Potentiation of Francisella resistance to conventional antibiotics through small molecule adjuvants†
Matthew D.
Stephens
a,
Veroncia B.
Hubble
a,
Robert K.
Ernst
b,
Monique L.
van Hoek
c,
Roberta J.
Melander
a,
John
Cavanagh
d and
Christian
Melander
*a
aDepartment of Chemistry, North Carolina State University, Raleigh, North Carolina 27695, USA. E-mail: ccmeland@ncsu.edu
bDepartment of Microbial Pathogenesis, University of Maryland – Baltimore, Baltimore, MD 21201, USA
cSchool of Systems Biology & National Center for Biodefense and Infectious Diseases, George Mason University, Manassas, VA 20110, USA
dDepartment of Molecular and Structural Biochemistry, North Carolina State University, Raleigh, North Carolina 27695, USA
First published on 30th September 2015
Abstract
A screen of 20 compounds identified small molecule adjuvants capable of potentiating antibiotic activity against Francisella philomiragia. Analogue synthesis of an initial hit compound led to the discovery of a potentially new class of small molecule adjuvants containing an indole core. The lead compound was able to lower the MIC of colistin by 32-fold against intrinsically resistant F. philomiragia.
Introduction
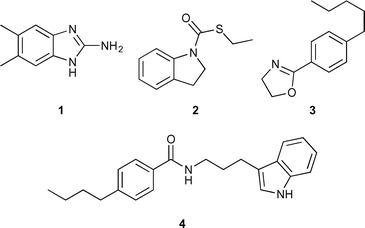
Francisella are small, facultative Gram-negative bacteria that are responsible for zoonotic disease in which humans are infected, usually from bites or contact with infected blood.1 The most common vectors of disease are ticks and wild rabbits.2Francisella can also be a highly infectious aerosol and has the potential to be weaponized.3 Encompassed under the Francisella genus are two species of consequence: Francisella philomiragia and Francisella tularensis. Very resilient, the bacteria are capable of surviving multiple weeks in the environment and have been found in water sources around the world.4 Although, not as virulent and somewhat rare in humans,5F. philomiragia is a potential model organism to study Francisella. Human cases of F. philomiragia are often associated with individuals with weakened immune systems or who have suffered from a near drowning experience.6F. philomiragia is also the causative agent of the disease francisellosis, most common among various species of fish.7 Francisellosis outbreaks have been responsible for mortality rates of up to 95% within farmed fish.8Our laboratory is interested in using small molecule adjuvants to potentiate antibiotic activity. We have shown that 2-aminoimidazoles (2-AIs) are capable of potentiating antibiotic activity against a wide variety of bacteria.9–13 We completed a screen of randomly chosen molecules from our in house library and identified four compounds, all of which, did not contain a 2-AI or 2-ABI core, but another nitrogen containing heterocycle (indole, indoline, or oxazoline) (Fig. 1) that were capable of potentiating antibiotic response in F. philomiragia. A structure activity relationship (SAR) study of compound 4 was carried out to discover potentially more potent leads and to better understand the structural requirements for activity. Herein, we report the discovery of the first example of a potentially new class of lead compounds containing an indole core capable of breaking F. philomiragia resistance to the antibiotic colistin.
Results and discussion
Adjuvant screen for MIC suppression of antibiotics against F. philomiragia
An initial pilot screen of 20 compounds from our in-house small molecule library14–18 was conducted by first determining the intrinsic antibiotic activity of each molecule by establishing its minimum inhibitory concentration (MIC) against F. philomiragia (Fig. S1, ESI†). Following our lab's previously reported screening protocol,9 the MIC of candidate conventional antibiotics were then determined in the absence or presence of each compound at 25% of the compound's MIC. Previous studies from our group has established that molecules from our internal library had little (if any) effect on bacterial growth at 25% MIC, allowing us to study non-microbicidal repotentiation of the conventional antibiotics under study. Streptomycin, an aminoglycoside, was chosen as the initial antibiotic to screen for potential adjuvants as it is considered to be the drug of choice for tularemia treatment.19 Initial testing with streptomycin revealed of the 20 compounds screened only compound 1 showed activity, a reduction in MIC of four-fold to 1 μg mL−1 (Table 1). Gentamicin, another aminoglycoside, has also been shown to be an effective treatment option for patients suffering from tularemia.20 Screening the same library for gentamicin repotentiation gave the same results for streptomycin, with compound 1 lowering the MIC four-fold from 1 μg mL−1 to 0.125 μg mL−1.| Compound | MIC (μM) | Concentration tested (μM) | Streptomycin (μg mL−1) | Gentamicin (μg mL−1) | Azithromycin (μg mL−1) | FR900098 (μg mL−1) | Colistin (μg mL−1) |
|---|---|---|---|---|---|---|---|
| — | — | — | 4 | 0.5 | 4 | 1024 | 256 |
| 1 | >200 | 50 | 1 | 0.125 | 4 | 1024 | 32 |
| 2 | >200 | 50 | 4 | 0.5 | 0.5 | 1024 | 256 |
| 3 | 200 | 50 | 8 | 1 | 2 | <64 | 512 |
| 4 | >200 | 50 | 4 | 0.25 | 2 | ND | 64 |
| 5 | >200 | 50 | 4 | 0.25 | 0.5 | 1024 | 8 |
Another class of antibiotics tested were macrolides, specifically azithromycin. Azithromycin is an attractive option for treatment because of its ability to concentrate within macrophages, where Francisella replicates at intracellular levels that are even greater than serum levels.21 Upon screening our library, compound 1 exerted no change in azithromycin activity. Another compound (2) displayed an eight-fold reduction in MIC, from 4 μg mL−1 to 0.5 μg mL−1). Compound 2 is of interest because unlike previously reported small molecule adjuvants from our lab it is neither a 2-AI nor 2-ABI, but rather contains an indoline core.
Our screen was further expanded to include a phosphonic acid antibiotic, FR900098. FR900098 has been shown to be effective at inhibiting Francisella sub-species.22 Compound 3 displayed a significant decrease in MIC of 16-fold from 1024 μg mL−1 to <64 μg mL−1.
Finally, we used colistin, a polymyxin antibiotic that is known to be ineffective at treating Francisella infections due to intrinsic resistance.23 The MIC of colistin alone was 256 μg mL−1. Library screening revealed an indole containing compound (4) that was also absent of a 2-AI or 2-ABI motif. The indole 4 exhibited a four-fold MIC reduction for colistin to 64 μg mL−1.
Library synthesis for structure activity relationship (SAR) study
Based on the results of our screen, availability of starting materials and cost of antibiotics, we opted to perform analogue synthesis of the indole 4. We were able to rapidly assemble an 11-membered analogue library of 4 (ESI† Table S1). The core is structurally similar to tryptamine with the only difference being a shorter 2-carbon linker. Direct reaction of tryptamine and derivatives with a variety of acylating agents in the presence of TEA gave a library of various structural motifs. Testing revealed that shortening the three-carbon linker to two carbons (5) drastically increased potency (Scheme 1), lowering the MIC of colistin 32-fold to 8 μg mL−1. The amide appears to be necessary as any change in functionality: amine, ester, or sulfonamide, results in complete loss in activity (ESI† Table S1). Alkylation or acylation of the indole nitrogen-1 position, also caused complete loss in MIC suppression. Changes in the alkyl tail length from butyl also gave disappointing results, as methyl, ethyl, and propyl derivatives had no impact on the antibiotic MIC.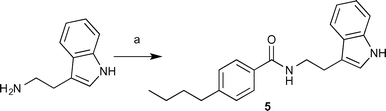 | ||
| Scheme 1 Synthesis of the most active compound (5). Reagents and conditions: (a) 4-butylbenzoyl chloride, TEA and DCM. | ||
Time kill curve of compound 5
In order to determine whether compound 5 was acting through a toxic or non-toxic mechanism, bacterial growth was measured as a function of time. F. philomiragia was grown with/without compound (50 μM) being present. Bacterial growth was checked at 8, 12, 16, 20, and 24 h time points (Fig. 2). Based on the analysis of the time kill curve, we observe minor growth delay at earlier time points; however growth is identical by 16 hours. | ||
| Fig. 2 Time kill curve of compound 5. Blue diamonds represent bacterial control. Red squares represent bacteria and compound 5 (50 μM). | ||
MIC determination in Francisella novicida
With lead compounds in hand, we then wanted to determine whether compounds retained activity against F. novicida, a sub-species of F. tularensis. As before, the MICs of the active compounds (1, 2, and 5) were determined and then antibiotic MICs were established in the absence or presence of each compound at 25% MIC. Compound 1 showed no change in MIC for any of the antibiotics tested (Table 2). The indole compound 5 displayed a two-fold reduction in MIC for colistin from 1024 μg mL−1 to 512 μg mL−1. The concentration of 5 was increased by 10 μM increments from 50 μM to 100 μM; however the increased concentration of 5 was unable to potentiate colistin in F. novicida further. Compound 2 was the only adjuvant tested to show any significant activity, reducing the MIC of azithromycin 16-fold fro 2 μg mL−1 to 0.125 μg mL−1.| Compound | MIC (μM) | Concentration tested (μM) | Gentamicin (μg mL−1) | Azithromycin (μg mL−1) | Colistin (μg mL−1) |
|---|---|---|---|---|---|
| — | — | — | 1 | 2 | 1024 |
| 1 | >200 | 50 | 1 | 2 | 1024 |
| 2 | >200 | 50 | 4 | 0.125 | 1024 |
| 5 | >200 | 50 | 4 | 1 | 512 |
Experimental section
Materials and methods
All reagents and solvents for synthesis were obtained from Sigma-Aldrich, St. Louis, MO, USA. Triethylamine was dried by refluxing CaH2, followed by distillation and storage over 4 Å molecular sieves. Deuterated solvents were acquired from Cambridge Isotope Laboratories (CIL). Purification was performed using 60 mesh standard silica from Sorbtech. 1H NMR (300 MHz) and 13C NMR (100 MHz) spectra were recorded at 25 °C on Varian Mercury spectrometers. Chemical shifts (δ) are given in ppm relative to TMS as an internal standard or the respective NMR solvent. Mass spectra were recorded on Thermo Fisher Scientific Exactive Plus MS (ESI).Synthesis
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 Hex/EtOAc to 2
1 Hex/EtOAc to 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 Hex/EtOAc to give a white solid (538 mg, 93% yield).
1 Hex/EtOAc to give a white solid (538 mg, 93% yield).
Biological
Conclusions
In summary, a pilot library screen resulted in the identification of several unique small molecule adjuvants, capable of potentiating various classes of antibiotics against F. philomiragia. Analogue synthesis of one lead (4) gave rise to compound 5, which displayed enhanced activity, culminating in a 32-fold MIC reduction to 8 μg mL−1 for colistin. Bacterial growth over time was measured to elucidate whether compound 5 was bactericidal. The CFUs measured for compound containing samples, correlate strongly with the control providing evidence 5 is acting through a non-toxic mechanism. This is the first example of a small molecule adjuvant able to potentiate F. philomiragia resistance to colistin, and 5 may represent the basis of a new class of small molecule adjuvants. Further exploration of this unique class of compound is currently underway in attempts to break F. novicida resistance.Acknowledgements
The authors would like to thank the National Institutes of Health (R01GM055769 to JC and CM) and DTRA (HDTRA1-11-1-0054 to MVH) for support.Notes and references
- J. M. Petersen, P. S. Mead and M. E. Schriefer, Vet. Res., 2009, 40, 7 CrossRef.
- R. J. Mani, J. A. Metcalf and K. D. Clinkenbeard, PLoS One, 2015, 10, e0130513 Search PubMed.
- D. L. Fritz, M. J. England, L. Miller and D. M. Waag, Comp. Med., 2014, 64, 341–350 CAS.
- R. R. Parker, E. A. Steinhaus, G. M. Kohls and W. L. Jellison, Bull. Natl. Inst. Health, 1951, 193, 1–161 CAS.
- T. L. Mailman and M. H. Schmidt, Can. J. Infect. Dis. Med. Microbiol, 2005, 16, 245–248 Search PubMed.
- J. D. Wenger, D. G. Hollis, R. E. Weaver, C. N. Baker, G. R. Brown, D. J. Brenner and C. V. Broome, Ann. Intern. Med., 1989, 110, 888–892 CrossRef CAS.
- A. B. Verhoeven, M. W. Durham-Colleran, T. Pierson, W. T. Boswell and M. L. Van Hoek, Biol. Bull., 2010, 219, 178–188 Search PubMed.
- R. S. Chern and C. B. Chao, Fish Pathol., 1994, 29, 61–71 CrossRef.
- S. A. Rogers, R. W. Huigens, 3rd, J. Cavanagh and C. Melander, Antimicrob. Agents Chemother., 2010, 54, 2112–2118 CrossRef CAS PubMed.
- T. L. Harris, R. J. Worthington and C. Melander, Angew. Chem., Int. Ed., 2012, 51, 11254–11257 CrossRef CAS PubMed.
- T. L. Harris, R. J. Worthington, L. E. Hittle, D. V. Zurawski, R. K. Ernst and C. Melander, ACS Chem. Biol., 2014, 9, 122–127 CrossRef CAS PubMed.
- R. J. Worthington, C. A. Bunders, C. S. Reed and C. Melander, ACS Med. Chem. Lett., 2012, 3, 357–361 CrossRef CAS PubMed.
- C. M. Brackett, R. J. Melander, I. H. An, A. Krishnamurthy, R. J. Thompson, J. Cavanagh and C. Melander, J. Med. Chem., 2014, 57, 7450–7458 CrossRef CAS PubMed.
- S. A. Rogers, D. C. Whitehead, T. Mullikin and C. Melander, Org. Biomol. Chem., 2010, 8, 3857–3859 CAS.
- R. W. Huigens, S. Reyes, C. S. Reed, C. Bunders, S. A. Rogers, A. T. Steinhauer and C. Melander, Bioorg. Med. Chem., 2010, 18, 663–674 CrossRef CAS PubMed.
- Z. Su, S. A. Rogers, W. S. McCall, A. C. Smith, S. Ravishankar, T. Mullikin and C. Melander, Org. Biomol. Chem., 2010, 8, 2814–2822 CAS.
- C. S. Reed, R. W. Huigens, 3rd, S. A. Rogers and C. Melander, Bioorg. Med. Chem. Lett., 2010, 20, 6310–6312 CrossRef CAS PubMed.
- S. A. Rogers and C. Melander, Angew. Chem., Int. Ed., 2008, 47, 5229–5231 CrossRef CAS PubMed.
- G. Enderlin, L. Morales, R. F. Jacobs and J. T. Cross, Clin. Infect. Dis., 1994, 19, 42–47 CrossRef CAS.
- A. Hassoun, R. Spera and J. Dunkel, Antimicrob. Agents Chemother., 2006, 50, 824 CrossRef CAS PubMed.
- S. Ahmad, L. Hunter, A. Qin, B. J. Mann and M. L. van Hoek, BMC Microbiol., 2010, 10, 123 CrossRef PubMed.
- E. S. McKenney, M. Sargent, H. Khan, E. Uh, E. R. Jackson, G. San Jose, R. D. Couch, C. S. Dowd and M. L. van Hoek, PLoS One, 2012, 7, e38167 CAS.
- A. C. Llewellyn, J. Zhao, F. Song, J. Parvathareddy, Q. Xu, B. A. Napier, H. Laroui, D. Merlin, J. E. Bina, P. A. Cotter, M. A. Miller, C. R. Raetz and D. S. Weiss, Mol. Microbiol., 2012, 86, 611–627 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5md00353a |
| This journal is © The Royal Society of Chemistry 2016 |