 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Protein-engineering of an amine transaminase for the stereoselective synthesis of a pharmaceutically relevant bicyclic amine†
Martin S.
Weiß
a,
Ioannis V.
Pavlidis
ab,
Paul
Spurr
c,
Steven P.
Hanlon
c,
Beat
Wirz
c,
Hans
Iding
*c and
Uwe T.
Bornscheuer
*a
aDept of Biotechnology and Enzyme Catalysis, Institute of Biochemistry, University of Greifswald, Felix-Hausdorff-Str. 4, D-17489 Greifswald, Germany. E-mail: uwe.bornscheuer@uni-greifswald.de
bGroup of Biotechnology, Dept of Biochemistry, University of Kassel, Heinrich-Plett-Str. 40, D-34132 Kassel, Germany
cProcess Research and Development, Biocatalysis, F. Hoffmann-La Roche Ltd., Grenzacher Str.124, 4070 Basel, Switzerland. E-mail: hans.iding@roche.com
First published on 7th October 2016
Abstract
Application of amine transaminases (ATAs) for stereoselective amination of prochiral ketones represents an environmentally benign and economically attractive alternative to transition metal catalyzed asymmetric synthesis. However, the restrictive substrate scope has limited the conversion typically to non-sterically demanding scaffolds. Recently, we reported on the identification and design of fold class I ATAs that effect a highly selective asymmetric synthesis of a set of chiral aromatic bulky amines from the corresponding ketone precursors in high yield. However, for the specific amine synthetic approach extension targeted here, the selective formation of an exo- vs. endo-isomer, these biocatalysts required additional refinement. The chosen substrate (exo-3-amino-8-aza-bicyclo[3.2.1]oct-8-yl-phenyl-methanone), apart from its pharmacological relevance, is a demanding target for ATAs as the bridged bicyclic ring provides substantial steric challenges. Protein engineering combining rational design and directed evolution enabled the identification of an ATA variant which catalyzes the specific synthesis of the target exo-amine with >99.5% selectivity.
Introduction
Transaminases are pyridoxal-5′-phosphate (PLP) dependent enzymes that catalyse the transfer of an amino group of a donor amine to the carbonyl carbon atom of an α-keto acid, ketones, or an aldehyde.1 In the past decade, amine transaminases (ATAs) came into focus as efficient biocatalysts for the preparation of optically pure amines which represent highly valuable key intermediates or products in the pharmaceutical, chemical, and agricultural sectors.2 However, for application as suitable biocatalysts for stereoselective amination, extensive protein engineering of ATAs is required in many cases, primarily due to their generally limited substrate scope. The most prominent example of such a successful ATA engineering resulted from a collaboration between Merck & Co. and Codexis which led to the realisation of an industrial process for the enzymatic asymmetric synthesis of the antidiabetic drug (R)-sitagliptin with >99.95% optical purity.3 Prior to this, access to (R)-sitagliptin was via enamine formation followed by asymmetric hydrogenation at high pressure (ca. 20 atm) using a rhodium-based chiral catalyst which rendered lower stereoselectivity and a rhodium contaminated product stream.3 The biocatalytic approach using the engineered variant ATA-117 represents therefore a prime example of green chemistry in terms of overall waste reduction, lower energy consumption and avoidance of hazardous heavy metal catalyst waste treatment.3-Amino-8-aza-bicyclo[3.2.1]oct-8-yl-phenyl-methanone (2) (Scheme 1) represents a model building block for pharmaceutically active ingredients and thus an attractive target for biocatalytic selective synthesis mediated by transaminases. The 8-azabicyclo[3.2.1]octane motif constitutes a structural element within many neuroactive compounds, among which cocaine and atropine4 and various derivatives thereof have been patented e.g. as modulators of chemokine receptor activity,5 an important target in the treatment of a number of diseases. Besides its pharmacological relevance, amine 2 is also a challenging compound for protein engineering of transaminases due to its bicyclic and bridged ring system.
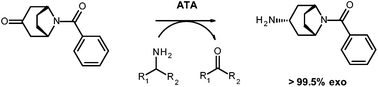 | ||
| Scheme 1 Selective synthesis of exo-3-amino-8-aza-bicyclo[3.2.1]oct-8-yl-phenyl-methanone 2via reductive amination of ketone 1 catalysed by the ATA 3FCR and its variants. Isopropylamine or alanine can be applied as amine donors. | ||
Recently, we succeeded in identifying and evolving ATAs of the fold class I ((S)-selective ATAs) that accept a broad spectrum of sterically demanding, but planar substrates.6 The best ATA variant enabled asymmetric synthesis of the target amines on a preparative scale with essentially perfect optical purities. Enzymatic routes to access the target amine 2 (Scheme 1) have not been described before and the bicyclic bridged ring system poses an unaddressed challenge. Thus, we herein describe the protein engineering of the (S)-selective amine transaminase from Ruegeria sp. TM1040 (3FCR)6,7 to transform the ketone 1 selectively to the desired exo-amine 2.
Results and discussion
Protein engineering of 3FCR
In order to identify the optimal starting scaffold for engineering we first screened our in-house transaminases. Among them, the wild-type transaminase from Ruegeria sp. TM1040 (pdb-code: 3FCR7) and its mutants 3FCR_DM and 3FCR_QM from our recent study6 were found to possess low activity (Table 1). Interestingly, the single mutation Y59W reduced the wild-type's activity, while mutation T231A exhibited doubled activity. These results prompted us to apply saturation mutagenesis at the four positions already identified within 3FCR_QM. The variant 3FCR_QM exhibited perfect stereoselectivity towards the exo-isomer in the reductive transamination while the variants without the Y152F mutation had only moderate selectivity (<40% exo to endo preference). This revealed that at three of the four positions (87, 152, 231), the optimal amino acids were already incorporated into the starting scaffold. However, we had to replace tryptophan in position 59. Saturation of this position resulted in a leucine substitution which provided a 3-fold increase of activity (145 mU mg−1) compared to the 3FCR_QM scaffold (45 mU mg−1) (Table 1).Having achieved this increase in activity, further rationally guided protein engineering was hampered by the fact that the substrate of interest does not allow following the typical transaminase design concepts. As the active site of ATAs is composed of a small and a large binding pocket, the common aim of most studies is to widen the small binding pocket. For the bicyclic and bridged ketone 1, its orientation in the active site is difficult to model due to its “three-dimensional bulkiness”. For this reason, parallel to the optimization of the four aforementioned positions, we performed directed evolution of the 3FCR scaffold using error-prone PCR in combination with our recently developed high-throughput glycine oxidase assay.8 Accordingly, the assay was adapted for liquid phase analysis in microtiter plates using the racemic amine 2 as an amine donor and glyoxylate as the acceptor. Our direct photometric assay9 could not be used in this case as ketone 1 and amine 2 possess virtually identical spectrophotometric properties.
Screening of ca. 4600 variants from two different error-prone PCR libraries based on the 3FCR_DM scaffold with the glycine oxidase agar plate assay provided a variant with increased activity and three amino acid exchanges. Characterization of the respective single mutants identified mutation I234F to be responsible for a 3.5-fold increased activity in the 3FCR_DM (data not shown). Further investigations by saturation of this position revealed that both phenylalanine and methionine are the most promising residues at this position in the kinetic resolution of amine 2. The beneficial effect of these two mutations was confirmed both in 3FCR_DM and 3FCR_QM. When the specific activity of the 3FCR_QM was only 45 mU mg−1, the 3FCR_QM/I234F exhibited 80 mU mg−1 and the 3FCR_QM/I234M 116 mU mg−1 while maintaining absolute stereoselectivity (Table 1).
These two new variants were applied in the selective synthesis (Scheme 1) using either alanine or isopropylamine (IPA) as an amine donor (Table 2), and they both proved to be beneficial. 3FCR_QM/I234M provided the best conversions, independently of the amine donor. However, in terms of catalytic efficiency 3FCR_QM/I234F appeared to be more appropriate, as it led to conversions comparable to 3FCR_QM/I234M despite the fact that 8-fold less enzyme was applied. For unclear reasons, introduction of mutation I234F resulted to significantly lower expression yield and hence this variant was not considered further. 3FCR_QM/I234M was subjected to a second round of error-prone PCR (∼8000 variants screened) revealing position 382 as a new hot spot. Its subsequent NNK saturation-mutagenesis identified L382M as the most promising variant (Table 3). In parallel, via rational design (see in silico analysis paragraph) we decided to mutate S86 to alanine to provide a little more flexibility in the loop where F87 is located. However, this mutation did not have the desired outcome and even decreased the activity when incorporated into 3FCR_QM/I234M/L382M.
| Variant | Conversion [%] (% exo/endo excess) | |
|---|---|---|
| With IPA | With alanine | |
| a Conditions for isopropylamine experiments: 2 mM 1, 0.2 M IPA, HEPES buffer (pH 8.0, 50 mM), 1 mM PLP, 5% DMSO, 30 °C, 600 rpm, 20 h. Conditions for alanine experiments: 8 mM 1, 0.2 M L-alanine, HEPES buffer (pH 8.0, 50 mM), 1 mM PLP, 20% DMSO, 5 mM NADH, 25 mM glucose, 0.05 mg mL−1 GDH, 5 μL mL−1 LDH, 30 °C, 600 rpm, 20 h. b n.d. = not determined. c Lower conversions as only small amounts of enzyme could be applied due to expression issues, see text. | ||
| 3FCR_DM | 0 (n.d.b) | 4 (n.d.b) |
| 3FCR_QM | 10 (>99) | 35 (>99) |
| 3FCR_QM/I234M | 19 (>99) | 66 (>99) |
| 3FCR_QM/I234Fc | 8 (>99) | 40 (>99) |
| Variant | Specific activity [mU mg−1] | Conversion [%, exo] |
|---|---|---|
| a Conditions for kinetic resolution: 1 mM rac-amine 2, 2 mM glyoxylate, CHES buffer (50 mM, pH 9.5), 37 °C, measurement for 40 min; conditions for asymmetric synthesis experiments: 2 mM prochiral ketone 1, 0.2 M IPA, HEPES buffer (pH 8.0, 50 mM), 1 mM PLP, 0.5 mg mL−1 of the respective variant, 5% DMSO, 30 °C, 600 rpm, 45 h. | ||
| 3FCR_QM | 45 | 23 (>99) |
| 3FCR_QM/Y59L | 173 | 96 (>99) |
| 3FCR_QM/I234M | 137 | 55 (>99) |
| 3FCR_QM/I234M/L382M | 198 | 60 (>99) |
| 3FCR_QM/S86A/I234M/L382M | 185 | 51 (>99) |
| 3FCR_QM/Y59L/S86A/I234M/L382M | 295 | 94 (>99) |
When we investigated the catalytic efficiency of all interesting variants for selective synthesis, the results were quite different from those observed in the kinetic resolution mode. Although all three mutations (namely Y59L, I234M and L382M) were beneficial for kinetic resolution, it seems that the key mutation for efficiency is Y59L. The other two mutations did lead to an improvement compared to the scaffold (3FCR_QM) but not to the same extent. Nevertheless, all variants presented in Table 3 maintained perfect exo/endo-selectivity (>99%) in the formation of the exo-amine 2.
Finally, a non-optimized small scale preparative synthesis with 3FCR_QM/I234M was performed using isopropylamine as an amine donor, which provided 75% conversion (60% isolated yield) and >99.5% selectivity for the exo-amine as determined by HPLC analysis.
In silico analysis
In order to discern the underlying reasons for the improved activity of the best variants towards 1, we performed an in silico analysis to identify possible interactions with the sterically demanding target compound. As seen in Fig. 1, position 59 can directly interact with the quinonoid of 2. Here, mutation to leucine provides significantly more space than tryptophan or tyrosine and it increases the hydrophobicity of the binding pocket. Position 87 seems to interact with the keto group of the substrate and thus the removal of the hydroxyl group of the tyrosine is beneficial. In order to provide more space, we aimed at introducing additional flexibility for residue 87 by mutating the neighboring positions. By sequence alignment, we observe that in several other ATAs, such as the one from Vibrio fluvialis, the adjacent position 86 is an alanine, while in 3FCR it is a serine. This mutation was investigated but as shown in Table 3, the mutants did not exhibit improved activity. Another significant mutation is Y152F. As described earlier,6 this mutation significantly stabilizes the 3FCR scaffold and thus facilitates the asymmetric synthesis performance which requires transaminases being stable for several hours, or even days. In this case another benefit arose: the removal of the hydroxyl group generated more volume for the bicyclic substrate, with its five-membered ring facing residue 152 and hence favoring formation of the exo-amine 2 (Fig. 1). All variants bearing mutation Y152F exhibited perfect selectivity producing as such the exo-amine exclusively. The variants without mutation Y152F displayed only moderate selectivity (<40% exo/endo excess). A reason might be the interaction between the hydroxyl group of the tyrosine and the nitrogen atom in the bicyclic ring structure, which enables the binding of the endo-isomer.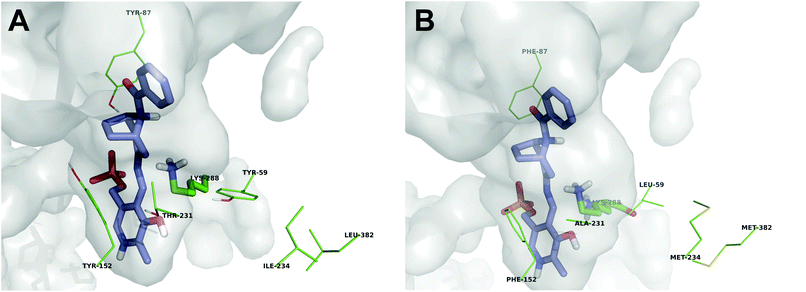 | ||
| Fig. 1 In silico analysis of the quinonoid of exo-3-amino-8-aza-bicyclo[3.2.1]oct-8-yl-phenyl-methanone 1 (blue sticks) in the active site of (A) 3FCR wild-type and (B) 3FCR_QM/Y59L/I234M/L382M. The side chains of the interesting positions are presented with green lines. The catalytic lysine (K288) is also shown (green sticks). It is obvious that in the variant (B) the binding pocket is significantly enlarged, mainly due to the Y59L and the Y152F mutations. It can also be seen that residues in positions 234 and 382 do not directly interact with the quinonoid, but they appear to have a secondary interaction with residue 59. | ||
As expected, the mutations identified within the error-prone PCR-derived libraries are not present in the first sphere of the interactions of the quinonoid with the enzyme and thus it is also more difficult to understand their effects. Both residues 234 and 382 are located behind residue 59 (Fig. 1). These mutations might stabilize residue 59 in a catalytically favored orientation, while residue 234 could affect via the backbone the orientation of the neighboring residue 231, which also has a significant impact on the catalytic activity.
Experimental
Materials
Ketone 1 and amine 2 as a mixture of exo- and endo-isomers as well as the individual isomers were made available by F. Hoffmann-La Roche (Scheme S1†). All other reagents were of analytical grade. Recombinant expression of the (S)-selective ATA from Ruegeria sp. TM1040 and all variants were performed as described previously, see also below for details.6 The glycine oxidase was produced also as described previously.8Directed evolution libraries
For the preparation of error-prone PCR libraries, the genes encoding the respective variants of the ATA from Ruegeria sp. TM1040 were amplified using the GeneMorph II Random Mutagenesis Kit from Agilent Technologies according to the manual instructions and using the following flanking primers: ePCR_Left_pET22b (GTTTAACTTTAAGAAGGAGATATACATATGC); ePCR_Right_3FCR (GTGGTGGTGATGGTGATGTGAACC). In order to adjust the mutation frequency, the amount of the parental plasmid template was varied between 25–50 ng of the pET22b plasmid template in a 50 μL reaction volume and 30 cycles of mutagenesis amplification were performed. Usually a set of libraries with different amounts of the parental plasmid template were prepared to cover variants with higher and lower mutation frequencies. For the preparation of the error-prone PCR libraries based on the 3FCR_DM, 25 ng or 50 ng template and for the library based on the 3FCR_QM/I234M 35 ng or 45 ng template were applied. For the latter libraries a random sample of 3 variants was sent for sequencing and revealed an average mutation frequency of 5 (35 ng template) and 2–5 (45 ng template) nucleotide exchanges per gene. The resulting PCR products were purified using the NucleoSpin® Gel and PCR Clean-up Kit from Macherey & Nagel according to the manual instructions, but with 50% diluted NTI buffer. The sample was eluted in 25 μL for application as megaprimers in a MEGAWHOP (mega primers and whole plasmid PCR). The MEGAWHOP reaction mixture (50 μL reaction volume) was prepared as follows: Pfu buffer, 0.3 mM dNTPs, 1–1.5 ng μL−1 parental plasmid, 5 μL purified megaprimers from the ep-PCR and 0.3 μL of Pfu Plus! DNA polymerase. The amplification was performed as follows: (a) 94 °C, 2 min; (b) 28 cycles: 94 °C, 30 s; 55 °C or 60 °C, 30 s; 72 °C, 7:05 min; (c) 72 °C, 14 min. The PCR product was digested with DpnI (20 μL mL−1) for 2 h at 37 °C and the restriction enzyme was inactivated by incubation at 80 °C for 20 min. 6 μL of the DpnI digested sample was transformed in 50 μL of electro-competent TOP10 cells (1.5 kV, 5 ms). After transformation, 1 mL LB-SOC was added and after 1 h incubation at 37 °C and 800 rpm a sample of 40 μL was plated out on an LB-amp (100 μg mL−1 ampicillin) and the remaining sample was used for inoculation of a 50 mL LB-amp (100 μg mL−1 ampicillin) overnight culture. After overnight incubation the colonies were counted on the agar plate to calculate the number of variants in the library (5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000–20
000–20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 transformants were usually obtained). The overnight culture was then applied for plasmid isolation. Electro-competent E. coli BL21(DE3) cells containing the compatible pCDF-1 glycine oxidase coding plasmid were transformed with 30 ng of the library plasmid DNA as described above. After incubation at 37 °C different amounts (30–60 μL) were plated out on amp/spec plates (100 μg mL−1 ampicillin, 50 μg mL−1 spectinomycin) for screening on nitrocellulose membranes as described further below.
000 transformants were usually obtained). The overnight culture was then applied for plasmid isolation. Electro-competent E. coli BL21(DE3) cells containing the compatible pCDF-1 glycine oxidase coding plasmid were transformed with 30 ng of the library plasmid DNA as described above. After incubation at 37 °C different amounts (30–60 μL) were plated out on amp/spec plates (100 μg mL−1 ampicillin, 50 μg mL−1 spectinomycin) for screening on nitrocellulose membranes as described further below.
Point and site-saturation mutagenesis
All variants were prepared using a modified version of the QuikChange PCR method. Primers were designed with the desired mismatches to provide the desired mutations. For site-saturation mutagenesis primers with NNK in the desired region were applied. For each reaction Pfu buffer, 0.2 mM dNTPs, 0.2 ng μL−1 parental plasmid, 0.2 μM of each primer and 0.2 μL of Pfu Plus! DNA polymerase were applied. 3% (v/v) DMSO was applied only for reactions using primers without NNK-codons. The amplification was performed as follows: (a) 94 °C, 2 min; (b) 25 cycles (QuikChange)/30 cycles (site-saturation): 94 °C, 30 s; 55 °C or 60 °C, 30 s; 72 °C, 7:05 min (c) 72 °C, 14 min. The PCR product was digested with DpnI (20 μL mL−1) for 2 h at 37 °C and the restriction enzyme was inactivated by incubation at 80 °C for 20 min. Chemo-competent Top10 E. coli cells were transformed with the PCR product. After confirmation of codon distribution in the case of a site-saturation mutagenesis or after confirmation of the correct sequence for QuikChange variants, the PCR products were transformed in chemo-competent BL21(DE3) E. coli cells for expression.Protein expression and purification
The glycine oxidase was expressed and purified as described previously.8 As the expression medium, TB-medium (Terrific Broth) instead of LB-medium was applied.The expression of the transaminase variants was carried out in baffled 250 mL flasks using 50 mL TB-medium containing 100 μg mL−1 ampicillin. Inoculation was carried out using 1 mL of an overnight culture of a microbial colony of E. coli BL21 (DE3) transformed with the plasmid containing the gene of interest. The E. coli strain was incubated at 37 °C with shaking at 180 rpm to grow to an optical density at 600 nm (OD600) of 0.6 to 0.8. The flasks were moved to a 20 °C shaker and after 10 min of incubation with shaking at 180 rpm isopropyl-β-D-thiogalactoside (IPTG) was added to a final concentration of 0.5 mM to induce the expression of the transaminase. Incubation was continued overnight (ca. 16 h) at 20 °C with shaking. Cells were harvested by centrifugation (4500g, 15 min, 4 °C) and the supernatant was discarded. The cell pellets were stored at −20 °C or were directly used for cell disruption and purification. For purification the cell pellets were resuspended in 6 mL pre-cooled (4 °C) lysis buffer (HEPES buffer 50 mM, pH 7.5 containing 0.1 mM PLP, 0.5 M NaCl, 10 mM imidazole and 50 μg mL−1 DNAse). The cells were disrupted via ultrasonication (Bandelin Sonopuls HD2070, Bandelin UW2070, MS 73 sonotrode) at 50% cycle and 55% power 5 minutes on ice, twice handled. The suspension was transferred to 2 mL tubes. To remove the cell debris, the tubes were centrifuged for 20 min at 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g and 4 °C. The supernatant was applied for protein purification via metal affinity chromatography using Roti®garose-His/Co Beads self-packed columns (∼1.5 mL bed volume). The clear lysate was loaded onto the columns and after 5 min of incubation the cap was opened and the flow-through was discarded. The column was washed 3 times with application buffer (HEPES buffer 50 mM, pH 7.5 containing 0.1 mM PLP, 0.5 M NaCl, 10 mM imidazole) and the flow-through was discarded. The cap was closed and 2.5 mL of elution buffer (HEPES buffer 50 mM, pH 7.5 containing 0.1 mM PLP, 0.5 M NaCl, 350 mM imidazole) was applied onto the columns. After 5 min of incubation the cap was opened and the protein solution was gathered for desalting using GE Healthcare PD10 desalting columns and desalting buffer (HEPES buffer 50 mM, pH 7.5 containing 0.1 mM PLP) according to the manual of instructions. Fig. S2† shows an SDS-PAGE of the purified variants. Protein concentrations were determined via the Pierce BCA Protein Assay Kit. The purified enzyme solutions were stored at +4 °C, or at −20 °C in 30% glycerol. For asymmetric synthesis experiments the desalting buffer was adjusted to pH 8.0 instead of pH 7.5. For preparative scale asymmetric synthesis experiments the transaminase variants were prepared as described previously.6
000g and 4 °C. The supernatant was applied for protein purification via metal affinity chromatography using Roti®garose-His/Co Beads self-packed columns (∼1.5 mL bed volume). The clear lysate was loaded onto the columns and after 5 min of incubation the cap was opened and the flow-through was discarded. The column was washed 3 times with application buffer (HEPES buffer 50 mM, pH 7.5 containing 0.1 mM PLP, 0.5 M NaCl, 10 mM imidazole) and the flow-through was discarded. The cap was closed and 2.5 mL of elution buffer (HEPES buffer 50 mM, pH 7.5 containing 0.1 mM PLP, 0.5 M NaCl, 350 mM imidazole) was applied onto the columns. After 5 min of incubation the cap was opened and the protein solution was gathered for desalting using GE Healthcare PD10 desalting columns and desalting buffer (HEPES buffer 50 mM, pH 7.5 containing 0.1 mM PLP) according to the manual of instructions. Fig. S2† shows an SDS-PAGE of the purified variants. Protein concentrations were determined via the Pierce BCA Protein Assay Kit. The purified enzyme solutions were stored at +4 °C, or at −20 °C in 30% glycerol. For asymmetric synthesis experiments the desalting buffer was adjusted to pH 8.0 instead of pH 7.5. For preparative scale asymmetric synthesis experiments the transaminase variants were prepared as described previously.6
Quantification of activity in liquid phase assay
To investigate our variants for activity in kinetic resolution, the glycine oxidase coupled assay8 was adapted for the application in microtiter plates. The assay was optimized for a reaction volume of 150 μL and to have a linear range under the further below specified conditions on the condition that the detected slope is below 0.8 OD h−1. For any samples providing higher slopes the assay was repeated with diluted samples in order to have a linear relationship between the activity and detected slope. Thus, we standardized protein concentrations to 0.25–1.0 mg mL−1 depending on the respective activity using desalting buffer (HEPES buffer 50 mM, pH 7.5, containing 0.1 mM PLP). To certify that the assay was working accordingly, an additional 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 dilution of each sample was prepared. To run the assay, 20 μL of each sample and each dilution were added in triplicates in a microtiter plate. The reaction was started by adding 130 μL of the mastermix solution and the increase of absorbance was followed at 498 nm for at least 40 min at 37 °C. The final assay concentrations were as follows: CHES buffer (pH 9.5, 50 mM), 1 mM rac-amine 2 (3-amino-8-aza-bicyclo[3.2.1]oct-8-yl-phenyl-methanone), 2 mM glyoxylate, 0.17 mg mL−1 glycine oxidase from Geobacillus kaustophilus, 5 U mL−1 horseradish peroxidase, 3.9 mM vanillic acid, 0.5% (v/v) methanol, 1.2 mM 4-aminoantipyrine and 0.08% ethanol (v/v). To guarantee the best comparability as possible, we always compared the activity with the parental mutant which was purified in parallel and treated identically with the variants of interest.
5 dilution of each sample was prepared. To run the assay, 20 μL of each sample and each dilution were added in triplicates in a microtiter plate. The reaction was started by adding 130 μL of the mastermix solution and the increase of absorbance was followed at 498 nm for at least 40 min at 37 °C. The final assay concentrations were as follows: CHES buffer (pH 9.5, 50 mM), 1 mM rac-amine 2 (3-amino-8-aza-bicyclo[3.2.1]oct-8-yl-phenyl-methanone), 2 mM glyoxylate, 0.17 mg mL−1 glycine oxidase from Geobacillus kaustophilus, 5 U mL−1 horseradish peroxidase, 3.9 mM vanillic acid, 0.5% (v/v) methanol, 1.2 mM 4-aminoantipyrine and 0.08% ethanol (v/v). To guarantee the best comparability as possible, we always compared the activity with the parental mutant which was purified in parallel and treated identically with the variants of interest.
To quantify the specific enzymatic activity, a calibration curve was recorded by using sequential dilutions of an aqueous 1 M glycine stock solution. 20 μL were applied in triplicates to the microtiter plate and 130 μL of the mastermix solution described above, were added. After incubation at 37 °C, an endpoint measurement at 498 nm was made. Linear regression provided a slope of 12.44 AU μmol−1 of glycine in 150 μL that allowed quantifying the reaction velocities from the slopes obtained in the kinetic assays (Fig. S1†).
Solid-phase assay screening of error-prone PCR libraries
The screening procedure of the error-prone PCR libraries was performed as described previously.8 The pH of the assay plates was pH 9.25 (first screening round) and pH 9.5 (second screening round) and the assay plates had the following final composition: CHES buffer (pH 9.5/9.25, 50 mM), 100 mM isopropylamine, 10 mM glyoxylate, in 3.9 mM vanillic acid, 0.5% (v/v) methanol, 1.2 mM 4-aminoantipyrine and 0.08% ethanol (v/v), and 1% (w/w) agarose. 100 U of horseradish peroxidase dissolved in 100 μL MilliQ water were spread on the surface of each plate directly before placing the membranes on top of the plates for screening. After incubation of the assay plates at 37 °C the most colored colonies were picked from each plate for expression in 96-well deep-well blocks and screening in microtiter plates as described previously.8Analytical reductive amination experiments
Reductive amination reactions in the analytical scale were performed in a total volume of 400 μL in glass vials using a glass vial shaker from Eppendorf. Detailed final concentrations and conditions are given in the main text. For HPLC analysis 50 μL of the sample were mixed with 50 μL of acetonitrile including 0.1% diethylamine that was passed through a tip filter before injection into HPLC.Preparative reductive amination
Into 82 ml reaction buffer (HEPES 50 mM; 2 mM PLP; 0.2 M 2-propylamine hydrochloride; pH 7.5) the purified mutant transaminase 3FCR Y59W/Y87F/Y152F/T231A/I234M solution (in total 78 mL, protein 1.1 mg mL−1) and 100 mg 8-benzoyl-8-aza-bicyclo[3.2.1]octan-3-one (1) dissolved in 20 ml DMSO were added under stirring at 30 °C. The reaction was allowed to proceed for 9 d (non-optimized) enabling a conversion of 75 area% (IPC-HPLC). The reaction mixture was acidified to pH 2.0 to precipitate the enzyme and stirring was continued for 15 min. Subsequently, the reaction mixture was filtered through a 25 g filter aid (Dicalite) bed and extracted twice with 50 mL dichloromethane to remove the remaining ketone 1. The combined organic phases were dried over anhydrous MgSO4, filtered and evaporated under vacuum at 40 °C yielding 37 mg of (HPLC: 97 area%) ketone 1 as a slightly yellow oil. The pH of the aqueous phase was adjusted to 12 using 2 N NaOH and extracted four times with 50 mL dichloromethane. The combined organic phases were dried over anhydrous MgSO4, filtered, evaporated under vacuum at 40 °C and dried 36 h under high vacuum at 60 °C yielding 60 mg of target amine 2 (60%) as a yellow oil. Optimization and scale-up of this proof-of-concept small scale preparative reaction to reduce reaction times and replace dichloromethane for work-up is the subject of future studies.HPLC methods for analytical scale reactions
HPLC-analysis of analytical scale reactions was performed via direct injection of the 20 μL sample on a 150 × 4.6 analytical OD-RH column from Chiralcel running 27% acetonitrile and 73% water, 0.1% diethylamine at 30 °C and 0.55 mL min−1 flow. Detection of the analytes took place at 230 nm.Analysis of preparative scale reaction product 2
Chemical purity HPLC (see also Fig. S3†): 98.9 area% [210 nm; X-Bridge C8; 50 × 2.1 mm, 2.5 μm, flow 1 mL min−1, 40 °C, A: 90% 10 mmol ammonium acetate in H2O/ACN (95/5), B: ACN, 10%]; chiral SFC: 100% exo [210 nm; Chiralpak AD-3; 150 × 4.6 mm, 5 μm; flow 3 mL min−1; left 40 °C; right 42 °C; A: 82% CO2, B: 18% methanol with 0.2% 2-propylamine]; 1H NMR (600 MHz, DMSO-d6, 120 °C) δ ppm 7.44 (s, 5 H), 4.29 (br s, 2 H), 3.16 (dt, J = 11.1, 5.5 Hz, 1 H), 1.86–2.00 (m, 3 H), 1.79 (ddd, J = 13.2, 5.1, 3.0 Hz, 3 H), 1.67–1.75 (m 3 H), 1.34–1.47 (m, 2 H); LC-MS: 231 (M + H)+. CAS registry number: 637018-99-6.Conclusions
Several variants of a (S)-selective transaminase were identified enabling an enzyme-catalyzed synthesis of the pharmaceutically relevant exo-3-amino-8-aza-bicyclo[3.2.1]oct-8-yl-phenylmethanone in good yield and excellent optical purity via reductive amination. The key mutations were identified by (a) saturation of the positions known to play a role in the accommodation of bulky substrates, and (b) two rounds of directed evolution. The results represent the first example of the transaminase-catalysed synthesis of an amine containing a bicyclic bridged moiety. Our study also shows that a combination of rational protein design with random mutagenesis by error-prone PCR was required to identify the most suitable variants, similar to the findings reported by Savile et al. for the development of an (R)-transaminase for Sitagliptin synthesis.3 Furthermore, our careful analysis of the performance of the 3FCR variants confirms that enzymes found to be active in kinetic resolution must not be suitable for asymmetric synthesis as published earlier for a mutant of the ATA mutant from Vibrio fluvialis.10Acknowledgements
The authors thank I. Duffour for developing the analysis methods, J. Joerger for the preparative separation of the exo- and endo-amines, C. Wyss-Gramberg for the NOESY NMR analysis, P. Meier for performing the preparative enzymatic experiments and K.-J. Gutmann for the synthesis of the requisite organic materials.References
- F. Steffen-Munsberg, C. Vickers, H. Kohls, H. Land, H. Mallin, A. Nobili, L. Skalden, T. van den Bergh, H.-J. Joosten, P. Berglund, M. Höhne and U. T. Bornscheuer, Biotechnol. Adv., 2015, 33, 566–604 CrossRef CAS PubMed.
- (a) D. Koszelewski, K. Tauber, K. Faber and W. Kroutil, Trends Biotechnol., 2010, 28, 324–332 CrossRef CAS PubMed; (b) M. Fuchs, J. E. Farnberger and W. Kroutil, Eur. J. Org. Chem., 2015, 6965–6982 CrossRef CAS PubMed.
- C. K. Savile, J. M. Janey, E. C. Mundorff, J. C. Moore, S. Tam, W. R. Jarvis, J. C. Colbeck, A. Krebber, F. J. Fleitz, J. Brands, P. N. Devine, G. W. Huisman and G. J. Hughes, Science, 2010, 329, 305–309 CrossRef CAS PubMed.
- A. Korolkovas, in Essentials of Medicinal Chemistry, ed. A. Korolkovas, Wiley Interscience, New York, 2nd edn, 1988, pp. 195–283 Search PubMed.
- J. Cumming, Int. Pat., WO2003080574A1, 2003 Search PubMed.
- I. V. Pavlidis, M. S. Weiß, M. Genz, P. Spurr, S. P. Hanlon, B. Wirz, H. Iding and U. T. Bornscheuer, Nat. Chem., 2016 DOI:10.1038/NCHEM.2578.
- F. Steffen-Munsberg, C. Vickers, A. Thontowi, S. Schätzle, T. Tumlirsch, M. Svedendahl Humble, H. Land, P. Berglund, U. T. Bornscheuer and M. Höhne, ChemCatChem, 2013, 5, 150–153 CrossRef CAS.
- M. S. Weiß, I. V. Pavlidis, C. Vickers, M. Höhne and U. T. Bornscheuer, Anal. Chem., 2014, 86, 11847–11853 CrossRef PubMed.
- S. Schätzle, M. Höhne, E. Redestad, K. Robins and U. T. Bornscheuer, Anal. Chem., 2009, 81, 8244–8248 CrossRef PubMed.
- A. Nobili, F. Steffen-Munsberg, H. Kohls, I. Trentin, C. Schulzke, M. Höhne and U. T. Bornscheuer, ChemCatChem, 2015, 7, 757–760 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6ob02139e |
| This journal is © The Royal Society of Chemistry 2016 |
