Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Opportunities for glyconanomaterials in personalized medicine
Niels-Christian
Reichardt
*ab,
Manuel
Martín-Lomas
a and
Soledad
Penadés
a
aCIC biomaGUNE, Glycotechnology Laboratory, Paseo Miramón 182, 20009 San Sebastian, Spain. E-mail: nreichardt@cicbiomagune.es
bCIBER BBN, Paseo Miramón 182, 20009 San Sebastian, Spain
First published on 31st August 2016
Abstract
In this feature article we discuss the particular relevance of glycans as components or targets of functionalized nanoparticles (NPs) for potential applications in personalized medicine but we will not enter into descriptions for their preparation. For a more general view covering the preparation and applications of glyconanomaterials the reader is referred to a number of recent reviews. The combination of glyco- and nanotechnology is already providing promising new tools for more personalized solutions to diagnostics and therapy. Current applications relevant to personalized medicine include drug targeting, localized radiation therapy, imaging of glycan expression of cancer cells, point of care diagnostics, cancer vaccines, photodynamic therapy, biosensors, and glycoproteomics.
Introduction
Traditional medicine at the beginning of the 19th century was deeply rooted in two major beliefs: (1) the body was viewed as a collection of parts that were all intimately connected and interlaced, leading to a systemic vision of every illness as opposed to a localized model of disease causation and (2) the body was seen as an equilibrated system of input and output in constant interaction with its environment and illness was a state of imbalance which had to be readjusted by diet, excretion, perspiration, ventilation or blood-letting. Consequently, among the preferred options for treatment were often those that produced strong and noticeable effects on the patient's body like e.g. mercury chloride (calomel), which was used as a purgative and diuretic, opium to relieve pain or stop diarrhea, and camphor to induce perspiration.1Lacking the molecular markers and in vivo imaging methods of today's medical practice, the physician's diagnosis and prognosis of an illness was based easily detectable symptoms, experience and perhaps more importantly a continued dialogue with the patient and his family.
The implementation of Descartes' scientific methodology not only revolutionized the natural sciences but with its reductionist approach also transformed medicine into an evidence based and analytical discipline.2 Applying this approach, clear disease categories were defined for the first time based on a set of common symptoms, reproducible methods for their analysis, and selective treatment usually of the single most dominant factor. The general success of this approach, e.g. in the tremendously efficient treatment of devastating bacterial infections with antibiotics or their prevention by immunization with vaccines clearly justified this idea.
Not all types of diseases however are equally well described and treated by this reductionist approach and a singular focus on a defined disease ignores the often remarkable differences among patients in response to a particular treatment (Table 1). The efficacy for most prescription drugs is rather moderate and lies between 30 and 70% depending on the treated disease.3 For some treatments like chemotherapy or anti-depressive drugs, efficacy can even fall below 25%.4 The result of these one-fits-all therapies has been an unnecessarily high expenditure for prescription drugs due to overtreatment but more importantly, an unjustified large number of patients suffering common drug side effects.
| Therapeutic area | Efficacy rate (%) |
|---|---|
| Alzheimer's disease | 30 |
| Analgesics (Cox-2) | 80 |
| Asthma | 60 |
| Cardiac arrhythmias | 60 |
| Depression (SSRI) | 62 |
| Diabetes | 57 |
| HCV | 47 |
| Incontinence | 40 |
| Migraine (acute) | 52 |
| Migraine (prophylaxis) | 50 |
| Oncology | 25 |
| Osteoporosis | 48 |
| Rheumatoid arthritis | 50 |
| Schizophrenia | 60 |
Personalized medicine was proposed more than a decade ago as a possible solution to overcome some of the shortcomings of current medical practice. It consists largely of an improved patient stratification process to select subpopulations that have a higher probability to benefit from a particular drug therapy (…the right drug for the right person…) and the exclusion of high risk individuals from treatment to reduce the number of patients suffering from severe side effects. The emphasis in today's personalized medicine approach from an industrial perspective is on diagnostics and risk assessment to improve the choice among existing treatment options rather than the development of individual treatment options for very small patient populations which are commercially unattractive as seen by low industry interest in rare diseases. Currently, personalized medicine is focused largely on genomic markers for patient stratification. While genomic disease markers are efficient in classifying patients based on hereditary risk factors and traits, they are less indicative in describing more dynamic risk factors which are influenced by the environment, diet, habits or the patient's gender or age. Here, the profiling of other biomolecules like oligosaccharides, lipids or metabolites with a generally significantly smaller repertoire of relevant structures than proteins, genes or RNA, integrated in an omics-wide analysis of a disease is likely to provide a clearer picture of the physiological condition that relates to a differentiated drug response, and hence allow patient classification into subgroups for personalized treatment (see Fig. 1).
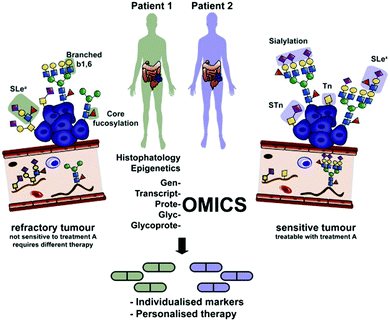 | ||
| Fig. 1 Integration of glycomics and glycoproteomics for the detection of aberrant glycosylation, e.g. in cancer into an omics-wide approach will be critical for improving diagnosis and personalized treatment. Reprinted from ref. 5 with permission from Elsevier. | ||
As biomarkers, glycans present a number of advantages over other biomolecules:6 Being a non-template controlled process, glycosylation is more strongly affected by environmental factors and physiological changes than e.g. protein expression. Glycan biosynthesis is also spatially limited to the Golgi and endoplasmic reticulum (ER), resulting in more uniform changes in glycosylation with disease on a cellular level. Finally, the usually on average higher molecular weight of glycans compared to metabolites and a generally higher structural homogeneity present advantages for the analysis and quantification of glycans.
As an example of a glycan biomarker, the DG9-glycan index, defined as the ratio of fucosylated and nonfucosylated triantennary glycans, provided optimal discrimination between two groups of subjects with different diabetes types according to a recent study.7 Efforts are also underway to integrate disease related information from biomarker studies into general glycan databases like UniCarbKB.8
In this feature article, we discuss the particular relevance of glycans as components or targets of functionalized nanoparticles (NPs) for potential applications in personalized medicine but we will not enter into descriptions for their preparation. For a more general view covering the preparation and applications of glyconanomaterials the reader is referred to a number of recent reviews.9–11 The combination of glyco- and nanotechnology is already providing promising new tools for more personalized solutions to diagnostics and therapy12–15 Current applications relevant to personalized medicine include drug targeting, localized radiation therapy, imaging of glycan expression on cancer cells, point of care diagnostics, cancer vaccines, photodynamic therapy, biosensors, glycoproteomics.
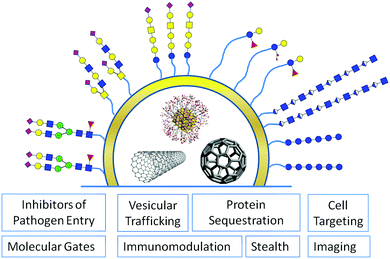
Glycans are ubiquitously distributed in all body tissues, and have important functions at a cellular level. Hence, any artificial nano-sized system for in vivo drug delivery or imaging should take advantage of the remarkable properties glycans offer to improve its biocompatibility or to facilitate tissue selective targeting.9 To fully understand the potential of glycans for functionalisation of nanomaterials it is important to recall the multiple functions glycans exert in nature on a cellular level (Fig. 2).
With over 200 glycosyltransferases involved in the biosynthesis of our glycome, defined as the collection of glycans found in one single species, our body invests heavily in the continued production of glycans underlining the importance of this class of biomolecules.16
Attached to a peptide or lipid backbone glycans interact specifically with carbohydrate specific receptors termed lectins on other cells facilitating reversible and tuneable cell–cell adhesion, a process that is important, e.g. for leukocyte targeting to infected tissue, in sperm-egg fertilization, and also in the infection through pathogens. This dense glycan coat on the extracellular surface is for other cells, parasites, viruses or bacteria, the first line of contact with a mammalian host, and in a constant co-evolutionary struggle pathogens have developed a number of ways to exploit glycans to evade recognition by the immune system or to overcome the glycan shield for infection.17,18
Targeting antigen presenting cells
Antigen presenting cells (APCs) like macrophages, dendritic cells or Langerhans cells are the sentinels of the human immune system and an important link between innate and adaptive immunity. They express a considerable number of carbohydrate binding surface receptors called C-type lectins (CLRs) that internalise pathogenic and also endogenous glycoconjugates and cross-present them to naïve T cells. As an example, DC-SIGN, a CLR that preferentially binds fucosylated and high mannose structures has been shown to be an entry portal for viruses like HIV, hepatitis, Marburg or dengue into dendritic cells.19 The clustering of DC-SIGN into lipid raft microdomains facilitates binding to large antigens with viral dimensions through a strong multivalent interaction, e.g. with the envelope glycoproteins20 (Fig. 3).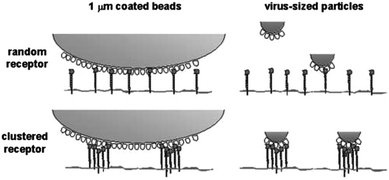 | ||
| Fig. 3 DC-SIGN receptor clustering in microdomains enhances the binding of virus-sized particles with respect to isolated DC-SIGN molecules. Reproduced from ref. 20 with permission by Rockefeller University Press. | ||
Consequently, blocking and sequestration of DC-SIGN cell surface receptors were accomplished using multivalent gold glyconanoparticles that had been functionalized with high mannose glycans of varying complexity mimicking the glycan shield of the gp120 HIV envelope protein and efficiently inhibiting the entry of HIV intro dendritic cells.21,22
Immunomodulation
Some pathogens express host-like glycan structures on their cells in a form of molecular mimicry to evade recognition by the host immune system.23 Helminthic parasites, for example, can target specific receptors located on host immune cells with host-like glycans to dampen the host immune response for their own benefit.24 Within their often complex life cycle which can span several different hosts, parasites adjust to the changing environmental conditions by dynamic changes in their cell surface glycosylation.25 The dynamic and varied structural expression of glycans is also a result of the million years of co-evolution between pathogens and theirs hosts and a strategy to counterbalance the rapid adaption of pathogens to a changing environment by unchallenged faster replication cycles.17 In light of the ubiquitous presence of carbohydrate binding proteins on immune cells, the selection of glycan structures for protecting nanomaterials for in vivo use should be guided taking into account the binding specificities of these lectins to either avoid a rapid decrease of circulating nanoparticles or rather to exploit the glycan-mediated internalisation of nanoparticles for immune cell targeting. Penades's group employed glucose as a major non-immunogenic carbohydrate component responsible for excellent water solubility in the design of their glyconanoparticles.26,27 On the other hand, the systemic administration of glyconanoparticles functionalised with the readily available monosaccharides galactose, fucose, N-acetylglucosamine or mannose has been shown to give rise to a tissue-dependant biodistribution.28 Also, glucose-functionalised particles can be concentrated in tissue overexpressing glucose transporters thereby leading to an uneven biodistribution of particles. This interaction of systemically administered glucose/folate functionalised gold nanoparticles with membrane bound glucose transporters and folate receptors that are both overexpressed in cancer, was exploited for targeted drug delivery to ovarian tumours.29 Likewise, glucose-functionalised gold nanoparticles also in combination with a neuropeptide ligand have been shown to enhance the crossing of the blood brain barrier presumably via a glucose transporter mediated uptake mechanism.27 These examples show that even sugars with an apparently low profile in molecular recognition processes like glucose can be used for directed targeting or in other terms should be assessed for potential off-target effects.Tumour targeting
Cell surface glycosylation is ubiquitous, but species and tissue specific. In addition, changes in glycosylation like an increase in sialylation or fucosylation of cell surface glycans are a hallmark of many cancer cells and are thought to play a functional role in metastasis, the migration of individual cancer cells from the tumour to other tissues. Consequently, targeting of cell surface glycans is a promising strategy for concentration of nanomaterials in tumor tissue or specific organs for imaging or therapeutic purposes.In a form of leukocyte mimicry, metastasizing tumour cells are thought to target and adhere to endothelial tissue far away from the parent tumour via selectin mediated binding to the same glycan sialyl-LeX.30 The overexpression of a 2,6 sialyltransferase in human breast cancer cells has been associated with an increased adhesion to and passage through the blood brain barrier possibly through P- and E-selectin mediated cell–cell adhesion. E and P selectins are overexpressed on inflamed vascular tissue and an early marker e.g. in brain lesions, that can be imaged by MRI using silica core shell superparamagnetic and iron oxide dextran coated nanoparticle based contrast agents carrying sialyl-LeX ligands, the natural selectin ligand.31,32 Likewise, the interaction of hyaluronic coated iron oxide nanoparticles with the cell surface receptor CD44 over-expressed on cancer cells has been exploited in imaging of tumours by MRI.33
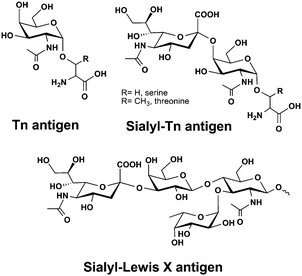
Mucins are large gel-forming and heavily glycosylated proteins with primary functions in lubrication and protection of epithelial tissues but they are also involved in cell differentiation, signalling and cell adhesion.34 Mucins are overexpressed in a large number of cancers and accompanied by an aberrant glycosylation profile presenting unique truncated structures like sialyl-Tn antigen (Fig. 4). Aberrant mucin glycosylation is currently being developed for improved cancer diagnostics and has been a molecular target for a large number of immunotherapeutic approaches including several anti-cancer vaccines.35 The current dogma of glycan based immunisation requires small carbohydrate antigens to be coupled to a peptide or protein carrier for the effective cross-presentation to naïve T cells by major histocompatibility complex (MHC) I or II leading to clonal expansion of T cells. A recent example however shows that a selective immune response that is cross-reactive to natural mucins can also be elicited with a peptide free system. Parry et al. attached Tn-antigen functionalized polymers of defined length and carbohydrate copy number onto gold nanoparticles to produce a highly multivalent antigen presentation. After being injected into rabbits, some of these constructs were able to elicit a Tn-antigen specific immune response that was cross-reactive with natural mucin, albeit less effective than constructs attached to a T-helper peptide or carrier toxin.36 Nevertheless, the failure of a large phase III sialyl-Tn/KLH vaccine trial to treat breast cancer highlighted the strong variation of sialyl-Tn expression among patients. A prior stratification of patients according to their sialyl-Tn expression level would most likely increase the number of patients benefitting from these types of therapies.37
Where the analysis of cell surface receptor density is an analytical challenge or not possible, theranostic nanoparticles14,38 that release their drug or vaccine cargo only upon encountering the minimal threshold density of surface receptors required for efficacy could minimize side effects and increase the number of responders to a particular treatment. For such an in vivo diagnostics and treatment platform, drug release mechanisms that respond to a receptor density threshold have to be developed. Multivalent effects between glycan functionalized nanoparticles and protein receptors could provide such a general mechanism for selective adhesion to cells presenting a specific surface receptor at a minimal density.39
Inhibitors of pathogen adhesion
The dense cloak of glycans is also frequently an attachment point of pathogens like bacteria, parasites or viruses initiating the infection. The interaction with host cell surface glycans is mediated by specific carbohydrate binding receptors termed lectins which are often found to be concentrated on the pili of bacteria mediating adhesion.40Pseudomonas aeruginosa, a bacterial pathogen primarily causing lung damage, invades host cells by a lipid zipper mechanism that operates via a clustering of the host cell surface glycolipid Gb3 by the bacterial lectin LecA.41Numerous examples of glyconanomaterials that selectively bind to uropathogenic Escherichia coli with potential for pathogen identification and as antiadhesives to prevent infection and biofilm formation have been described.42–44 More recently, trimeric cluster thiomannosides conjugated to diamond nanoparticles have been shown to inhibit type 1 fimbriae-mediated E. coli adhesion and biofilm formation45 and mannose-coated singlewalled carbon nanotubes (SWCNTs) were able to reduce the colony formation of the E. coli strain ORN178 by 10%.46
The carbohydrate–lectin mediated entry of pathogens has spurred the development of a large number of multivalent carbohydrate display systems including glyconanomaterials as potential inhibitors of this adhesion process. Recent examples include a trideca-fullerene functionalised with 120 mannose residues as a nanomolar inhibitor of virus adhesion in an Ebola infection model47 or mannose functionalised glyconanoparticles interfering with LecB binding of Pseudomonas aeruginosa, an important process in the formation of biofilms,48 and inhibiting the HIV trans infection of dendritic cells to T cells.21
The influenza virus makes extensive use of sialic acid derived glycans on the host cell surface in its replication cycle. It first attaches to the host lung tissue rich in alpha 2,3-sialosides via the envelope protein hemagglutinin (HA) initiating the endocytosis of the virus. After replication in the cell the budding virus detaches from the host cell again now cutting the binding host sialic acids with a specific sialidase to be able to move on for the next infection.49 For some time, a number of groups have been developing sialic acid (SA) modified polymers and nanoparticles to intervene in the HA–SA acid interaction and protect against influenza infection.50,51
Stealthing
Moving to their more intriguing roles in cellular communication, cell surface glycans also behave as an efficient physical barrier against the entry of bacterial or viral pathogens, protect proteins against protease degradation and can increase protein solubility significantly by reducing non-specific interactions with other proteins.Glycosylated plasma proteins like haptoglobin or antitrypsin with a high carbohydrate content and homogenous glycan distribution are most effectively protected against adsorption on hydrophobic interfaces.52
Surface glycosylation has consequently been employed in many materials to enhance biocompatibility, solubility and to add targeting functionality. This has been particularly important for enabling the use of highly hydrophobic carbon allotropes graphene, fullerenes or carbon nanotubes for biological and biomedical applications.10 The functionalisation of nanomaterials produced for in vivo applications in imaging, radiation therapy or drug delivery with glycans is also an efficient strategy to reduce the non-specific adsorption of plasma proteins on the nanomaterial surface, which can alter significantly their physiological response affecting agglomeration, cellular uptake, or reduce their circulatory half life by opsonisation.53
The spontaneous, dynamic and largely unpredictable adsorption of plasma proteins to surfaces termed protein corona formation has been recognized as the single most important hurdle for clinical applications of nanomaterials and strategies for reducing non-specific adsorption are in high demand.54 Complement plasma proteins and IgGs adsorb to insufficiently protected non-stealth particles and label them for rapid removal within seconds by macrophages of the mononuclear phagocytic system (opsonisation).55 This process poses a major challenge for the development of drug delivery systems with increased circulatory half life based on polymeric nanoparticles. Adsorption of proteins like fibrinogen or ribonuclease A to nanomaterials can also lead to destabilisation of their structure, e.g. by partial unfolding altering their function and promoting particle aggregation.56–58 This does not seem to be a universal process. For instance, protein adsorption on nanoparticles with high curvature actually improves protein stability and retains enzymatic functions, for example, for soy bean peroxidase, lysozyme and human carbonic anhydrase.56 Pre-coating of nanoparticles with a single protein-like albumin has been shown to increase colloidal stability and reduce binding of opsonins (C3, IgGs), consequently reducing nanoparticle uptake by macrophages.59 Protein coating of nanomaterials can also be further exploited for the attachment and proteolytic release of drugs.60,61
An increasing number of studies have revealed that the composition and presence of a protein corona is dependent on the nanoparticle size, charge, shape, surface functionalization and incubation time.54,62–71 While most studies have suggested a limited complexity of protein coronas of several dozens of proteins, a recent quantitative proteomic study identified up to 300 different proteins in the protein corona of silica and polystyrene nanoparticles.68 Protein adsorption from serum occurred within minutes and the corona composition changed little with prolonged exposure time. In addition, the authors observed that the particle net charge after corona formation was negative independent of the initial particle charge.68,69 The great majority of studies have analysed protein corona formation in vitro largely due to the difficulties involved in isolating nanoparticles after in vivo exposure.54 In a recent paper, however, the authors reported on the use of iron oxide nanoparticles that were isolated form rat sera with the help of a strong magnetic field to analyse and compare in vitro and in vivo protein corona formation. Their findings suggest significant differences in the protein corona composition highlighting the limitations of in vitro assays for correct simulation of this biomedically relevant process.72
Heparin is an inhibitor of the complement system and was covalently linked to polymethylmethacrylate (PMMA) nanoparticles to enhance the circulatory half-life of nanoparticles and circumvent opsonisation.73
The majority of serum proteins are glycosylated and as mentioned above glycosylation affects the circulatory half-life, stability and function. It therefore comes as no surprise that glycosylation also has a major impact on the colloidal stability of nanoparticles coated with a protein corona and affects the interaction between cells and nanoparticles, as a recent report by Wan et al. shows.74 Protein corona coated nanoparticles were incubated with a mixture of glycosidases for varying time lengths and the impact of reduced protein glycosylation on colloidal stability and cell nanoparticle interaction was measured. Partial removal of glycans from the hard protein corona led to a progressive decrease in the colloidal stability most probably due to the loss of the repellent functions of the outer glycan coating facilitating non-specific interactions between protein interfaces that ultimately destabilize the nanoparticle–protein complexation.74 The deglycosylated corona nanoparticles also adhered more strongly to macrophage membranes leading to increased NP uptake. In addition, the uptake of deglycosylated corona NPs by both M1 and M2 macrophages led to an increased expression of the pro-inflammatory cytokines hIL-1β and hTNF-α compared to the fully glycosylated corona NPs. In a recent example the Penadés and Liz-Marzan groups assessed the protective effect of glycan ligands for suppressing protein corona formation on gold nanorods. Lactose functionalised nanorods, GlcNAc functionalised gold NPs and PEG-functionalised nanomaterials showed a similar strongly reduced protein adsorption compared to charged citrate stabilised nanoparticles and rods, which rapidly formed complex protein coronas, highlighting the excellent repellent properties of glycans in protein rich media.64 A macrophage uptake assay comparing lactose and PEG functionalization showed a rapid uptake of PEGylated gold nanorods while lactose efficiently prevented phagocytosis (Fig. 5).64 These findings seem to be in line with a recent report by Schöttler et al. showing a stealth protection of PEGylated nanostructures against cellular uptake only in the presence of proteins adsorbed onto the PEG-layer.75 An analysis of the protein corona of PEGylated nanoparticles after exposure to plasma proteins showed selective adhesion of a high amount of clustering an apolipoprotein that reduced uptake by macrophages by up to 75%. These examples also demonstrate that certain glycans unlike PEG or other polymers used for the protection of nanomaterials against non-specific protein adsorption do not require a specific protein coating to exert their stealth effect.
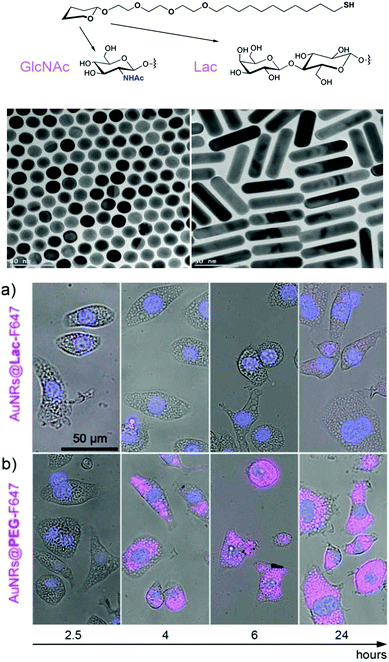 | ||
| Fig. 5 TEM images of GlcNAc-coated spherical gold nanoparticles AuNPs@GlcNAc and lactose-coated gold nanorods (AuNRs@Lac). Illustrative examples of the behaviour of fluorescently-labelled lactose- and polyethylene glycol-coated gold nanorods (AuNRs@Lac-F647 and AuNRs@PEG-F647) internalized by macrophage-like J774 cells over time at 37 °C, 5% CO2. The pink colour indicates the presence of nanoparticles, whereas the blue colour denotes the nuclei. Adapted from ref. 64 with permission. Copyright public domain. | ||
The in situ formation of gold or silver nanoparticles with polysaccharides as a reduction and/or passivation agent can be regarded as an alternative and particularly green chemistry method for the functionalization of nanoparticles with glycans.76 To this end, various polysaccharides including starch, chitosan and carboxymethylcellulose or dextran have been employed in the preparation of noble metal nanoparticles77 with good stability, water solubility and biocompatibility. Single-walled carbon nanotubes, which are highly insoluble in aqueous media have been solubilised by wrapping them in polysaccharides like starch or Arabic gum.78,79 The iodine embedded inside a helical amylase polysaccharide was displaced by a single carbon nanotube or bundles of carbon nanotubes while the charged Arabic gum polysaccharide spontaneously coated the hydrophobic carbon nanotubes in a process that also involved the exfoliation of carbon nanoropes into single tubes.78
This early work on glycan functionalized nanomaterials demonstrates the potential of glycans and glycan mixtures for tissue targeting and for passivation against non-specific protein adsorption. Many approaches to protect nanomaterials against agglomeration and non-specific protein adsorption employ polyethylene glycol, a material with generally low toxicity. As the extended use of polyethylene glycol as a common additive in cosmetics and drugs has been recently found to cause an anti-PEG directed immune response in nearly a quarter of the population80 the use of PEG-based surfactants in the preparation of nanomaterials for in vivo use should at least be reconsidered.
Vesicular trafficking
Glycosylation plays an important role in trafficking proteins to apical and basolateral membranes and recent studies also suggest their involvement in protein trafficking to exosomes.81Exosomes are 40–100 nm sized vesicles secreted by a wide range of mammalian cells as vehicles for intercellular trafficking of cellular components including miRNA, mRNA, proteins, metabolites, etc. Glycan analysis of exosomes and source tissues revealed a conserved glycan repertoire for exosomes which could help guide exosomes between different tissues via carbohydrate–lectin interactions.81 Isolated and modified exosomes have been proposed as vehicles for drug and gene delivery but the vesicle loading with cargo is still insufficient.82 The formation of vesicles by self-assembly approaches with natural or synthetic lipids and block polymers circumvents the cargo loading step as drugs or genes for delivery are encapsulated in the assembly step. In addition, exosome mimetics prepared in this manner allow at least theoretically a defined functionalization of the exosome surface with targeting and stealth ligands.83,84
Extracellular matrix glycans for sequestering and storing proteins
Glycosaminoglycans (GAGs) like heparan sulfate, chondroitin and dermatan sulfate are highly sulfated linear polysaccharides located within the extracellular space. Apart from physical functions in tissue hydration as lubricants or protein stabilizers, many GAGs are also key biological response modifiers85 that can engage in specific interactions with growth factors, cytokines and enzymes, act as signalling molecules in wound repair, tumorigenesis and infection and facilitate host cell invasion by pathogenic microbes.86 Linhardt's group prepared gold and silver nanoparticles via reduction of silver and gold salts in the presence of heparin and hyaluronic acid both as coating and reducing agents. Hyaluronic acid and heparin coatings led to the stabilization of nanoparticles in physiological media while maintaining their anticoagulant and anti-inflammatory properties.87 Proteoglycans like aggrecan can sequester and stabilize proteins like growth factors by specific interactions between proteins and charged polysaccharides. A recent report by Kipper's group describes the use of heparin functionalised nanoparticles as vehicles for growth factor delivery. Mimicking natural aggrecan, the heparin functionalized nanoparticles readily complexed and stabilized growth factors like FGF1 for delivery and continued slow release for as long as a month.88Molecular gates
Glycans have also been employed as molecular gates for the enzymatically stimulated release of cargo from mesoporous nanoparticles. Agostini et al. developed mesoporous silica nanoparticles functionalized with galactooligosaccharides (GOS) as functional gates to close the pores and retain a fluorophore cargo within the nanostructure.89 Specific enzymatic hydrolysis of the GOS gates by beta-galactosidase opened the pores and released the trapped fluorophore from the mesoporous particles (Fig. 6). High beta galactosidase activity is characteristic for senescent cells, which are cells that have reached their limit of possible divisions and due to their changed phenotype constitute a threat for the growth and development of surrounding healthy cells. Agostini et al. exploited the beta-galactosidase stimulated release of the gated fluorophore from mesoporous particles to detect senescent cells in vitro. This principle of cell-type specific cargo release might be further developed for the release of a telomere reactivation or a cytotoxic drug for in vivo therapy. A similar approach for the release of a toxic drug from its nanoparticle protection was developed by the Duncan group. The neurotoxicity of phospholipase A2, a snake venom, that had shown otherwise good efficacy in phase 1 clinical trials for the treatment of breast cancer was significantly reduced by covalent coupling to dextran nanoparticles.90 The masked toxin was liberated by the enzymatic hydrolysis of the dextran carrier through alpha amylase which is overexpressed in tumour cells.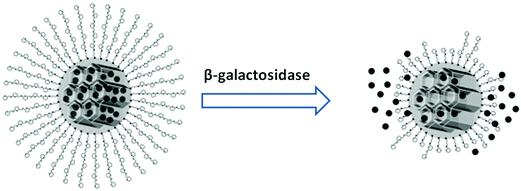 | ||
| Fig. 6 Release of fluorophores from galacto-oligosaccharide gated mesoporous nanoparticles. Reproduced from ref. 89 with permission from John Wiley and Sons. | ||
Imaging
Patient stratification into responders and non-responders is a key principle of personalized medicine that requires sensitive diagnostic methods. In vivo imaging techniques like magnetic resonance imaging (MRI), positron emission tomography (PET) and fluorescence imaging are non-invasive and could be harnessed to provide a spatio-temporal distribution of analytes in contrast to the measurement of isolated single parameters (biomarkers). Due to their optical and magnetic properties, small size that allows penetration into most tissues and their large aspect ratios for the attachment of functional molecules with high density, metal nanomaterials are ideally suited as contrast agents in many imaging modalities.91 Lectins have been used for many years in the histological examination of tissue samples, e.g. to detect and quantify changes in glycosylation in tumours.92 Recently, lectins have also made their way into nanoparticle based in vivo imaging of the glycan distribution of mice brains.93 Gao et al. encapsulated CdSe/ZnS quantum dots into PEG-PLA nanoparticles and functionalized them covalently with wheat germ agglutinin, a lectin recognizing GlcNAc and sialic acid for improved delivery to the brain after administration through the nasal mucus which is rich in sialic acid presenting glycans. The particles were efficiently delivered to the brain, allowing imaging for over 4 h and were nearly completely cleared after 8 h suggesting a good safety profile for their use as imaging agents. The cytotoxicity and often promiscuous binding patterns of many lectins have limited their widespread use for in vivo imaging applications. The discovery of highly selective lectins capable of recognizing larger glycan structures94 and the recombinant engineering of non-toxic lectins95 are addressing these current limitations opening the door to a new generation of in vivo glycan imaging agents.Glyconanotechnology as an enabling technology for personalized medicine
As we have seen, glycans are ubiquitously distributed in all body tissues and have important functions at a cellular level. Hence, any artificial nanosized system for in vivo drug delivery or imaging should take advantage of the remarkable properties glycans offer to improve biocompatibility and facilitate tissue selective targeting. Here, we have focused on the applications of glyconanotechnology which have made or are likely to make an impact on the development of more personalized diagnostics and therapies, features that make nanomaterials fit for purpose.The combination of glyco- and nanotechnology is already providing promising new tools for more personalized solutions to diagnostics and therapy. Glycans can play a central role as antigens or targeting probes for nanoparticle applications in oncology and immunology, and also open up opportunities in other areas where the unparalleled stealthing properties and usually low immunogenicity of glycans may be valuable assets for any nanomaterial designed for biomedical use.
Acknowledgements
We acknowledge generous funding from the Spanish Ministry of Economy and Competiveness MINECO (grant number: CTQ2014-58779-R). This project has received funding from the European Union's Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement No. 642870 (ETN-Immunoshape).References
- C. E. Rosenberg, Milbank Meml. Fund Q., 2002, 80, 237–260 CrossRef.
- A. C. Ahn, M. Tewari, C.-S. Poon and R. S. Phillips, PLoS Med., 2006, 3, e208 Search PubMed.
- S. Bates, Drug Discovery Today, 2010, 15, 115–120 CrossRef PubMed.
- B. B. Spear, M. Heath-Chiozzi and J. Huff, Trends Mol. Med., 2001, 7, 201–204 CrossRef CAS PubMed.
- A. Almeida and D. Kolarich, Biochim. Biophys. Acta, 2016, 1860, 1583–1595 CrossRef CAS PubMed.
- C. B. Lebrilla and H. J. An, Mol. BioSyst., 2009, 5, 17–20 RSC.
- G. Thanabalasingham, J. E. Huffman, J. J. Kattla, M. Novokmet, I. Rudan, A. L. Gloyn, C. Hayward, B. Adamczyk, R. M. Reynolds and A. Muzinic, et al. , Diabetes, 2013, 62, 1329–1337 CrossRef CAS PubMed.
- M. P. Campbell and N. H. Packer, Biochim. Biophys. Acta, 2016, 1860, 1669–1675 CrossRef CAS PubMed.
- N. C. Reichardt, M. Marttín-Lomas and S. Penadés, Chem. Soc. Rev., 2013, 42, 4358–4376 RSC.
- B. K. Gorityala, J. Ma, X. Wang, P. Chen and X.-W. Liu, Chem. Soc. Rev., 2010, 39, 2925–2934 RSC.
- K. El-Boubbou and X. Huang, Curr. Med. Chem., 2011, 18, 2060–2078 CrossRef CAS PubMed.
- L. J. Peek, C. R. Middaugh and C. Berkland, Adv. Drug Delivery Rev., 2008, 60, 915–928 CrossRef CAS PubMed.
- H. Lee, K. Lee, I. K. Kim and T. G. Park, Biomaterials, 2008, 29, 4709–4718 CrossRef CAS PubMed.
- J. H. Ryu, S. Lee, S. Son, S. H. Kim, J. F. Leary, K. Choi and I. C. Kwon, J. Controlled Release, 2014, 190, 477–484 CrossRef CAS PubMed.
- H.-L. Zhang, X.-L. Wei, Y. Zang, J.-Y. Cao, S. Liu, X.-P. He, Q. Chen, Y.-T. Long, J. Li and G.-R. Chen, et al. , Adv. Mater., 2013, 25, 4097–4101 CrossRef CAS PubMed.
- A. S. Opat, C. van Vliet and P. A. Gleeson, Biochimie, 2001, 83, 763–773 CrossRef CAS PubMed.
- A. Varki, Cell, 2006, 126, 841–845 CrossRef CAS PubMed.
- J. R. Bishop and P. Gagneux, Glycobiology, 2007, 17, 23R–34R CrossRef CAS PubMed.
- T. B. Geijtenbeek, D. S. Kwon, R. Torensma, S. J. van Vliet, G. C. van Duijnhoven, J. Middel, I. L. Cornelissen, H. S. Nottet, V. N. KewalRamani, D. R. Littman, C. G. Figdor and Y. van Kooyk, Cell, 2000, 100, 587–597 CrossRef CAS PubMed.
- A. Cambi, F. de Lange, N. M. van Maarseveen, M. Nijhuis, B. Joosten, E. M. H. P. van Dijk, B. I. de Bakker, J. A. M. Fransen, P. H. M. Bovee-Geurts, F. N. van Leeuwen, N. F. Van Hulst and C. G. Figdor, J. Cell Biol., 2004, 164, 145–155 CrossRef CAS PubMed.
- O. Martínez-Avila, L. M. Bedoya, M. Marradi, C. Clavel, J. Alcamí and S. Penadés, ChemBioChem, 2009, 10, 1806–1809 CrossRef PubMed.
- B. Arnáiz, O. Martínez-Ávila, J. M. Falcon-Perez and S. Penadés, Bioconjugate Chem., 2012, 23, 814–825 CrossRef PubMed.
- E. Severi, D. W. Hood and G. H. Thomas, Microbiology, 2007, 153, 2817–2822 CrossRef CAS PubMed.
- I. van Die and R. D. Cummings, Glycobiology, 2010, 20, 2–12 CrossRef CAS PubMed.
- A. van Remoortere, C. H. Hokke, G. J. van Dam, I. van Die, A. M. Deelder and D. H. van den Eijnden, Glycobiology, 2000, 10, 601–609 CrossRef CAS PubMed.
- R. Ojeda, J. L. de Paz, A. G. Barrientos, M. Martín-Lomas and S. Penadés, Carbohydr. Res., 2007, 342, 448–459 CrossRef CAS PubMed.
- J. Frigell, I. Garcia, V. Gómez-Vallejo, J. Llop and S. Penadés, J. Am. Chem. Soc., 2013, 136, 449–457 CrossRef PubMed.
- K. El-Boubbou, D. C. Zhu, C. Vasileiou, B. Borhan, D. Prosperi, W. Li and X. Huang, J. Am. Chem. Soc., 2010, 132, 4490–4499 CrossRef CAS PubMed.
- M. H. El-Dakdouki, J. Xia, D. C. Zhu, H. Kavunja, J. Grieshaber, S. O’Reilly, J. J. McCormick and X. Huang, ACS Appl. Mater. Interfaces, 2014, 6, 697–705 CAS.
- I. P. Witz, Cancer Treat. Res., 2006, 130, 125–140 CAS.
- T. D. Farr, C.-H. Lai, D. Grünstein, G. Orts-Gil, C.-C. Wang, P. Boehm-Sturm, P. H. Seeberger and C. Harms, Nano Lett., 2014, 14, 2130–2134 CrossRef CAS PubMed.
- S. I. van Kasteren, S. J. Campbell, S. Serres, D. C. Anthony, N. R. Sibson and B. G. Davis, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 18–23 CrossRef CAS PubMed.
- Y. Lee, H. Lee, Y. B. Kim, J. Kim, T. Hyeon, H. Park, P. B. Messersmith and T. G. Park, Adv. Mater., 2008, 20, 4154–4157 CAS.
- M. Andrianifahanana, N. Moniaux and S. K. Batra, Biochim. Biophys. Acta, 2006, 1765, 189–222 CAS.
- D. Feng, A. S. Shaikh and F. Wang, ACS Chem. Biol., 2016, 11, 850–863 CrossRef CAS PubMed.
- A. L. Parry, N. A. Clemson, J. Ellis, S. S. Bernhard, B. G. Davis and N. R. Cameron, J. Am. Chem. Soc., 2013, 135, 9362–9365 CrossRef CAS PubMed.
- D. Miles, H. Roché, M. Martin, T. J. Perren, D. A. Cameron, J. Glaspy, D. Dodwell, J. Parker, J. Mayordomo, A. Tres, J. L. Murray and N. K. Ibrahim, Oncologist, 2011, 16, 1092–1100 CrossRef CAS PubMed.
- R. Kojima, D. Aubel and M. Fussenegger, Curr. Opin. Chem. Biol., 2015, 28, 29–38 CrossRef CAS PubMed.
- T. K. Dam and C. F. Brewer, Glycobiology, 2010, 20, 270–279 CrossRef CAS PubMed.
- A. Audfray, A. Varrot and A. Imberty, C. R. Chim., 2013, 16, 482–490 CrossRef CAS.
- T. Eierhoff, B. Bastian, R. Thuenauer, J. Madl, A. Audfray, S. Aigal, S. Juillot, G. E. Rydell, S. Müller, S. de Bentzmann, A. Imberty, C. Fleck and W. Römer, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 12895–12900 CrossRef CAS PubMed.
- K. El-Boubbou, C. Gruden and X. Huang, J. Am. Chem. Soc., 2007, 129, 13392–13393 CrossRef CAS PubMed.
- M. Hartmann, P. Betz, Y. Sun, S. N. Gorb, T. K. Lindhorst and A. Krueger, Chem. – Eur. J., 2012, 18, 6485–6492 CrossRef CAS PubMed.
- N. P. Pera and R. J. Pieters, MedChemComm, 2014, 5, 1027–1035 RSC.
- M. Khanal, F. Larsonneur, V. Raks, A. Barras, J.-S. Baumann, F. A. Martin, R. Boukherroub, J.-M. Ghigo, C. O. Mellet and V. Zaitsev, et al. , Nanoscale, 2015, 7, 2325–2335 RSC.
- J. C. Martín, M. Assali, E. Fernández-Garcia, V. Valdivia, E. Sánchez-Fernández, J. G. Fernández, R. Wellinger, I. Fernández and N. Khiar, J. Mater. Chem. B, 2016, 4, 2028–2037 RSC.
- A. Muñoz, D. Sigwalt, B. M. Illescas, J. Luczkowiak, L. Rodriguez-Pérez, I. Nierengarten, M. Holler, J.-S. Remy, K. Buffet and S. P. Vincent, et al. , Nat. Chem., 2016, 8, 50–57 CrossRef.
- M. Reynolds, M. Marradi, A. Imberty, S. Penadés and S. Pérez, Chem. – Eur. J., 2012, 18, 4264–4273 CrossRef CAS PubMed.
- M. von Itzstein, Nat. Rev. Drug Discovery, 2007, 6, 967–974 CrossRef CAS PubMed.
- M. Mammen, G. Dahmann and G. M. Whitesides, J. Med. Chem., 1995, 38, 4179–4190 CrossRef CAS PubMed.
- J. J. Landers, Z. Cao, I. Lee, L. T. Piehler, P. P. Myc, A. Myc, T. Hamouda, A. T. Galecki and J. R. Baker, J. Infect. Dis., 2002, 186, 1222–1230 CrossRef CAS PubMed.
- J. D. Andrade and V. Hlady, Ann. N. Y. Acad. Sci., 1987, 516, 158–172 CrossRef CAS PubMed.
- M. Rahman, S. Laurent, N. Tawil, L. Yahia and M. Mahmoudi, Protein-Nanoparticle Interactions, Springer, 2013, pp. 21–44 Search PubMed.
- S. Zanganeh, R. Spitler, M. Erfanzadeh, A. M. Alkilany and M. Mahmoudi, Int. J. Biochem. Cell Biol., 2016, 75, 143–147 CrossRef CAS PubMed.
- D. E. Owens and N. A. Peppas, Int. J. Pharm., 2006, 307, 93–102 CrossRef CAS PubMed.
- W. Shang, J. H. Nuffer, J. S. Dordick and R. W. Siegel, Nano Lett., 2007, 7, 1991–1995 CrossRef CAS PubMed.
- S. Dominguez-Medina, L. Kisley, L. J. Tauzin, A. Hoggard, B. Shuang, A. S. D. S. Indrasekara, S. Chen, L.-Y. Wang, P. J. Derry and A. Liopo, et al. , ACS Nano, 2016, 10, 2103–2112 CrossRef CAS PubMed.
- Z. J. Deng, M. Liang, M. Monteiro, I. Toth and R. F. Minchin, Nat. Nanotechnol., 2011, 6, 39–44 CrossRef CAS PubMed.
- V. Mirshafiee, R. Kim, S. Park, M. Mahmoudi and M. L. Kraft, Biomaterials, 2016, 75, 295–304 CrossRef CAS PubMed.
- A. Aires, S. M. Ocampo, D. Cabrera, L. de la Cueva, G. Salas, F. J. Teran and A. L. Cortajarena, J. Mater. Chem. B, 2015, 3, 6239–6247 RSC.
- M. Chanana, P. Rivera Gil, M. A. Correa-Duarte, L. M. Liz-Marzán and W. J. Parak, Angew. Chem., Int. Ed., 2013, 52, 4179–4183 CrossRef CAS PubMed.
- S. Winzen, S. Schoettler, G. Baier, C. Rosenauer, V. Mailaender, K. Landfester and K. Mohr, Nanoscale, 2015, 7, 2992–3001 RSC.
- S. Ju and W.-S. Yeo, Nanotechnology, 2012, 23, 135701 CrossRef PubMed.
- I. García, A. Sánchez-Iglesias, M. Henriksen-Lacey, M. Grzelczak, S. Penadés and L. M. Liz-Marzán, J. Am. Chem. Soc., 2015, 137, 3686–3692 CrossRef PubMed.
- I. Lynch and K. A. Dawson, Nano Today, 2008, 3, 40–47 CrossRef CAS.
- J. Klein, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 2029–2030 CrossRef CAS PubMed.
- M. P. Monopoli, C. Åberg, A. Salvati and K. A. Dawson, Nat. Nanotechnol., 2012, 7, 779–786 CrossRef CAS PubMed.
- S. Tenzer, D. Docter, J. Kuharev, A. Musyanovych, V. Fetz, R. Hecht, F. Schlenk, D. Fischer, K. Kiouptsi, C. Reinhardt, K. Landfester, H. Schild, M. Maskos, S. K. Knauer and R. H. Stauber, Nat. Nanotechnol., 2013, 8, 772–781 CrossRef CAS PubMed.
- A. M. Alkilany, P. K. Nagaria, C. R. Hexel, T. J. Shaw, C. J. Murphy and M. D. Wyatt, Small, 2009, 5, 701–708 CrossRef CAS PubMed.
- N. Khlebtsov and L. Dykman, Chem. Soc. Rev., 2011, 40, 1647–1671 RSC.
- D. Hühn, K. Kantner, C. Geidel, S. Brandholt, I. De Cock, S. J. H. Soenen, P. Rivera Gil, J.-M. Montenegro, K. Braeckmans, K. Müllen, G. U. Nienhaus, M. Klapper and W. J. Parak, ACS Nano, 2013, 7, 3253–3263 CrossRef PubMed.
- U. Sakulkhu, L. Maurizi, M. Mahmoudi, M. Motazacker, M. Vries, A. Gramoun, M.-G. O. Beuzelin, J.-P. Vallée, F. Rezaee and H. Hofmann, Nanoscale, 2014, 6, 11439–11450 RSC.
- C. Passirani, G. Barratt, J.-P. Devissaguet and D. Labarre, Life Sci., 1998, 62, 775–785 CrossRef CAS PubMed.
- S. Wan, P. M. Kelly, E. Mahon, H. Stöckmann, F. Caruso, K. A. Dawson, Y. Yan and M. P. Monopoli, ACS Nano, 2015, 9, 2157–2166 CrossRef CAS PubMed.
- S. Schöttler, G. Becker, S. Winzen, T. Steinbach, K. Mohr, K. Landfester, V. Mailänder and F. R. Wurm, Nat. Nanotechnol., 2016, 11, 372–377 CrossRef PubMed.
- C. Lemarchand, R. Gref and P. Couvreur, Eur. J. Pharm. Biopharm., 2004, 58, 327–341 CrossRef CAS PubMed.
- J. Virkutyte and R. S. Varma, Chem. Sci., 2011, 2, 837–846 RSC.
- R. Bandyopadhyaya, E. Nativ-Roth, O. Regev and R. Yerushalmi-Rozen, Nano Lett., 2002, 2, 25–28 CrossRef CAS.
- A. Star, D. W. Steuerman, J. R. Heath and J. F. Stoddart, Angew. Chem., Int. Ed., 2002, 41, 2508–2512 CrossRef CAS.
- R. P. Garay, R. El-Gewely, J. K. Armstrong, G. Garratty and P. Richette, Expert Opin. Drug Delivery, 2012, 9, 1319–1323 CrossRef CAS PubMed.
- B. S. Batista, W. S. Eng, K. T. Pilobello, K. D. Hendricks-Muñoz and L. K. Mahal, J. Proteome Res., 2011, 10, 4624–4633 CrossRef CAS PubMed.
- S. Kooijmans, P. Vader, S. M. van Dommelen, W. W. van Solinge and R. M. Schiffelers, Int. J. Nanomed., 2012, 7, e41 Search PubMed.
- M. Antonietti and S. Förster, Adv. Mater., 2003, 15, 1323–1333 CrossRef CAS.
- V. Percec, P. Leowanawat, H.-J. Sun, O. Kulikov, C. D. Nusbaum, T. M. Tran, A. Bertin, D. A. Wilson, M. Peterca, S. Zhang, N. P. Kamat, K. Vargo, D. Moock, E. D. Johnston, D. A. Hammer, D. J. Pochan, Y. Chen, Y. M. Chabre, T. C. Shiao, M. Bergeron-Brlek, S. André, R. Roy, H.-J. Gabius and P. A. Heiney, J. Am. Chem. Soc., 2013, 135, 9055–9077 CrossRef CAS PubMed.
- J. M. Trowbridge and R. L. Gallo, Glycobiology, 2002, 12, 117R–125R CrossRef CAS PubMed.
- M. A. Skidmore, S. E. Guimond, T. R. Rudd, D. G. Fernig, J. E. Turnbull and E. A. Yates, Connect. Tissue Res., 2008, 49, 140–144 CrossRef CAS PubMed.
- M. M. Kemp, A. Kumar, S. Mousa, T.-J. Park, P. Ajayan, N. Kubotera, S. A. Mousa and R. J. Linhardt, Biomacromolecules, 2009, 10, 589–595 CrossRef CAS PubMed.
- L. W. Place, M. Sekyi and M. J. Kipper, Biomacromolecules, 2014, 15, 680–689 CrossRef CAS PubMed.
- A. Agostini, L. Mondragón, A. Bernardos, R. Martínez-Máñez, M. D. Marcos, F. Sancenón, J. Soto, A. Costero, C. Manguan-García, R. Perona, M. Moreno-Torres, R. Aparicio-Sanchis and J. R. Murguía, Angew. Chem., Int. Ed., 2012, 51, 10556–10560 CrossRef CAS PubMed.
- E. L. Ferguson and R. Duncan, Biomacromolecules, 2009, 10, 1358–1364 CrossRef CAS PubMed.
- T. L. Doane and C. Burda, Chem. Soc. Rev., 2012, 41, 2885–2911 RSC.
- E. L. Bird-Lieberman, A. A. Neves, P. Lao-Sirieix, M. O’Donovan, M. Novelli, L. B. Lovat, W. S. Eng, L. K. Mahal, K. M. Brindle and R. C. Fitzgerald, Nat. Med., 2012, 18, 315–321 CrossRef CAS PubMed.
- X. Gao, J. Chen, J. Chen, B. Wu, H. Chen and X. Jiang, Bioconjugate Chem., 2008, 19, 2189–2195 CrossRef CAS PubMed.
- J.-H. Liao, C.-T. H. Chien, H.-Y. Wu, K.-F. Huang, I. Wang, M.-R. Ho, I.-F. Tu, I.-M. Lee, W. Li, Y.-L. Shih, C.-Y. Wu, P. A. Lukyanov, S.-T. D. Hsu and S.-H. Wu, J. Am. Chem. Soc., 2016, 138, 4787–4795 CrossRef CAS PubMed.
- F. Medina-Bolivar, R. Wright, V. Funk, D. Sentz, L. Barroso, T. D. Wilkins, W. Petri and C. L. Cramer, Vaccine, 2003, 21, 997–1005 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2016 |