Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
LabDisk with complete reagent prestorage for sample-to-answer nucleic acid based detection of respiratory pathogens verified with influenza A H3N2 virus†
F.
Stumpf‡
*a,
F.
Schwemmer‡
b,
T.
Hutzenlaub
a,
D.
Baumann
a,
O.
Strohmeier
a,
G.
Dingemanns
c,
G.
Simons
c,
C.
Sager
d,
L.
Plobner
e,
F.
von Stetten
ab,
R.
Zengerle
abf and
D.
Mark
a
aHahn-Schickard, Georges-Koehler-Allee 103, 79110 Freiburg, Germany. E-mail: Fabian.Stumpf@Hahn-Schickard.de
bLaboratory for MEMS Applications, IMTEK - Department of Microsystems Engineering, University of Freiburg, Georges-Koehler-Allee 103, 79110 Freiburg, Germany
cPathoFinder B.V., Randwycksingel 45, 6229 EG Maastricht, The Netherlands
dMedical Care Center, Dr. Stein and colleagues, Wallstraße 10, 41061 Moenchengladbach, Germany
eAgrobiogen GmbH Biotechnologie, Larezhausen 2-3, 86567 Hilgertshausen, Germany
fBIOSS – Centre for Biological Signalling Studies, University of Freiburg, 79110 Freiburg, Germany
First published on 26th November 2015
Abstract
Portable point-of-care devices for pathogen detection require easy, minimal and user-friendly handling steps and need to have the same diagnostic performance compared to centralized laboratories. In this work we present a fully automated sample-to-answer detection of influenza A H3N2 virus in a centrifugal LabDisk with complete prestorage of reagents. Thus, the initial supply of the sample remains the only manual handling step. The self-contained LabDisk automates by centrifugal microfluidics all necessary process chains for PCR-based pathogen detection: pathogen lysis, magnetic bead based nucleic acid extraction, aliquoting of the eluate into 8 reaction cavities, and real-time reverse transcription polymerase chain reaction (RT-PCR). Prestored reagents comprise air dried specific primers and fluorescence probes, lyophilized RT-PCR mastermix and stick-packaged liquid reagents for nucleic acid extraction. Employing two different release frequencies for the stick-packaged liquid reagents enables on-demand release of highly wetting extraction buffers, such as sequential release of lysis and binding buffer. Microfluidic process-flow was successful in 54 out of 55 tested LabDisks. We demonstrate successful detection of the respiratory pathogen influenza A H3N2 virus in a total of 18 LabDisks with sample concentrations down to 2.39 × 104 viral RNA copies per ml, which is in the range of clinical relevance. Furthermore, we detected RNA bacteriophage MS2 acting as internal control in 3 LabDisks with a sample concentration down to 75 plaque forming units (pfu) per ml. All experiments were applied in a 2 kg portable, laptop controlled point-of-care device. The turnaround time of the complete analysis from sample-to-answer was less than 3.5 hours.
Introduction
Respiratory tract infections were the fourth most common cause of death in 2012 according to the WHO, accounting for roughly 3.1 million deaths worldwide.1 Currently, nucleic acid detection is generally performed in central laboratories. However, analysis in centralized laboratories typically leads to turnaround times in the order of days. Due to the lack of these fast diagnostics, empirical antibiotic therapy is the common practice. Antibiotics are often used on suspicion with no effect on the course of the disease.2 Such unnecessary use of antibiotics is one of the leading reasons for the spread of multi-resistant pathogens, which have evolved to become a major threat to global public health.3Therefore there is an urgent need for a diagnostic system which is able to rapidly test for infections at the point-of-care.4 An ideal point-of-care system would be rapid, easy to use by non-experts with minimal hands on time and able to detect multiple pathogenic species in parallel in a small lightweight device. While a range of sample-to-answer systems for nucleic acid based detection of respiratory infections are commercially available, no system satisfies all the needs of such a point-of-care system (see Table 1). Most of the sample-to-answer systems listed in Table 1 are not suitable for point-of-care usage by non-experts due to their CLIA moderate complexity. The Alere™ i system (Alere Inc., USA) is the only CLIA waived product though it still requires multiple manual handling steps (e.g. manual washing of swabs). The Alere™ i system provides a fast time-to-result due to its isothermal amplification but is therefore limited in degree of multiplexing (two geometrical separated single-plex reactions). In fact, all systems are significantly limited in their degree of multiplexing (≤5-plex), with the exception of the FilmArray® system (BioFire Diagnostics, Inc., USA) which consists of 2 devices (loading station and FilmArray® instrument) and just as the 3M™ Integrated Cycler (Focus Diagnostics Inc., USA) system, doesn't provide prestorage of liquid reagents.5,6
| Company | Number of processing devices | Size and weight of devices | Degree of multiplexing | Complexity | Time-to-result | |
|---|---|---|---|---|---|---|
| a Laptop/desktop-PC needed for instrument control and data analysis. b Size and weight of second device of the FilmArray® system (loading station). c 1-plex per reaction cavity and geometric-multiplexing7 demonstrated, due to 8 reaction cavities potential for 8 × 4-plex = 32-plex in future developments. d Reduction of time-to-result was not object of this publication. | ||||||
| Alere Inc., Alere™ i | 1 | 14 × 15 × 21 cm3, 3 kg | 2-plex | CLIA waived | 15 min | |
| Cepheid Inc., GeneXpert® I | 1a | 10 × 29 × 30 cm3, 4 kg | 5-plex | CLIA moderate complexity | <120 min | |
| Roche Molecular Systems Inc., cobas® LiaT | 1 | 11 × 19 × 24 cm3, 3.76 kg | 2-plex | CLIA moderate complexity | 25–60 min | |
| Focus Diagnostics Inc., 3M™ Integrated Cycler | 1a + barcode reader | 21 × 31 × 31 cm3, 8 kg | 3-plex | CLIA moderate complexity | ∼60 min | |
| BioFire Diagnostics Inc., FilmArray® | 2a + barcode reader | 17 × 25 × 39 cm3, 9 kg4 × 11 × 18b cm3, 0.45b kg | 20-plex | CLIA moderate complexity | 60 min | |
| Hahn-Schickard, LabDisk | 1a | 15 × 18 × 28 cm3, 2 kg | 1-plexc | 1 manual step (not CLIA certified) | 210d min | |
The lack of liquid reagent prestorage results in additional handling steps which require additional hands-on time and training to use the system correctly. Thus, there is no point-of-care system offering truly minimal handling, while featuring a high degree of multiplexing as well as prestorage of all required liquid reagents. Lab-on-a-Chip devices are a promising candidate for point-of-care systems to detect nucleic acids. Different platforms with varying automation principles have been suggested for this task.8–11 Centrifugal microfluidics is especially suitable, due to its simple interfaces and the fact that it only requires a single actuator.12–15 Process chains demonstrated in centrifugal microfluidics are lysis,16–18 nucleic acid extraction19–25 and real-time amplification of nucleic acids.7,26–33 Due to recent advances in magnetic bead handling,24 valving,34–40 aliquoting41–43 and pumping44–47 integration of full sample-to-answer systems on the centrifugal microfluidic LabDisk platform now became feasible. Kim et al. presented an integrated centrifugal microfluidic disk for detection of food-borne pathogens. Valving was realized via laser actuated ferrowax valves. The laser diode used for actuation of valves was also used for thermal lysis and amplification via RPA. After RPA the amplification product is diluted and read-out is performed on a lateral flow strip.48 Jung et al. presented a Lab-on-a-Disk for detection of H1N1 from purified viral RNA or viral lysates. RNA was extracted via sequential flow of liquids over stationary microbeads.21 After extraction, the eluate was amplified via reverse transcription-LAMP. The chip did not include multiplexing or lysis.49 Roy et al. presented a centrifugal microfluidic disk for detection of Bacillus atrophaeus subsp. globigii spores. The disk includes mechanical cellular lysis, PCR amplification, digestion of amplicons and hybridization to a microarray. The disk is fabricated in thermoplastic elastomer, which allowed for sealing of the PCR chamber during PCR amplification by external pressure.50 G. Czilwik et al. presented a centrifugal microfluidic disk for neonatal sepsis. The disk extracts DNA from blood serum using a bind-wash-elute protocol automated using silica coated magnetic beads transported between adjacent chambers. The eluate is used to rehydrate a lyophilized polymerase pellet for pre-amplification of the target DNA. The pre-amplified target DNA is then pumped radially inwards by centrifugo-dynamic inward pumping and aliquoted into 13 reaction cavities for individual real-time PCR of the pathogens of interest. Detection of a sepsis panel has been demonstrated with four different bacterial pathogens.51
So far, no centrifugal microfluidic disk for nucleic acid sample-to-answer analysis with complete prestorage of reagents has been demonstrated. For the first time we demonstrate a fully automated sample-to-answer system – comprising prestorage of all required reagents – for detection of respiratory pathogens based on centrifugal microfluidics. The disk includes RNA extraction via chemical lysis and a bind-wash-elute protocol, based on transport of silica coated magnetic particles between adjacent chambers. The eluate is pumped to the aliquoting structure via centrifugo-dynamic inward pumping.52 The eluate is aliquoted into 8 × 10 μl aliquots, potentially allowing for 32-plex multiplexing (geometrical 8-plex distribution with up to 4-plex detection per reaction cavity using 4 different detection channels for real-time RT-PCR) with subsequent RT-PCR of the extracted RNA. All required reagents are prestored, either as dried reagents (magnetic beads, primer and fluorescence probes, lyophilized RT-PCR mastermix) or as liquid reagents using miniature stick-packs. Supply of the sample into the LabDisk remains the only manual handling step. The LabDisk is processed in a LabDisk player, a 15 cm × 18 cm × 28 cm small and 2 kg light device suitable for operation at the point-of-care. Functionality of the system was verified with an influenza real-time PCR assay using the respiratory pathogen influenza A H3N2 virus.
Materials and methods
LabDisk fabrication
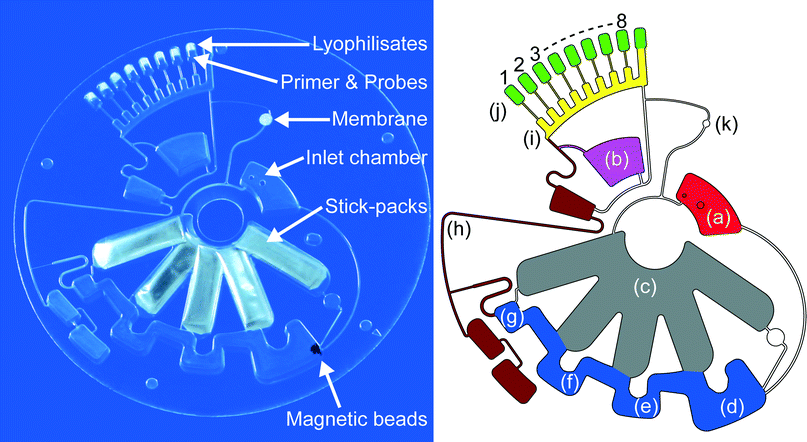
All microfluidic structures were designed as 3D-CAD drawing (SolidWorks, Dassault Systèmes SolidWorks Corp., France) and manufactured in-house by Hahn-Schickard Lab-on-a-Chip Design and Foundry Service (http://www.hahn-schickard.de/en/manufacturing/lab-on-a-chip-design-foundry-service) applying microthermoforming by soft lithography (μTSL)7 as rapid prototyping and scalable replication process.53 In brief, the microstructures were milled in a 4 mm thick polymethylmethacrylate (PMMA) substrate using an ultra-precision milling machine (Kern EVO, KERN Microtechnik GmbH, Germany). The PMMA substrate is cast with polydimethylsiloxane (PDMS) (Elastosil RT-607, Wacker Chemie AG, Germany) and cured at 80 °C for 2 hours before the PDMS master mold with the elevated microfluidic structures is post-cured at 200 °C for one hour in order to prevent outgassing of remaining monomers of the PDMS during the following thermoforming process. The cured PDMS master mold is placed into a hot embossing machine (HEX01, Jenoptik AG, Germany) for micro-thermoforming of the LabDisk (diameter = 130 mm) using 188 μm thick cyclic olefin polymer foil (COP, ZF14, TOPAS Advanced Polymers GmbH, Germany). COP acts as an appropriate fabrication material for LabDisk production7 due to its beneficial properties like mechanical and thermal stability, high transparency as well as biological inertness. Sample inlet and air vent were cut into the LabDisk using a CO2 laser (PLS 3.60, Universal Laser Systems, Inc., USA). The surfaces of the nucleic acid extraction chambers which later come in contact with extraction buffers and magnetic beads during bead transfer as well as the stick-pack chambers were hydrophobically coated by pipetting 120 μL of a 0.5% w/w Teflon AF solution (E. I. du Pont de Nemours and Company, USA) dissolved in Fluorinert FC-77 (3M Co., USA). Used surface coating does not have inhibiting effects on the assay performance.Reagent prestorage
Liquid buffers for nucleic acid extraction (Agrobiogen GmbH, Germany) are pre-stored in miniature stick-packs54 – 30 mm × 9 mm aluminum pouches allowing long-term storage and featuring frangible seals, which are opened due to liquid pressure under centrifugation at a very well defined spinning frequency. For liquid reagent prestorage, mass production of miniature stick-packs was applied on a commercial stick-packaging machine (SBL50, Merz-Verpackungsmaschinen GmbH, Germany) with a cycle time of 1.4 seconds per stick-pack. Modification of sealing parameters during stick-pack fabrication enables rotational frequency protocol defined on-demand release of 200 μl highly wetting binding buffer at the target release frequency of 70 Hz after the lysis procedure. All other liquid buffers (180 μl lysis buffer, 200 μl washing buffer 1, 200 μl washing buffer 2 and 120 μl elution buffer) are previously released at the target release frequency of 50 Hz. Oligonucleotide solutions – for multiplex detection of 22 pathogens (18 viruses and 4 atypical bacteria) which are generally recognized to cause a serious respiratory infection (RealAccurate Quadruplex Respiratory qPCR panels, PathoFinder B.V., The Netherlands; see ESI† Table S1 for detailed panel information) – consisting of primers and fluorescence probes were mixed with trehalose (Carl Roth GmbH & Co. KG, Germany) as stabilizing agent,55 pipetted into the reaction cavities of the LabDisk (for detailed allocation of the panels see Fig. 1 & 3) resulting in a 23-plex (18 viruses, 4 atypical bacteria and an internal control) detection by geometrical 8-plex distribution with up to 4-plex detection per reaction cavity using 4 different detection channels (FAM, Yakima Yellow, Texas Red and TYE665) for real-time RT-PCR. Subsequently magnetic beads (Agrobiogen GmbH, Germany) – including an excess of magnetic beads to prevent nucleic acid overloading – were mixed with trehalose and pipetted into the lysis chamber. Oligonucleotide solutions and magnetic beads were air dried for 4 hours at room temperature and ambient pressure (final trehalose concentration: 200 mM for magnetic beads, 56 mM for primer and fluorescence probes). Our previously reported protocol confirmed long-term functionality of air-dried oligonucleotide primers and fluorescent hydrolysis probes stabilized by trehalose for 1 year.55 After loading of stick-packs into the corresponding chambers, each of the reaction cavities was loaded with one bead of lyophilized polymerase (One-Step RT-PCR Master Lyophilisate, Jena Bioscience GmbH, Germany). These lyophilisates provide the RT-PCR mastermix after being dissolved by the RNA eluate and contain according to manufacturer SCRIPT Reverse Transcriptase, Hot Start Polymerase, dNTPs, reaction buffer, MgCl2 and stabilizers. After loading and drying of reagents the LabDisk was sealed with an adhesive foil (Polyolefin-foil #900320, HJ-Bioanalytik GmbH, Germany). A 0.2 μm pore size PTFE membrane filter (Merck Millipore KGaA, Germany) is attached to cover the air vent in order to prevent contamination by aerosols. Fixation and positioning holes were cut into the LabDisk using a CO2 laser (PLS 3.60, Universal Laser Systems, Inc., USA).Processing device
Processing of the LabDisk was conducted in a prototype LabDisk player (Qiagen Lake Constance GmbH, Germany) featuring PCR-thermocycling, four channel real-time fluorescence detection and the possibility to run predefined rotational frequency protocols consisting of a sequence of rotational frequencies (0–90 Hz), accelerations, decelerations (from 0.1 Hz s−1 through to 50 Hz s−1) and temperature variations (for 10 μl liquid: heating ramp 0.7 K s−1 and cooling ramp 1.0 K s−1). For fluorescence detection of the reaction cavities as well as magnetic bead transfer a precise positioning accuracy of 0.1° is provided. The LabDisk is mounted on an aluminum holder fixed to the rotational axis inside the processing chamber providing stationary magnets (#S-08-04-N and #S-06-03-N, Webcraft GmbH Supermagnete.de, Germany) on two radial positions for automated transfer of magnetic beads during nucleic acid extraction process.24 Temperature control of the processing chamber is realized by heating of circulating air with heating wires and valve-regulated influx of ambient air is used for cooling. Due to the size and weight of the portable processing device (15 cm × 18 cm × 28 cm; 2 kg) the LabDisk player is suitable for operation at the point-of-care.Microfluidic layout
The LabDisk for sample-to-answer nucleic acid based detection of respiratory pathogens consists of an inlet chamber for the sample which is connected to the lysis and binding chamber of the nucleic acid extraction structure. Stick-packs are pre-stored in chambers of 3.55 mm height. To save space, the chambers overlap at inner radii. Appropriate stick-pack chambers are connected to different chambers of the DNA extraction structure, the lysis and binding chamber, containing the prestored magnetic beads, followed by the washing chamber 1, the washing chamber 2 and the elution chamber. All these radially outward arranged chambers are interconnected by radially inwards placed bridging areas for the transfer of the magnetic beads. The elution chamber is connected to a fluidic network consisting of channels and pneumatic chambers52 for eluate transfer to the aliquoting structure, a sloping feed channel with branched off metering chambers which are connected to the cross contamination free7 reaction cavities via centrifugo-pneumatic valves.41 Prestored primer, fluorescence probes and RT-PCR mastermix lyophilisates will be rehydrated as soon as the reaction mix is transferred to the reaction cavities. The microfluidic layout of the LabDisk is shown in Fig. 1.Fluidic processing and RT-PCR based amplification
The LabDisk is mounted onto the holder in the LabDisk player and after supplying 200 μl of the sample into the inlet chamber, the automated microfluidic process flow (Fig. 2) begins by starting the rotational frequency protocol (see ESI† Table S2 for detailed protocol). Centrifugal forces are used for microfluidic unit operations like transport, mixing, valving and aliquoting. As initial sample material we used the RNA bacteriophage MS2 (DSM-13767, type: Levivirus, species: Enterobacteria phage MS2, DSMZ GmbH, Germany) acting as internal control of the used respiratory panels and was diluted with SM buffer, pH 7.5 (Alfa Aesar GmbH & Co KG, Germany) and applied with the concentration of 75 pfu ml−1. As respiratory pathogen sample we used the influenza A virus (H3N2) (ATCC® VR822™, classification: Orthomyxoviridae, influenza virus A; strain: A/Victoria/3/75, American Type Culture Collection (ATCC), USA) with a stock concentration of 2.39 × 106 viral RNA copies per ml (determined by digital PCR) which was diluted with 1× Tris-EDTA buffer (Sigma-Aldrich Co. LLC., USA) to a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 and a 1
10 and a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 dilution. After RNA extraction the eluate is transferred into the reaction cavities where the eluate rehydrates the prestored RT-PCR lyophilisates as well as the primer and fluorescence probes. Subsequently the RT-PCR with real-time fluorescent signal readout is performed.
100 dilution. After RNA extraction the eluate is transferred into the reaction cavities where the eluate rehydrates the prestored RT-PCR lyophilisates as well as the primer and fluorescence probes. Subsequently the RT-PCR with real-time fluorescent signal readout is performed.
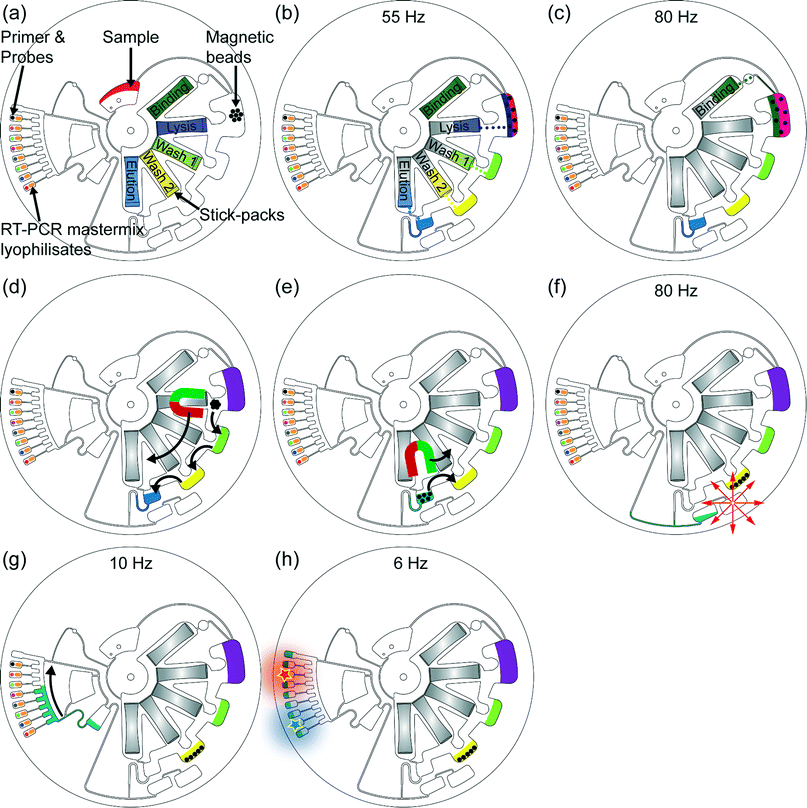 | ||
| Fig. 2 Centrifugal microfluidic process-flow: (a) the 200 μl sample is supplied into the LabDisk. Then, the automated rotational frequency protocol starts: the sample is pumped radially outwards into the lysis chamber by centrifugation and therein prestored magnetic beads get rehydrated. (b) At a frequency of 55 Hz the RNA extraction buffers (except the binding buffer) are released out of the stick-packs and transferred to the corresponding and adjacent chambers of the RNA extraction structure, magnetic beads get rehydrated and resuspended and sample lysis starts while applying shake mode mixing (frequency alteration between 2 Hz and 14 Hz). The chemical lysis was applied for 10 minutes. All application times during extraction procedure were applied as specified by the manufacturer of the reagents. (c) The binding buffer is added to the lysed sample at a frequency of 80 Hz enabling RNA binding to the magnetic beads. The binding step takes 5 minutes and is accompanied by shake mode mixing. (d) Magnetic beads are transferred through the washing buffers by magnetic actuation.24 In each washing buffer the magnetic beads were mixed for 3 minutes by magnet induced collection of the magnetic beads (0 Hz for 3 seconds) followed by resuspension due to acceleration to the frequency of 10 Hz (hold for 7 seconds). Finally, the magnetic beads are transferred into the elution buffer and the RNA is released of the beads in the following 5 minutes. (e) After RNA elution the magnetic beads are transferred into the previous washing chamber, preventing potential failures in the following microfluidic procedure induced by the magnetic beads. (f) The RNA eluate is transferred to the radially inward placed collection chamber by centrifugo-dynamic inward pumping.52 (g) By applying 10 Hz the RNA eluate is transferred into the aliquoting structure, preparing 10 μl aliquots. The excess volume of RNA eluate is collected in the adjacent waste reservoir located at the end of the aliquoting structure. At 20 Hz aliquots of 10 μl are centrifuged into each of the eight reaction cavities inducing rehydration of the therein prestored RT-PCR lyophilisates as well as of primers and fluorescent probes. (h) Finally, reverse transcription and PCR with real-time fluorescent readout are performed. | ||
Results and discussion
Microfluidic performance
The microfluidic process flow as illustrated in Fig. 2 was successfully demonstrated in 54 out of 55 LabDisks. In one experiment the eluate was not forwarded to the aliquoting structure due to a clogged channel of the air vent system. As there was no eluate transfer into the reaction cavities, false negative results were prevented.Liquid reagent release
Stick-pack reagent release frequency was optically determined using a stroboscopic image acquisition system (Biofluidix GmbH, Germany) with a total of 100 stick-packs (20 stick-packs per extraction buffer). For evaluation frequency was increased with an acceleration rate of 0.1 Hz s−1 and the results are stated in Table 2.| Binding buffer | Lysis buffer | Washing buffer 1 | Washing buffer 2 | Elution buffer | |
|---|---|---|---|---|---|
| Prestored volume [μm] | 200 | 180 | 200 | 200 | 200 |
| Mean release frequency [Hz], n = 20 | 70.5 | 51.7 | 49.8 | 49.7 | 48.8 |
| Standard deviation [Hz] | 1.2 | 0.7 | 1.0 | 0.8 | 0.8 |
| CV [%] | 1.7 | 1.4 | 2.0 | 1.6 | 1.6 |
The measured release frequencies demonstrate successful performance of on-demand frequency controlled reagent release. For ensuring the controlled reagent release in the LabDisk the parameters in the rotational frequency protocol were set to 60 seconds at 55 Hz for the lysis, washing 1, washing 2 and elution buffer release and 60 seconds at 80 Hz for the binding buffer release.
Assuming a normal distribution of release frequencies, we can estimate the error rates for the different stick-packs based on Table 2. The system will fail, if a) the stick-pack containing the binding buffer opens prematurely (release frequency ≤ 55 Hz, 12.9 sigma) or not at all (release frequency > 80 Hz, 7.9 sigma), b) the stick-pack containing the lysis buffer does not open at the initial 55 Hz rotation (4.7 sigma) or c) one of the other stick-packs does not open at the binding buffer release frequency of 80 Hz (>35 sigma). Based on the cumulated error rates for individual stick-packs, the error rate for full disks is 1 out of 825.000.
Preliminary detection of internal control of respiratory panels
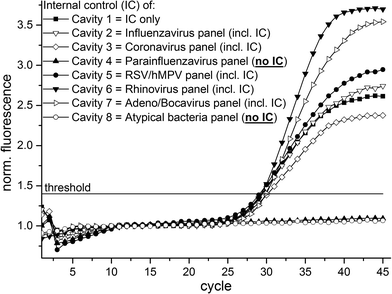
For initial experiments and demonstration of geometrical multiplexing capability the RNA bacteriophage MS2 – acting as the internal control (IC) of the respiratory panels – was used as sample. Due to the composition of the respiratory panels (see ESI† Table S1 for detailed panel information) the IC oligonucleotides was absent in two of the reaction cavities (detailed oligonucleotide solution allocation in Fig. 1 & 3) and thus no positive fluorescence signals were expected in these cavities. For this experiment we initially equipped chamber (b) (see Fig. 1) with 100 μl qScript One-Step RT-qPCR buffer (Quanta BioSciences, Inc., USA) instead of using the RT-PCR mastermix lyophilisates. The RT-PCR buffer is transferred to the reaction cavities (10 μl each) during microfluidic process steps (a) and (b), illustrated in Fig. 2. After supplying of 200 μl RNA bacteriophage MS2 (75 pfu ml−1) into the LabDisks the automated microfluidic processing (illustrated in Fig. 2) was started. The usage of the liquid RT-PCR buffer results in a final reaction volume of 20 μl in the reaction cavities. The subsequent real-time RT-PCR begins with the initial RT step (50 °C for 10 minutes) followed by initial denaturation at 95 °C for 1 minute and 45 thermocycles (95 °C for 20 seconds; 60 °C for 60 seconds) with real-time fluorescent signal readout. A total of three LabDisks were successfully processed and an exemplary result of one of the experiments is shown in Fig. 3. Positive amplification signals were detected by real-time RT-PCR in all six reaction cavities containing the IC oligonucleotide solution and, respectively, no amplification was detected in the two reaction cavities where IC oligonucleotide solution was absent, demonstrating sample-to-answer detection and geometrical multiplexing as success of the experiments. The time to result was less than 3 hours.Detection of influenza H3N2 virus
For demonstration of the fully automated sample-to-answer process with sample supply as the only manual handling step and detection of a respiratory pathogen we used influenza A virus (H3N2) (ATCC® VR822™) (American Type Culture Collection (ATCC), USA) as sample in a total of 18 LabDisks with prestorage of all reagents. Three virus concentrations were used as sample and the concentration of the stock solution was determined by digital PCR being 2.39 × 106 viral RNA copies per ml. Additionally the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 dilution (concentration approx.: 2.39 × 105 viral RNA copies per ml) and the 1
10 dilution (concentration approx.: 2.39 × 105 viral RNA copies per ml) and the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 dilution (concentration approx.: 2.39 × 104 viral RNA copies per ml) from stock were used as sample. The final reaction volume was 10 μl in each reaction cavity. The real-time RT-PCR begins with the initial RT step (50 °C for 30 minutes) followed by initial denaturation at 95 °C for 5 minutes and 45 thermocycles consisting of denaturation (94 °C for 5 seconds) as well as annealing and elongation steps (55 °C for 40 seconds) with real-time fluorescence signal readout. The fluorescence signals of all 18 experiments were identically normalized and plotted in Fig. 4. We achieved positive amplification signals in 18 out of 18 LabDisks especially emphasizing the positive amplification results in the 5 LabDisks with the 1
100 dilution (concentration approx.: 2.39 × 104 viral RNA copies per ml) from stock were used as sample. The final reaction volume was 10 μl in each reaction cavity. The real-time RT-PCR begins with the initial RT step (50 °C for 30 minutes) followed by initial denaturation at 95 °C for 5 minutes and 45 thermocycles consisting of denaturation (94 °C for 5 seconds) as well as annealing and elongation steps (55 °C for 40 seconds) with real-time fluorescence signal readout. The fluorescence signals of all 18 experiments were identically normalized and plotted in Fig. 4. We achieved positive amplification signals in 18 out of 18 LabDisks especially emphasizing the positive amplification results in the 5 LabDisks with the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 dilution as sample (concentration approx.: 2.39 × 104 viral RNA copies per ml). Literature describes the range of clinical relevance of pathogen detection for the influenza A virus in between of 105 viral RNA copies per ml56 and 106 viral RNA copies per ml.57 We detected the influenza A H3N2 virus with approximately fourfold lower concentration than clinical relevance and thus obtaining sufficient sensitivity for clinical relevance. The time-to-result was less than 3.5 hours. Clinical real patient samples like sputum are typically of varying physical properties like viscosity and homogeneity. Therefore, an integrated sample preparation unit for liquefaction and homogenization of clinical real patient samples will be implemented and evaluated onto the LabDisk in future projects.
100 dilution as sample (concentration approx.: 2.39 × 104 viral RNA copies per ml). Literature describes the range of clinical relevance of pathogen detection for the influenza A virus in between of 105 viral RNA copies per ml56 and 106 viral RNA copies per ml.57 We detected the influenza A H3N2 virus with approximately fourfold lower concentration than clinical relevance and thus obtaining sufficient sensitivity for clinical relevance. The time-to-result was less than 3.5 hours. Clinical real patient samples like sputum are typically of varying physical properties like viscosity and homogeneity. Therefore, an integrated sample preparation unit for liquefaction and homogenization of clinical real patient samples will be implemented and evaluated onto the LabDisk in future projects.
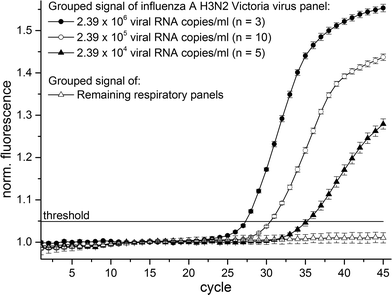 | ||
| Fig. 4 Combined fluorescence signals of 18 individual LabDisk experiments with different influenza A H3N2 virus concentrations as sample. | ||
Conclusion and outlook
The fully automated sample-to-answer process including chemical lysis, RNA extraction and subsequent real-time RT-PCR was demonstrated in the centrifugal LabDisk platform. Reduction to one initial manual handling step (sample supply) was realized by prestorage of all required liquid and solid reagents. The use of frequency dependent on-demand release of liquid buffers out of stick-packs facilitates handling and post-lysis addition of highly wetting binding buffer. Verification was performed with RNA bacteriophage MS2 as sample (75 pfu per ml) and additionally the influenza A H3N2 virus was detected with clinical relevant sensitivity. The space-saving microfluidic design leaves room for an improved LabDisk design containing more reaction cavities enabling the implementation of more pathogen panels and thus increases the degree of multiplexity. By further compression of the presented microfluidic design two identical structures could be implemented on one LabDisk. Furthermore the internal control might be prestored by air drying into the sample chamber preventing manual addition of the internal control to the sample.The next step is a clinical validation of all integrated pathogen panels which either requires off- or on-disk homogenization and liquefaction of crude clinical samples.58 Handling of viscous and inhomogeneous crude clinical samples, such as sputum or tracheal and bronchial secretion is a challenging task which is not solved in any of the discussed commercially available systems (Table 1). Handling of inhomogeneous sample material may be addressed in further developments by adding a defined inlet port with an integrated sample preparation unit onto the LabDisk. In addition we will significantly reduce the time-to-result in further developments by shortening of the nucleic extraction protocol and reverse transcription process time or the usage of time-saving isothermal amplification methods e.g. recombinase polymerase amplification (RPA) or loop mediated isothermal amplification (LAMP). Geometrical multiplexing41 guarantees high degree of multiplexing even by applying isothermal amplification methods. Furthermore, the aliquoting structure in the current disk could be replaced with centrifugal step emulsification,59 which would allow for fully integrated sample-to-digital-answer disposables.
Overall, the LabDisk features 1) a disposable with only one structured part and a flat sealing foil, 2) a simple reader with a rotational motor, heating unit and fluorescent detector, and 3) the possibility to use standard PCR assays without microarrays. This could be very attractive features when commercializing the system.
Acknowledgements
The authors like to thank for financial support from the EU FP7-HEALTH program (research project PARCIVAL – “Partner Network for a Clinically Validated Multi-Analyte Lab-on-a-Chip Platform”, grant agreement No. GA 278090).Notes and references
- World Health Organisation, http://www.who.int/mediacentre/factsheets/fs310/en/ (accessed 07-22-2015, 2015).
- C. C. Butler, K. Hood, T. Verheij, P. Little, H. Melbye, J. Nuttall, M. J. Kelly, S. Mölstad, M. Godycki-Cwirko, J. Almirall, A. Torres, D. Gillespie, U. Rautakorpi, S. Coenen and H. Goossens, BMJ, 2009, 338, b2242 CrossRef CAS PubMed.
- World Health Organisation, Antimicrobial resistance: global report on surveillance, World Health Organization, 2014 Search PubMed.
- The White House National Action Plan for Combating Antibiotic-Resistant Bacteria, 2015, https://www.whitehouse.gov/sites/default/files/docs/national_action_plan_for_combating_antibotic-resistant_bacteria.pdf (accessed 07-22-2015, 2015) Search PubMed.
- K. H. Rand, H. Rampersaud and H. J. Houck, J. Clin. Microbiol., 2011, 49, 2449–2453 CrossRef PubMed.
- M. Xu, X. Qin, M. L. Astion, J. C. Rutledge, J. Simpson, K. R. Jerome, J. A. Englund, D. M. Zerr, R. T. Migita, S. Rich, J. C. Childs, A. Cent and M. A. Del Beccaro, Am. J. Clin. Pathol., 2013, 139, 118–123 CrossRef PubMed.
- M. Focke, F. Stumpf, B. Faltin, P. Reith, D. Bamarni, S. Wadle, C. Müller, H. Reinecke, J. Schrenzel, P. Francois, D. Mark, G. Roth, R. Zengerle and F. von Stetten, Lab Chip, 2010, 10, 2519–2526 RSC.
- R. H. Liu, J. Yang, R. Lenigk, J. Bonanno and P. Grodzinski, Anal. Chem., 2004, 76, 1824–1831 CrossRef CAS PubMed.
- J. Pipper, M. Inoue, L. F.-P. Ng, P. Neuzil, Y. Zhang and L. Novak, Nat. Med., 2007, 13, 1259–1263 CrossRef CAS PubMed.
- S. Schumacher, J. Nestler, T. Otto, M. Wegener, E. Ehrentreich-Förster, D. Michel, K. Wunderlich, S. Palzer, K. Sohn, A. Weber, M. Burgard, A. Grzesiak, A. Teichert, A. Brandenburg, B. Koger, J. Albers, E. Nebling and F. F. Bier, Lab Chip, 2012, 12, 464–473 RSC.
- G. Xu, T.-M. Hsieh, D. Y. S. Lee, E. M. Ali, H. Xie, X. L. Looi, E. S.-C. Koay, M.-H. Li and J. Y. Ying, Lab Chip, 2010, 10, 3103–3111 RSC.
- D. Mark, S. Haeberle, G. Roth, F. von Stetten and R. Zengerle, Chem. Soc. Rev., 2010, 39, 1153–1182 RSC.
- O. Strohmeier, M. Keller, F. Schwemmer, S. Zehnle, D. Mark, F. von Stetten, R. Zengerle and N. Paust, Chem. Soc. Rev., 2015, 44, 6187–6229 RSC.
- J. Ducrée, S. Haeberle, S. Lutz, S. Pausch, F. von Stetten and R. Zengerle, J. Micromech. Microeng., 2007, 17, S103–S115 CrossRef.
- L. X. Kong, A. Perebikovsky, J. Moebius, L. Kulinsky and M. Madou, J. Lab. Autom., 2015 DOI:10.1177/2211068215588456.
- H. Kido, M. Micic, D. Smith, J. Zoval, J. Norton and M. Madou, Colloids Surf., B, 2007, 58, 44–51 CrossRef CAS PubMed.
- J. Kim, S. Hee Jang, G. Jia, J. V. Zoval, N. A. Da Silva and M. J. Madou, Lab Chip, 2004, 4, 516–522 RSC.
- J. Siegrist, R. Gorkin, M. Bastien, G. Stewart, R. Peytavi, H. Kido, M. Bergeron and M. Madou, Lab Chip, 2010, 10, 363–371 RSC.
- Y.-K. Cho, J.-G. Lee, J.-M. Park, B.-S. Lee, Y. Lee and C. Ko, Lab Chip, 2007, 7, 565–573 RSC.
- J. Hoffmann, D. Mark, S. Lutz, R. Zengerle and F. von Stetten, Lab Chip, 2010, 10, 1480–1484 RSC.
- J. H. Jung, B. H. Park, Y. K. Choi and T. Seo, Lab Chip, 2013, 13, 3383–3388 RSC.
- A. Kloke, A. R. Fiebach, S. Zhang, L. Drechsel, S. Niekrawietz, M. M. Hoehl, R. Kneusel, K. Panthel, J. Steigert, F. von Stetten, R. Zengerle and N. Paust, Lab Chip, 2014, 14, 1527–1537 RSC.
- B. H. Park, J. H. Jung, H. Zhang, N. Y. Lee and T. S. Seo, Lab Chip, 2012, 12, 3875–3881 RSC.
- O. Strohmeier, A. Emperle, G. Roth, D. Mark, R. Zengerle and F. von Stetten, Lab Chip, 2013, 13, 146–155 RSC.
- O. Strohmeier, S. Keil, B. Kanat, P. Patel, M. Niedrig, M. Weidmann, F. Hufert, J. Drexler, R. Zengerle and F. von Stetten, RSC Adv., 2015, 5, 32144–32150 RSC.
- M. Amasia, M. Cozzens and M. J. Madou, Sens. Actuators, B, 2012, 161, 1191–1197 CrossRef CAS.
- P.-A. Auroux, Y. Koc, A. deMello, A. Manz and P. J. R. Day, Lab Chip, 2004, 4, 534–546 RSC.
- M. Focke, F. Stumpf, G. Roth, R. Zengerle and F. von Stetten, Lab Chip, 2010, 10, 3210–3212 RSC.
- J. H. Jung, S. J. Choi, B. H. Park, Y. K. Choi and T. S. Seo, Lab Chip, 2012, 12, 1598–1600 RSC.
- S. Lutz, P. Weber, M. Focke, B. Faltin, J. Hoffmann, C. Müller, D. Mark, G. Roth, P. Munday, N. Armes, O. Piepenburg, R. Zengerle and F. von Stetten, Lab Chip, 2010, 10, 887–893 RSC.
- O. Strohmeier, S. Laßmann, B. Riedel, D. Mark, G. Roth, M. Werner, R. Zengerle and F. von Stetten, Microchim. Acta, 2014, 181, 1681–1688 CrossRef CAS.
- O. Strohmeier, N. Marquart, D. Mark, G. Roth, R. Zengerle and F. von Stetten, Anal. Methods, 2014, 6, 2038–2046 RSC.
- M. M. Hoehl, M. Weißert, A. Dannenberg, T. Nesch, N. Paust, F. Stetten, R. Zengerle, A. H. Slocum and J. Steigert, Biomed. Microdevices, 2014, 16, 375–385 CAS.
- M. M. Aeinehvand, F. Ibrahim, S. W. Harun, W. Al-Faqheri, T. H. G. Thio, A. Kazemzadeh and M. Madou, Lab Chip, 2014, 14, 988–997 RSC.
- W. Al-Faqheri, F. Ibrahim, T. H. G. Thio, J. Moebius, K. Joseph, H. Arof and M. Madou, PLoS One, 2013, 8, e58523 CAS.
- R. Gorkin III, C. E. Nwankire, J. Gaughran, X. Zhang, G. G. Donohoe, M. Rook, R. O'Kennedy and J. Ducrée, Lab Chip, 2012, 12, 2894–2902 RSC.
- D. J. Kinahan, S. M. Kearney, N. Dimov, M. T. Glynn and J. Ducrée, Lab Chip, 2014, 14, 2249–2258 RSC.
- B. S. Lee, Y. U. Lee, H.-S. Kim, T.-H. Kim, J. Park, J.-G. Lee, J. Kim, H. Kim, W. G. Lee and Y.-K. Cho, Lab Chip, 2011, 11, 70–78 RSC.
- Y. Ouyang, S. Wang, J. Li, P. S. Riehl, M. Begley and J. P. Landers, Lab Chip, 2013, 13, 1762–1771 RSC.
- F. Schwemmer, S. Zehnle, D. Mark, F. von Stetten, R. Zengerle and N. Paust, Lab Chip, 2015, 15, 1545–1553 RSC.
- D. Mark, P. Weber, S. Lutz, M. Focke, R. Zengerle and F. von Stetten, Microfluid. Nanofluid., 2011, 10, 1279–1288 CrossRef CAS.
- P. Andersson, G. Jesson, G. Kylberg, G. Ekstrand and G. Thorsén, Anal. Chem., 2007, 79, 4022–4030 CrossRef CAS PubMed.
- F. Schwemmer, T. Hutzenlaub, D. Buselmeier, N. Paust, F. von Stetten, D. Mark, R. Zengerle and D. Kosse, Lab Chip, 2015, 15, 3250–3258 RSC.
- K. Abi-Samra, L. Clime, L. Kong, R. Gorkin III, T.-H. Kim, Y.-K. Cho and M. Madou, Microfluid. Nanofluid., 2011, 11, 643–652 CrossRef CAS.
- N. Godino, R. Gorkin III, A. V. Linares, R. Burger and J. Ducrée, Lab Chip, 2013, 13, 685–694 RSC.
- R. Gorkin, L. Clime, M. Madou and H. Kido, Microfluid. Nanofluid., 2010, 9, 541–549 CrossRef.
- M. C. R. Kong and E. D. Salin, Anal. Chem., 2010, 82, 8039–8041 CrossRef CAS PubMed.
- T.-H. Kim, J. Park, C.-J. Kim and Y.-K. Cho, Anal. Chem., 2014, 86, 3841–3848 CrossRef CAS PubMed.
- J. H. Jung, B. H. Park, S. J. Oh, G. Choi and T. S. Seo, Biosens. Bioelectron., 2015, 68, 218–224 CrossRef CAS PubMed.
- E. Roy, G. Stewart, M. Mounier, L. Malic, R. Peytavi, L. Clime, M. Madou, M. Bossinot, M. G. Bergeron and T. Veres, Lab Chip, 2015, 15, 406–416 RSC.
- G. Czilwik, T. Messinger, O. Strohmeier, S. Wadle, F. von Stetten, N. Paust, G. Roth, R. Zengerle, P. Saarinen, J. Niittymäki, K. McAllister, O. Sheils and D. Mark, Lab Chip, 2015, 15, 3749–3759 RSC.
- S. Zehnle, F. Schwemmer, G. Roth, F. von Stetten, R. Zengerle and N. Paust, Lab Chip, 2012, 12, 5142–5145 RSC.
- M. Focke, D. Kosse, D. Al-Bamerni, S. Lutz, C. Müller, H. Reinecke, R. Zengerle and F. von Stetten, J. Micromech. Microeng., 2011, 21, 115002 CrossRef.
- T. van Oordt, Y. Barb, J. Smetana, R. Zengerle and F. von Stetten, Lab Chip, 2013, 13, 2888–2892 RSC.
- M. Rombach, D. Kosse, B. Faltin, S. Wadle, G. Roth, R. Zengerle and F. von Stetten, BioTechniques, 2014, 57, 151–155 CAS.
- R. R. Jansen, J. Wieringa, S. M. Koekkoek, C. E. Visser, D. Pajkrt, R. Molenkamp, M. D. de Jong and J. Schinkel, J. Clin. Microbiol., 2011, 49, 2631–2636 CrossRef PubMed.
- A. Franz, O. Adams, R. Willems, L. Bonzel, N. Neuhausen, S. Schweizer-Krantz, J. U. Ruggeberg, R. Willers, B. Henrich, H. Schroten and T. Tenenbaum, J. Clin. Virol., 2010, 48, 239–245 CrossRef PubMed.
- P.-H. Huang, L. Ren, N. Nama, S. Li, P. Li, X. Yao, R. A. Cuento, C.-H. Wei, Y. Chen, Y. Xie, A. A. Nawaz, Y. G. Alevy, M. J. Holtzman, J. P. McCoy, S. J. Levine and T. J. Huang, Lab Chip, 2015, 15, 3125–3131 RSC.
- F. Schuler, F. Schwemmer, M. Trotter, S. Wadle, R. Zengerle, F. von Stetten and N. Paust, Lab Chip, 2015, 15, 2759–2766 RSC.
Footnotes |
| † Electronic supplementary information (ESI) available: Details of respiratory qPCR panels and rotational frequency protocol. See DOI: 10.1039/c5lc00871a |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2016 |