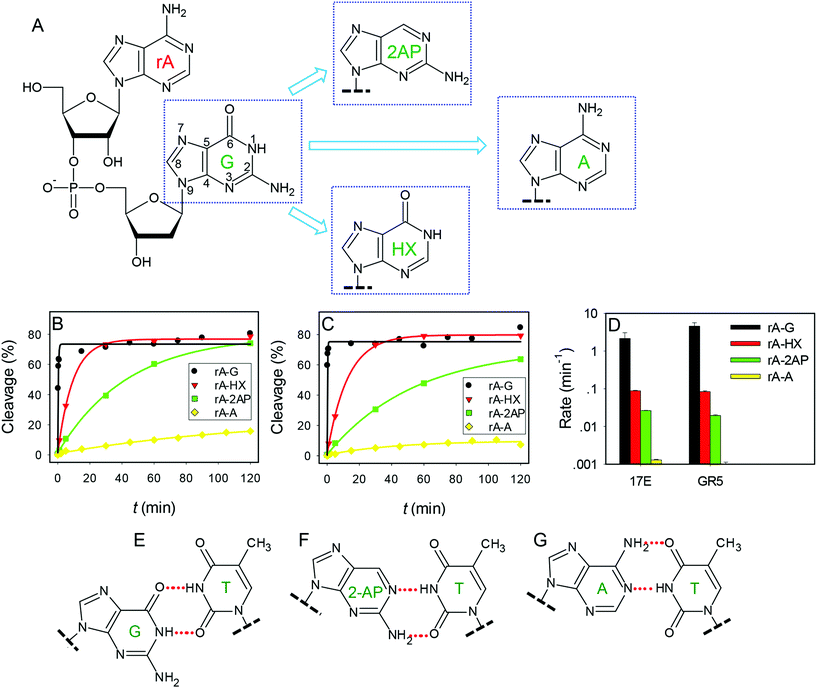
The above work focused only on the enzyme strands. Previous biochemical assays indicated that the composition of the chimeric dinucleotide junction of the substrate (e.g. rA18·G1.1) is also important.38,45 In addition to the four highly conserved nucleotides in 17E, the ones marked in red are also conserved (T2.1, T12, A15 and A15.0) to effectively cleave the rA18·G1.1 junction. With other cleavage junctions, these nucleotides may need to be changed accordingly to maintain optimal activity.38 Therefore, if the enzyme core sequence is fixed, changing the dinucleotide junction composition may have a huge influence on enzyme activity. This might be useful for further comparing these two DNAzymes.While the structure of G and A are quite similar (both are purines), direct switching from G to A has obscured the exact chemical role of the functional groups of G (Fig. 4A). Therefore, in this work, we mutated G to hypoxanthine (HX) and 2-aminopurine (2AP) to probe the functional groups one at a time. HX differs from guanine by the 2-amino group, while 2AP differs from guanine by lacking the 6-keto group. The assays were carried out at pH 6.0 to obtain measurable rates for the wild-type enzymes.32 The original rA·G junction has the highest rate for both DNAzymes (Fig. 3B and C, black traces). The rates of rA·HX were more than one order of magnitude slower compared to the original rA·G junction (Fig. 4D), and rA·2AP was even slower in both cases. Finally, the rates were the slowest with the rA·A junction, dropping by over 3 orders of magnitude compared to the original rA·G junction. These identical patterns further suggest a similar mechanism of these two DNAzymes, since all the modifications brought about the same trend in activity change.
Two reasons may explain the guanine mutation activity pattern: either disruption of metal binding or the enzyme structure. Pb2+ has well-defined coordination sites in the nucleobases. As far as guanine is concerned, the N7 and O6 positions are important for Pb2+ binding.52,53 Since adenine and guanine have drastically different rates (>1000-fold), this argues against the importance of the N7 position (both have the N7 position available). Since the HX mutant is only slightly more active than the 2AP mutant, it is unlikely that the O6 oxygen is involved in Pb2+ binding. Otherwise, a much more significant change is expected.36,56 Therefore, this guanine should play more of a structural role. For example, it may base pair with other nucleotides to stabilize secondary or tertiary structures for catalysis.
This guanine can formally pair with the thymine in the 17E to form a wobble pair, which has been confirmed to be crucial for cleaving the rA·G junction.6,37,38 Other 17E mutants have been evolved to cleave other types of junctions, which lack the wobble pair.38 Therefore, this wobble is not absolutely conserved, which also confirms its structural role.
In a typical wobble pair, the O6 and N1 positions in guanine are hydrogen bonded with thymine (Fig. 4E). By changing the guanine to HX, the wobble formation is maintained. The drop of rate by over 10-fold suggests that the amino group might be involved in additional stabilization roles. With the 2AP junction, the same hydrogen bonding can still be formed with the wobble face of the thymine (Fig. 4F), but this requires slightly more structural changes explaining the drop of rates by ∼100–300 fold. On the other hand, with adenine (which confers the cumulative effect of both the HX and 2-AP mutations), the Watson–Crick face of the thymine has to be used (Fig. 4G), and the structure perturbation is more significant, explaining its lowest rate.
It is interesting to note that GR5 has exactly the same trend as 17E for the junction mutations (Fig. 4D). Based on the above discussions, if the activity for different cleavage junctions is mainly related to the T2.1 in 17E, there might be a corresponding thymine in GR5. Interestingly, only one thymine, T5, resides in the GR5 enzyme loop. A careful examination of the aligned sequences resulting from the Pb2+ selection in the original paper, reveals that this thymine is conserved in 70% of the sequences.13 However, T5 in GR5 is quite far away from the cleavage junction based on the secondary structure.
Other mutations to GR5
To test the function of T5 in GR5, a few more mutants were studied. In 17E almost all the nucleotides in the substrate strand are base paired with the enzyme, while in GR5, three free nucleotides exist on the 3′ side of rA. We also want to test the importance of these unpaired nucleotides in GR5. First, we gradually pair them up by extending the enzyme. In the box marked 1 in Fig. 5A, we added T or TC. Both the extended GR5 mutants still retained high activity (Fig. 5B), suggesting that the unpaired nucleotides might be useful for providing flexibility but without a specific chemical role. We then gradually deleted the nucleotides before T5 (the nucleotides in box 2), and they also did not significantly affect the activity, unless all the four nucleotides in box 2 were deleted (Fig. 5B). Finally, T5 in box 3 was mutated to other nucleotides and the activity was not much affected by this as well (Fig. 5B). These experiments suggest that T5 itself is not critically important, thus arguing against the speculation that T5 in GR5 plays a similar role to T2.1 in 17E. Therefore, some other nucleotides in GR5 might interact with the cleavage site guanine in the substrate, and this will be a topic of further studies.Further discussions
GR5 and 17E are two highly efficient and widely used Pb2+-dependent DNAzymes. Both have the highest activity with Pb2+,13,33,37 and they share the same substrate. In this work, we performed mutation studies on both DNAzymes. Our data suggests that these two Pb2+-specific DNAzymes have the same catalytic mechanism in the presence of Pb2+. Aside from these similarities, a number of differences also exist. For example, 17E is also active with many other metal ions, but GR5 is active only with Pb2+.Using Pb2+ for RNA-cleaving ribozymes has been reported since early 1990s,57–59 and the study of Pb2+ for RNA hydrolysis can be dated back to an even earlier time.52 This might be the reason for Breaker and Joyce to pick Pb2+ for their first DNAzyme selection,13 from which GR5 was reported. Under physiological conditions, the available free Pb2+ concentration is close to zero and Mg2+ is the most important cation. To target RNA, most subsequent selections were carried out with Mg2+ or other physiologically relevant metals. Under various conditions, 17E was the main outcome (e.g. with Mg2+,6,48 Zn2+,16 Mg2+/Mn2+,45 Cd2+,50 or Mg2+/Mn2+/Cu2+.49). It is easy to understand that GR5 did not appear again, since GR5 is completely inactive with these metals.
A related question is whether 17E can appear in Pb2+-dependent selections. 17E is also highly active with Pb2+ and it appears to be a good solution to such a selection. In particular, 17E has appeared in a number of different selections. The reasons for 17E being so popular were attributed to its small size and tolerance to mutations.38,45 It has not been reported whether any 17E motifs occurred in the original Pb2+-dependent selection.13 The paper presented 20 sequences and only one of them 5′CTGCTACCAGCGGTACGAAATAGT3′, seems to be falling under the observed structural variations for 17E with the 5′rAG3′ cleavage junction as defined by Li and co-workers.45
If we define GR5 to be containing the five conserved nucleotides as discussed above, the possibility of having it in a 15-nucleotide stretch is (¼)5. The actual possibility is higher since the five nucleotides in the middle can be at different positions. On the other hand, at least 8 nucleotides have to be in fixed positions in 17E. Apart from the 4 for activity, the other four are required for matching with the substrate cleavage junction (e.g. those in red in Fig. 1A). A few others serve the so-called ‘chaperone’ role to assist DNAzyme folding.51 Essentially, most nucleotides in 17E have their own role and position.36–38,51 Therefore, the chance of having the right sequence is maximally (¼)8, which is more than 64-fold lower in possibility than GR5. This can explain the much higher appearance of GR5 instead of 17E in the Pb2+ selection. It is interesting to note that GR5 was originally selected in the presence of 50 mM Mg2+ and 1 mM Pb2+. Therefore, in theory, 17E would have had the opportunity to be selected (it is Mg2+ dependent). This is another evidence that GR5 has a higher chance to be selected.
17E is a special DNAzyme that can utilize many metal ions for catalysis. The recurrence of 17E in many selections indicates that very few solutions are available for DNAzymes to catalyze RNA hydrolysis in the presence of these metals (e.g. 17E appears to be the main solution). Therefore, 17E is optimized for other metal ions. Since 17E happens to contain AG and CG conserved nucleotides, it affords high activity with Pb2+. On the other hand, GR5 is optimized for Pb2+ and has no activity with other metals. We recently performed a new in vitro selection experiment in the presence of Pb2+, and many new DNAzymes were discovered.15 The ones that are active with Pb2+ are found to be inactive with Mg2+. This further supports the more stringent sequence requirements for Mg2+. In a sense, we may consider 17E to be a special form of GR5. More experiments are needed to test this hypothesis and to fully elucidate their relation.51 For example, we need to first precisely define GR5 in terms of its primary and secondary structural features. To do this, more comprehensive mutation studies are needed.
In addition to these two Pb2+-specific DNAzymes, a few other RNA-cleaving enzymes are also known. For example, the UO22+-dependent DNAzyme has some Pb2+ activity at pH 7, although the activity at pH 5.5 is very low.35 We recently reported a lanthanide-dependent DNAzyme that is also active with Pb2+.21 In the well-known leadzyme (a ribozyme), the enzyme strand contains only two unpaired nucleotides (AG) and it is quite active with Pb2+.58 Therefore, Pb2+ is a highly efficient metal to assist RNA cleavage.
So then what is so special about Pb2+? Pb2+ has long been known to be highly effective in RNA hydrolysis. The hydrated Pb2+ has a pKa value of 7.2–7.8 and its deprotonated species can interact with the 2′-OH on the rA sugar ring to assist the nucleophilic attack reaction.53 For comparison, other metals have quite different pKa values (8.5 for Eu3+, 9.0–9.6 for Zn2+, and 11.4 for Mg2+).60,61 This special property has rendered Pb2+ to be highly effective for RNA cleavage and the DNAzyme provides a scaffold to efficiently utilize Pb2+ for this purpose.
While the main goal of this work is to gain the fundamental understanding of GR5, it may also have implications for practical applications. We now have a much better understanding on the sequence/activity relationship of GR5. Although none of the mutants appear to be faster than the original GR5, such knowledge may provide freedom for biosensor design and the development of aptazymes.
Conclusions
In summary, we performed a side-by-side comparison of two Pb2+-dependent DNAzymes: GR5 and 17E. They share the same substrate sequence and have similar conserved nucleotides in the enzyme catalytic core. By performing mutations in the enzyme core, we identified the similar roles of four highly conserved nucleotides in both enzymes and they share the same activity pattern. By mutating guanine at the cleavage junction to adenine, HX and 2AP, we further confirmed that these two enzymes have the same mechanism in the presence of Pb2+. This guanine has been suggested to play a structural role instead of a metal binding one. In addition, we compared Mg2+ and Pb2+ for the activity of 17E as a function of enzyme mutants. This study indicates that the nucleotide important for Mg2+ catalysis is also important for the Pb2+-dependent activity. Based on this study, we rationalized the general activity of Pb2+ in many different DNAzymes and proposed that 17E may be considered to be a special form of GR5, explaining the very high activity of 17E in the presence of Pb2+.Acknowledgements
Funding for this work is from the Natural Sciences and Engineering Research Council of Canada (NSERC).References
- R. R. Breaker, Nat. Biotechnol., 1997, 15, 427–431 CrossRef CAS PubMed.
- D. Sen and C. R. Geyer, Curr. Opin. Chem. Biol., 1998, 2, 680–687 CrossRef CAS PubMed.
- Y. Lu, Chem. – Eur. J., 2002, 8, 4588–4596 CrossRef CAS.
- K. Schlosser and Y. F. Li, Chem. Biol., 2009, 16, 311–322 CrossRef CAS PubMed.
- S. K. Silverman, Angew. Chem., Int. Ed., 2010, 49, 7180–7201 CrossRef CAS PubMed.
- S. W. Santoro and G. F. Joyce, Proc. Natl. Acad. Sci. U. S. A., 1997, 94, 4262–4266 CrossRef CAS.
- Y. Lu and J. Liu, Acc. Chem. Res., 2007, 40, 315–323 CrossRef CAS PubMed.
- S. K. Silverman, Acc. Chem. Res., 2009, 42, 1521–1531 CrossRef CAS PubMed.
- J. Liu, Z. Cao and Y. Lu, Chem. Rev., 2009, 109, 1948–1998 CrossRef CAS PubMed.
- X.-B. Zhang, R.-M. Kong and Y. Lu, Annu. Rev. Anal. Chem., 2011, 4, 105–128 CrossRef CAS PubMed.
- W. L. Ward, K. Plakos and V. J. DeRose, Chem. Rev., 2014, 114, 4318–4342 CrossRef CAS PubMed.
- R. K. O. Sigel and A. M. Pyle, Chem. Rev., 2007, 107, 97 CrossRef CAS PubMed.
- R. R. Breaker and G. F. Joyce, Chem. Biol., 1994, 1, 223–229 CrossRef CAS PubMed.
- J. Li and Y. Lu, J. Am. Chem. Soc., 2000, 122, 10466–10467 CrossRef CAS.
- R. Saran, Q. Chen and J. Liu, J. Mol. Evol., 2015, 1–10 Search PubMed.
- J. Li, W. Zheng, A. H. Kwon and Y. Lu, Nucleic Acids Res., 2000, 28, 481–488 CrossRef CAS PubMed.
- S. W. Santoro, G. F. Joyce, K. Sakthivel, S. Gramatikova and C. F. Barbas III, J. Am. Chem. Soc., 2000, 122, 2433–2439 CrossRef CAS PubMed.
- J. Liu, A. K. Brown, X. Meng, D. M. Cropek, J. D. Istok, D. B. Watson and Y. Lu, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 2056–2061 CrossRef CAS PubMed.
- J. Liu and Y. Lu, Angew. Chem., Int. Ed., 2007, 46, 7587–7590 CrossRef CAS PubMed.
- M. Hollenstein, C. Hipolito, C. Lam, D. Dietrich and D. M. Perrin, Angew. Chem., Int. Ed., 2008, 47, 4346–4350 CrossRef CAS PubMed.
- P.-J. J. Huang, J. Lin, J. Cao, M. Vazin and J. Liu, Anal. Chem., 2014, 86, 1816–1821 CrossRef CAS PubMed.
- P.-J. J. Huang, M. Vazin, Ż. Matuszek and J. Liu, Nucleic Acids Res., 2015, 43, 461–469 CrossRef CAS PubMed.
- P.-J. J. Huang, M. Vazin and J. Liu, Anal. Chem., 2014, 86, 9993–9999 CrossRef CAS PubMed.
- S.-F. Torabi, P. Wu, C. E. McGhee, L. Chen, K. Hwang, N. Zheng, J. Cheng and Y. Lu, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 5903–5908 CrossRef CAS PubMed.
- Z. Liu, S. H. J. Mei, J. D. Brennan and Y. Li, J. Am. Chem. Soc., 2003, 125, 7539–7545 CrossRef CAS PubMed.
- P.-J. J. Huang and J. Liu, Anal. Chem., 2014, 86, 5999–6005 CrossRef CAS PubMed.
- J. Liu and Y. Lu, J. Am. Chem. Soc., 2003, 125, 6642–6643 CrossRef CAS PubMed.
- Y. Xiao, A. A. Rowe and K. W. Plaxco, J. Am. Chem. Soc., 2007, 129, 262 CrossRef CAS PubMed.
- H. Wang, Y. Kim, H. Liu, Z. Zhu, S. Bamrungsap and W. Tan, J. Am. Chem. Soc., 2009, 131, 8221–8226 CrossRef CAS PubMed.
- J. Liu and Y. Lu, J. Am. Chem. Soc., 2005, 127, 12677–12683 CrossRef CAS PubMed.
- X. H. Zhao, R. M. Kong, X. B. Zhang, H. M. Meng, W. N. Liu, W. H. Tan, G. L. Shen and R. Q. Yu, Anal. Chem., 2011, 83, 5062–5066 CrossRef CAS PubMed.
- P.-J. J. Huang and J. Liu, Chem. Commun., 2014, 50, 4442–4444 RSC.
- T. Lan, K. Furuya and Y. Lu, Chem. Commun., 2010, 46, 3896–3898 RSC.
- P. Bruesehoff, J. J. Li, A. J. Augustine and Y. Lu, Comb. Chem. High Throughput Screening, 2002, 5, 327–335 CrossRef CAS PubMed.
- A. Brown, in Chemistry, University of Illinois at Urbana-Champaign, Urbana, 2006 Search PubMed.
- A. Peracchi, M. Bonaccio and M. Clerici, J. Mol. Biol., 2005, 352, 783–794 CrossRef CAS PubMed.
- A. K. Brown, J. Li, C. M. B. Pavot and Y. Lu, Biochemistry, 2003, 42, 7152–7161 CrossRef CAS PubMed.
- K. Schlosser, J. Gu, L. Sule and Y. F. Li, Nucleic Acids Res., 2008, 36, 1472–1481 CrossRef CAS PubMed.
- G. S. Sekhon and D. Sen, Biochemistry, 2010, 49, 9072–9077 CrossRef CAS PubMed.
- J. Liu and Y. Lu, J. Am. Chem. Soc., 2002, 124, 15208–15216 CrossRef CAS PubMed.
- H. K. Kim, I. Rasnik, J. W. Liu, T. J. Ha and Y. Lu, Nat. Chem. Biol., 2007, 3, 762–768 CrossRef PubMed.
- H.-K. Kim, J. Liu, J. Li, N. Nagraj, M. Li, C. M. B. Pavot and Y. Lu, J. Am. Chem. Soc., 2007, 129, 6896–6902 CrossRef CAS PubMed.
- H.-K. Kim, J. Li, N. Nagraj and Y. Lu, Chem. – Eur. J, 2008, 14, 8696–8703 CrossRef CAS PubMed.
- D. Mazumdar, N. Nagraj, H.-K. Kim, X. Meng, A. K. Brown, Q. Sun, W. Li and Y. Lu, J. Am. Chem. Soc., 2009, 131, 5506–5515 CrossRef CAS PubMed.
- R. P. G. Cruz, J. B. Withers and Y. Li, Chem. Biol., 2004, 11, 57–67 CrossRef CAS PubMed.
- K. Schlosser and Y. Li, ChemBioChem, 2010, 11, 866–879 CrossRef CAS PubMed.
- A. Peracchi, J. Biol. Chem., 2000, 275, 11693–11697 CrossRef CAS PubMed.
- D. Faulhammer and M. Famulok, Angew. Chem., Int. Ed., 1996, 35, 2837–2841 CrossRef CAS.
- K. Schlosser and Y. Li, Biochemistry, 2004, 43, 9695–9707 CrossRef CAS PubMed.
- A. Kasprowicz, K. Stokowa-Soltys, J. Wrzesinski, M. Jezowska-Bojczuk and J. Ciesiolka, Dalton Trans., 2015, 44, 8138–8149 RSC.
- B. Wang, L. Cao, W. Chiuman, Y. Li and Z. Xi, Biochemistry, 2010, 49, 7553–7562 CrossRef CAS PubMed.
- R. S. Brown, B. E. Hingerty, J. C. Dewan and A. Klug, Nature, 1983, 303, 543–546 CrossRef CAS PubMed.
- R. K. O. Sigel and H. Sigel, Acc. Chem. Res., 2010, 43, 974–984 CrossRef CAS PubMed.
- C. P. Da Costa and H. Sigel, Inorg. Chem., 2000, 39, 5985–5993 CrossRef CAS PubMed.
- M. Zuker, Nucleic Acids Res., 2003, 31, 3406–3415 CrossRef CAS PubMed.
- A. R. Fersht and L. Serrano, Curr. Opin. Struct. Biol., 1993, 3, 75–83 CrossRef CAS.
- T. Pan and O. C. Uhlenbeck, Biochemistry, 1992, 31, 3887–3895 CrossRef CAS PubMed.
- T. Pan and O. C. Uhlenbeck, Nature, 1992, 358, 560–563 CrossRef CAS PubMed.
- T. Pan, B. Dichtl and O. C. Uhlenbeck, Biochemistry, 1994, 33, 9561–9565 CrossRef CAS PubMed.
- D. T. Richens, The Chemistry of Aqua Ions, John Wiley and Sons, New York, 1997 Search PubMed.
- J. Burgess, Metal ions in solution, Ellis Horwood Ltd., Chichester, 1978 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Mfold predicted secondary structures of the GR5 DNAzyme and mutants. See DOI: 10.1039/c5qi00125k |
| This journal is © the Partner Organisations 2016 |