Research highlights: functions of the drinking water microbiome – from treatment to tap
C. Kimloi
Gomez-Smith
*a,
David T.
Tan
b and
Danmeng
Shuai
c
aDepartment of Civil, Environmental, and Geo-Engineering, University of Minnesota, 500 Pillsbury Drive SE, Minneapolis, USA. E-mail: gomez224@umn.edu
bMalaysia Innovation Hub, Level 19, Wisma R & D Universiti Malaya, Jalan Pantai Baharu, 59990 Kuala Lumpur, Malaysia
cDepartment of Civil and Environmental Engineering, The George Washington University, 800 22nd St NW Suite 3530, Science and Engineering Hall, Washington, DC 20052, USA
First published on 2nd March 2016
Abstract
Maintaining drinking water safety from treatment to point-of use is a critical health priority. Growth and proliferation of opportunistic pathogens in premise plumbing is a well-known concern that can be mitigated by controlling water heater temperatures and water stagnation patterns. However, there is growing evidence that upstream processes, beginning with choice of treatment methods, have significant influences on premise plumbing microbial communities. Here, we highlight four papers that explore the roles of microbial communities in drinking water quality, and how design and treatment choices shape these roles.
Delivery of safe drinking water to point of use is the primary goal for drinking water utilities. Although water utilities utilize residual disinfectants and nutrient-limited environments to prevent the regrowth of microbes following water treatment, microbial communities can and do colonize, persist, and grow within drinking water systems – primarily in the form of biofilms. These drinking water communities are functionally active, diverse and can exert significant effects on drinking water quality. Critical functions in drinking water microbiomes include nitrification, which causes the decay of the chloramine disinfection residual, and protective functions against disinfection. It is impractical to eradicate microbes in drinking water distribution systems. Nonetheless, understanding the microbiome composition and function can enable strategies for monitoring and protecting end-point water quality. Here, we highlight determinants and functions of the drinking water microbiome in water treatment processes, distribution system biofilms, and premise (household/building) plumbing components.
Upstream drinking water processes determine community composition in water heaters
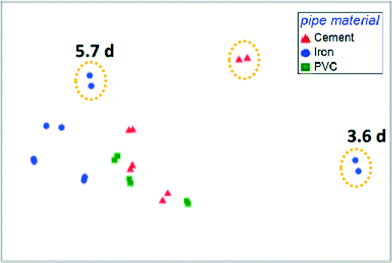
The presence of opportunistic pathogens within premise plumbing environments is a substantial health risk. These organisms are well-adapted to harsh drinking water environments, can flourish in premise plumbing, and are linked to occurrences of infection in hospitals and homes. Certain conditions in premise plumbing, such as water heater temperatures or water stagnation patterns, can be optimized to minimize the proliferation of these organisms. However, other upstream parameters may play important roles. Identifying key determinants of the premise plumbing microbiome is often complicated by the heterogeneity of full-scale systems. To overcome this challenge, Wang et al. (DOI: 10.1021/acs.est.5b01538) used simulated distribution systems and premise plumbing water heaters to evaluate the effect of upstream pipe materials, disinfectant type, water age, and stagnation regimes on the community composition and opportunistic pathogen abundance.A critical finding by Wang et al. was that upstream drinking water conditions and community composition exerted a greater influence over the water heater microbiome than the operating conditions and stagnation patterns within the heaters themselves (Fig. 1). Specifically, disinfectant type and concentration, water age, and pipe materials in the upstream distribution system substantially affected the presence and quantity of opportunistic pathogen gene markers. Furthermore, the distinctive effect of disinfectant dose and type on community structure persisted even after the disinfectant residual had decayed. Nitrification in water heaters resulted in the rapid decay of chloramine and, consequently, regrowth of total bacteria. Evidence that the premise plumbing heater microbiome was seeded and governed by upstream drinking water quality is significant, as opportunistic pathogen proliferation has largely been attributed to conditions within premise plumbing instead. These findings demonstrate the importance of considering upstream processes when selecting strategies for controlling opportunistic pathogens in premise plumbing.
Metabolic functions of drinking water treatment filters – implications for downstream seeding
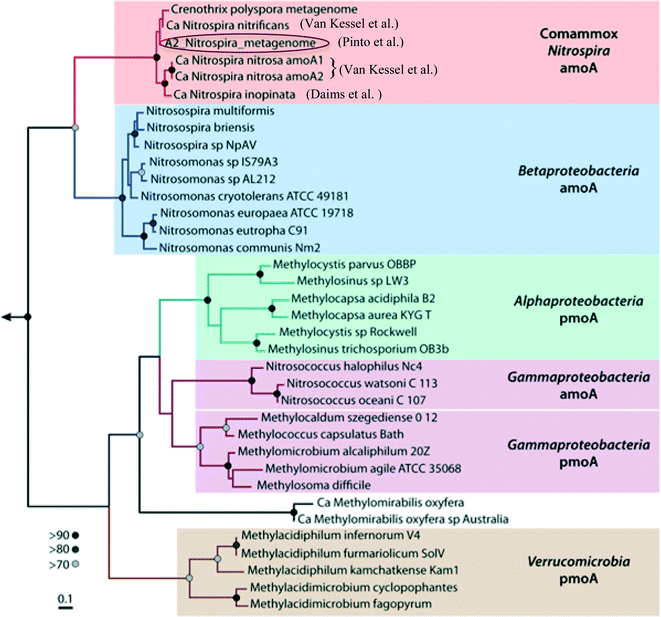
Water treatment processes are a major driver for community differences in distribution systems, even in premise plumbing as shown above. Accordingly, the choice of treatment systems can have a large role in determining the functions of drinking water microbial communities. Pinto et al. (DOI: 10.1021/es302042t) previously found evidence that the microbial communities of biologically-active drinking water treatment filters have a substantial role in seeding and shaping the downstream distribution system microbiome. In order to characterize potential functions that may be seeded downstream, Pinto et al. (DOI: 10.1128/mSphere.00054-15) have now used shotgun sequencing to elucidate potential functions in the microbiome of biologically-active, drinking water treatment filters, and, in this process, assembled a draft genome of a Nitrospira-like bacterium.Nitrospira are known nitrite-oxidizing bacteria (NOB); however, in addition to genes specific to nitrite-oxidation functions, the assembled Nitrospira draft genome also contained all of the genes necessary for ammonia oxidation functions (ammonia monooxygenase and hydroxylamine oxidoreductase genes). The placement and sequences of the ammonia oxidation genes, as well as other gene clusters, in the draft genome shared high similarity to genes of the Nitrospira strains recently identified by Daims et al. (DOI: 10.1038/nature16461) and van Kessel et al. (DOI: 10.1038/nature16459) (Fig. 2) that can perform complete nitrification (comammox) – oxidation of ammonia to nitrate. Although further studies are needed to link the genetic potential in this draft genome with functional capabilities, these findings may explain discrepancies observed by LaPara et al. (DOI: 10.1128/AEM.01692-15) and Shaw et al. (DOI: 10.1128/AEM.01297-15) between the abundances of ammonia-oxidation genes and known ammonia-oxidizing bacteria in drinking water systems. This study provided new metagenomic evidence that Nitrospira-like bacteria that are potentially capable of performing complete nitrification were present in the water treatment filters. The potential seeding of comammox Nitrospira into downstream drinking water distribution systems is a significant concern, as they could catalyze the rapid decay of chloramine disinfectants and thus degrade water quality.
GSH metabolism: a key protective function against disinfection residuals for drinking water biofilms
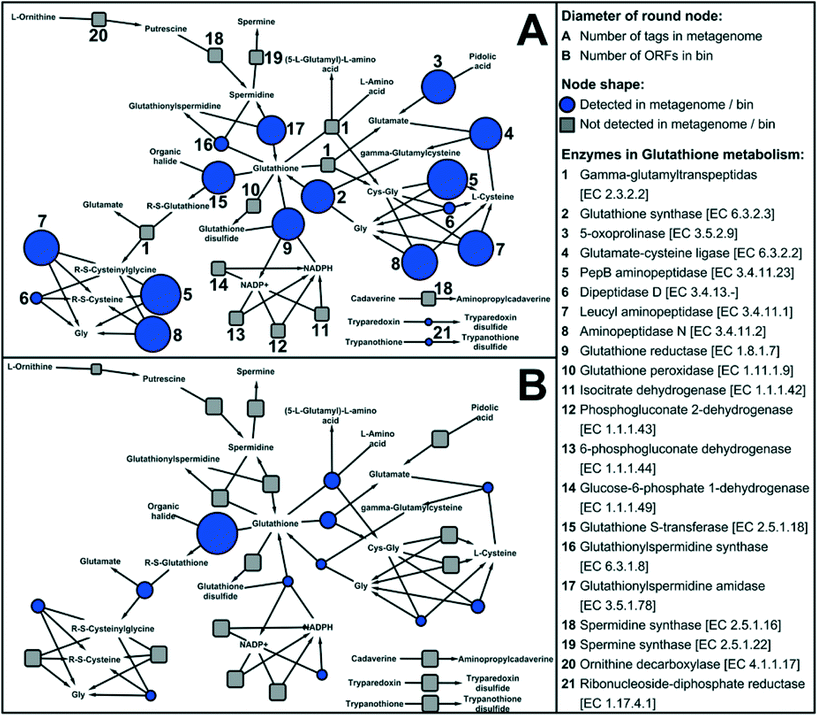
Disinfection processes during water treatment select for resistant bacteria, which may then seed downstream drinking water microbiomes as observed by Wang et al. (DOI: 10.1021/es402636u). The surviving microbes have protective functions including oxidative stress response systems that protect against reactive chemicals or repair oxidative damage. These protective functions provide an important competitive advantage in the presence of a disinfectant residual, as seen by the dominance of disinfectant resistant microbes in drinking water distribution systems. Vulnerable microbes, however, can survive by taking advantage of the protective functions of biofilm communities. One potentially important protective function is glutathione (GSH) metabolism. GSH has previously been shown by Chao et al. (DOI: 10.1038/srep03550) and Gomez-Alvarez et al. (DOI: 10.1186/1471-2180-12-122) to enhance disinfection resistance in drinking water microbiomes, and may have a regulatory role in other oxidative stress response systems such as SoxRS, OxyR, and RpoS. Chao et al. (DOI: 10.1038/srep10044) applied shotgun metagenomic analyses to simulated drinking water biofilms in order to explore metabolic functions of the microbiome, including the metabolic functions for protective mechanisms.There are three known enzymes, representing different pathways, for GSH synthesis. Genes encoding for all three were abundant in the biofilm, suggesting robust GSH synthesis by diverse pathways. For GSH degradation, the most abundant pathway reflected in the metagenome was glutathione S-transferase (GST), which can catalyze the reaction of GSH with the substrates of toxic compounds (e.g., the residual chlorine in drinking water). Among the oxidative stress response systems potentially linked to GSH, SoxRS and OxyR oxidative stress response systems were also relatively abundant, but RpoS system genes were relatively rare. Strong stimulation of GSH synthesis in response to disinfectant and starvation stresses may explain the overall disinfectant-resistant characteristics of chlorinated drinking water microbiomes. The large number of genes for GSH metabolism identified in the biofilm metagenome (Fig. 3) suggests the important role of this protein for biofilm disinfection resistance.
Nitrification metabolic functions of premise plumbing biofilms
Drinking water environments with extended water residence times, such as water towers and premise plumbing, are particularly vulnerable to nitrification of chloramine. In premise plumbing, warmer temperatures, overnight periods of stagnation, and reactions with various pipe materials, can contribute to decreases in chloramine residuals. Additionally, systems in which nitrifying organisms become established can experience persistent, re-occurring nitrification events despite mitigation measures. This is a concern as adequate disinfectant concentrations are important in suppressing the growth of opportunistic pathogens – such as Legionella pneumophila – that can otherwise proliferate in this environment. Santillana et al. (DOI: 10.1039/C5EW00273G) investigated the nitrification functions of simulated premise plumbing communities grown on different pipe materials.Nitrification occurred more quickly on polyvinyl chloride (PVC) as opposed to copper materials, possibly due to the higher abundance of archaea on the PVC materials (Fig. 4). Ammonia oxidation in this system was attributed to ammonia oxidizing archaea as no bacterial ammonia-oxidation genes were present in the communities. Sequencing of 16S rRNA and archaeal ammonia-oxidation genes indicated similarities to genes of the archaeon Candidatus Nitrosotenuis uzonensis. Quantification of nitrite-oxidizing genes (nxrA, nxrB) indicated the presence of known nitrite-oxidizing genera – Nitrobacter and Nitrospira. Species identification using 16S rRNA genes should be interpreted with caution; however, the similarity of these Nitrospira to strict nitrite-oxidizing species (N. japonica), in conjunction with the absence of bacterial ammonia-oxidation genes, suggests that these Nitrospira do not have the ability to completely nitrify, unlike the comammox strains described above. This study indicates that nitrification in some premise plumbing systems may occur as a result of archaeal–bacterial metabolic interactions. These findings emphasize the importance of studying inter-kingdom interactions in order to characterize the functional capabilities of the drinking water microbiome.
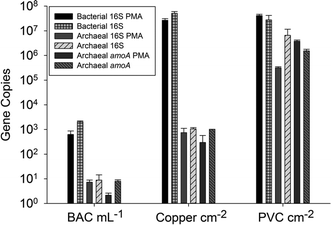 | ||
| Fig. 4 16S rRNA and amoA gene copy numbers from qPCR for influent water filtered by biologically active carbon (BAC) and biofilm from the copper and PVC reactor. BAC gene copy numbers are per mL, and biofilm samples per cm2. Samples are paired with or without the treatment of propidium monoazide (PMA) for the removal of DNA from cells with damaged membranes. Reproduced from DOI: 10.1039/C5EW00273G with permission from the Royal Society of Chemistry. | ||
Outlook
These studies illustrate the influence that upstream processes have on the drinking water microbiome and how functions in these microbiomes can impact end-point water quality. Drinking water treatment approaches such as disinfection select for resistant microbes, while biologically active filters may inadvertently seed the distribution system and premise plumbing with nitrifying organisms, leading to re-occurring nitrification events that enable the spread of opportunistic pathogens from protected biofilms into the bulk water. New approaches targeting upstream systems, such as limiting ammonia in water treatment filters to inhibit the growth of nitrifying organisms and minimize the seeding of these problematic microbes into the distribution system, should be explored. A challenging question that needs to be addressed in future studies regards the applicability of these and other similar findings from controlled environments to real-world systems. Real drinking water systems are diverse, with unique source water microbial communities and chemistries, differing water treatment operations, as well as varying hydraulic regimes, pipe materials, and conditions within the aging infrastructure of the distribution system. Understanding these factors will further advance the development and optimization of targeted microbial control for safe drinking water from treatment to tap.| This journal is © The Royal Society of Chemistry 2016 |