Open Access Article
Open Access ArticleComposite bottlebrush mechanics: α-internexin fine-tunes neurofilament network properties†
M.
Kornreich‡
a,
E.
Malka-Gibor‡
b,
A.
Laser-Azogui
a,
O.
Doron
a,
H.
Herrmann
c and
R.
Beck
*a
aThe Raymond and Beverly Sackler School of Physics and Astronomy, Tel-Aviv University, 69978 Tel Aviv, Israel. E-mail: roy@post.tau.ac.il
bSackler Faculty of Medicine, Tel-Aviv University, Tel-Aviv, 69978, Israel
cDepartment of Molecular Genetics, German Cancer Research Center, D-69120 Heidelberg, Germany
First published on 16th June 2015
Abstract
Neuronal cytoplasmic intermediate filaments are principal structural and mechanical elements of the axon. Their expression during embryonic development follows a differential pattern, while their unregulated expression is correlated to neurodegenerative diseases. The largest neurofilament proteins of medium (NF-M) and high molecular weight (NF-H) were shown to modulate the axonal architecture and inter-filament spacing. However, the individual roles of the remaining α-internexin (α-Inx) and neurofilament of low molecular weight (NF-L) proteins in composite filaments remained elusive. In contrast to previous predictions, we show that when co-assembled with NF-M, the shortest and the least charged α-Inx protein increases inter-filament spacing. These findings suggest a novel structural explanation for the expression pattern of neurofilament proteins during embryonic development. We explain our results by an analysis of ionic cross-links between the disordered polyampholytic C-terminal tails and suggest that a collapsed conformation of the α-Inx tail domain interferes with tail cross-linking near the filament backbone.
1 Introduction
The cytoskeleton is composed of three interconnected structures: the actin microfilaments, microtubules and intermediate filaments (IFs). The 10 nm diameter of IFs is “intermediate” between actin microfilaments (8 nm) and microtubules (26 nm).1,2 Different from microfilaments and microtubules, IF expression is very complex. In human, 70 genes are expressed encoding IF proteins in various tissues and cells types in routes parallel to embryonic differentiation.2,3Here we focus on IF proteins expressed in the mammalian nerve system. This group includes the three neurofilament (NF) triplet subunits NF-L (62 kDa), NF-M (103 kDa) and NF-H (117 kDa), as well as α-internexin (α-Inx, 66 kDa), which was only recently identified as the fourth neuronal IF.4,5 In addition to the IF proteins mentioned, GFAP, vimentin, nestin and peripherin are major components of cytoskeletons in different neural cells.
IF expression in neural cells follows a sequential pattern.6 In the prenatal stage, neurons predominately express α-Inx, while in mature neurons, α-Inx expression levels decrease and the NF triplet proteins expression levels increase. NF-M is the first NF protein to be simultaneously expressed with α-Inx.7 This results in filaments composed of the NF triplet proteins as well as α-Inx.5 The reasons for this developmental specificity of neurons with regards to IF proteins are not entirely understood. It is hypothesized that the multitude of IF proteins in the central nervous system is the basis an intricate fine-tuning of structures and functions at the cellular and tissue levels. While the late introduction of the larger NF-M and NF-H proteins in neurons is explained by the requirement for enhanced radial growth of the axonal caliber, the prenatal preference of α-Inx over NF-L as precursor protein is unclear and will be addressed here.
All cytoplasmic IF proteins have a common tripartite domain organization: a central α-helical rod domain of about 310 amino acids is flanked by non-structured N-terminal head and C-terminal tail domains of varying lengths (Fig. 1A). The main difference between the neuronal IF proteins lies within the polyampholytic, intrinsically disordered C-terminal domain whose length ranges between 91 to 679 amino acids in mammals (Fig. 1 and Table S1, ESI†). The head and the rod domains of the proteins make the backbone of the filament, while the C-terminal segments, i.e. the tails, protrude outside, forming a bottlebrush shape8–10 (Fig. 1B). The four proteins are divided into two groups based on tail-length and assembly properties. The first group comprises α-Inx and NF-L, whose tail-lengths in bovine are 91 and 158, respectively, self-assemble into 10 nm filaments in vitro. The second group comprises NF-M with a tail of 514 and NF-H with a tail of 679 amino acids; and, only assembles into heteropolymer filaments with either α-Inx or NF-L.
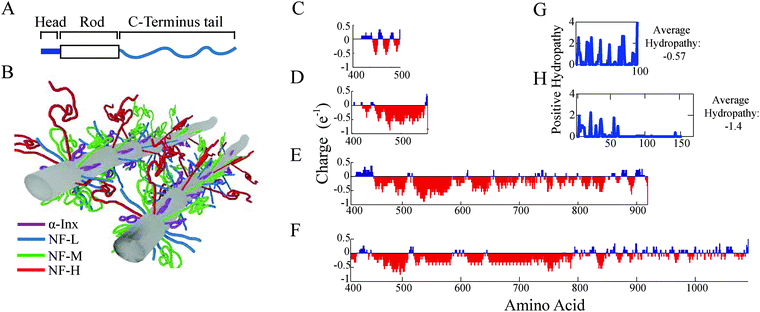 | ||
| Fig. 1 Subunit proteins form filaments that interact via polyampholyte C-terminal tails. (A) An illustration of a neural intermediate filament subunit includes a well-conserved central α-helical rod domain flanked by intrinsically disordered N-terminal head and C-terminal tail domains. (B) A schematic of interacting bottlebrush IFs composed of four different neuronal subunit proteins. (C–F) Tail charge distributions of (C) un-phosphorylated α-Inx and phosphorylated (D) NF-L, (E) NF-M and (F) NF-H are calculated at pH = 6.8 and averaged over a 5-amino acid window. The positive values of (G) α-Inx and (H) NF-L tail hydrophobicity index, summed with a 3-amino acid window, are plotted next to the overall hydrophobicity score, −0.57 and −1.4, respectively.17 | ||
At high densities, filaments interact via their protruding C-terminus tails to form hydrogel networks. This provides the mechanical and structural properties to neuronal cells in myelinated fibers.11–15 NF-M and NF-H tails are considerably longer and contain a higher number of charges than NF-L and α-Inx tails (Fig. 1). Accordingly, the interactions between the long-tailed proteins are considered to be the decisive factors in setting the traits of the NF network. The shorter tails are expected to remain closer to the filament backbone, support filament assembly and hinder aggregation.16 Consequently, the short-tailed proteins are not assumed to be significantly involved in inter-filament interactions.
Since either NF-L or α-Inx is sufficient for filament formation with NF-M and NF-H,18–21 the simultaneous expression of both NF-L and α-Inx in mature neurons presumably presents some functional redundancy or a gain of additional undetermined function. The simultaneous expression may suggest some synergistic adjustment of the inter-filament distance due to inter- or intra-filament interactions by NF-L and α-Inx. For example, it was theoretically predicted that the conformation of the long-tails is affected by NF-L and α-Inx ratios.22
In order to investigate the role of α-Inx and NF-L in NF complexes, we structurally characterize hydrogel networks of various subunit protein compositions and osmotic pressures. We find that the two short-tailed α-Inx and NF-L equally serve as a substrate for assembly with NF-M and NF-H. While it is thought that adjacent filaments interact via their protruding long tails, we will show below that the short tailed subunits significantly alter the structural properties of networks of composite filaments. We will also show that the effect is synergistic as it also depends on the identity of the long-tailed partner, either NF-M or NF-H. In networks composed of filaments self-assembled from three and four subunit proteins, the absence of one protein, either long or short-tailed, does not dramatically modify the network properties. As a potential mechanism explaining our experimental results, we suggest ionic cross-bridging interactions between the polyampholytic tails.
2 Materials and methods
2.1 Native NF purification
NF triplet subunits (NF-L, NF-M and NF-H) were purified from bovine spinal cord using a modification of an earlier protocol.23,24 Spinal cords were homogenized in an equal volume of buffer A (0.1 M MES, 1 mM EGTA, 1 mM MgCl2, 0.02% (w/v) sodium azide, pH 6.8 with NaOH) with 1% (w/v) Triton X-100 and 1 mM phenylmethylsulfonyl fluoride. The homogenates were centrifuged at 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 RPM (Beckman rotor type 45-Ti) for 70 min at 4 °C. An equal volume of glycerol was added to the supernatant and incubated overnight. A pellet of NFs was recovered from the glycerol solution by precipitation at 40 K RPM (Beckman rotor type 45-Ti) for 90 min at 4 °C. The pellet was homogenized in buffer A with 0.8 M sucrose and clarified by spinning through a step gradient of 0.8 M sucrose buffer (0.8 M sucrose in buffer A) layered on top of 1.5 M sucrose buffer (1.5 M sucrose in buffer A) for 4 h at 55
000 RPM (Beckman rotor type 45-Ti) for 70 min at 4 °C. An equal volume of glycerol was added to the supernatant and incubated overnight. A pellet of NFs was recovered from the glycerol solution by precipitation at 40 K RPM (Beckman rotor type 45-Ti) for 90 min at 4 °C. The pellet was homogenized in buffer A with 0.8 M sucrose and clarified by spinning through a step gradient of 0.8 M sucrose buffer (0.8 M sucrose in buffer A) layered on top of 1.5 M sucrose buffer (1.5 M sucrose in buffer A) for 4 h at 55![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 RPM (Beckman rotor type 70-Ti). The pellet was homogenized in buffer B (0.1 M potassium phosphate and 0.1% (v/v) 2-mercaptoethanol in 8 M urea, pH 6.5), and applied to a DEAE sepharose column (DEAE Sepharose fast flow column, GE Healthcare). The column was rinsed with buffer B containing 55 mM NaCl which eluted NF-H and protein contaminates. The next elution step, performed with buffer B at pH 7 containing 200 mM NaCl, eluted NF-L and NF-M. Using hydroxylapatite (HT) column chromatography (hydroxylapatite bio gel HT gel, Bio-Rad), the contaminants were removed from the NF-H fractions. NF-L and NF-M were separated by HT column with a gradient of 0.1 to 0.4 M potassium phosphate pH 7.0. Purity and separation were verified by SDS-PAGE (Fig. S1, ESI†).
000 RPM (Beckman rotor type 70-Ti). The pellet was homogenized in buffer B (0.1 M potassium phosphate and 0.1% (v/v) 2-mercaptoethanol in 8 M urea, pH 6.5), and applied to a DEAE sepharose column (DEAE Sepharose fast flow column, GE Healthcare). The column was rinsed with buffer B containing 55 mM NaCl which eluted NF-H and protein contaminates. The next elution step, performed with buffer B at pH 7 containing 200 mM NaCl, eluted NF-L and NF-M. Using hydroxylapatite (HT) column chromatography (hydroxylapatite bio gel HT gel, Bio-Rad), the contaminants were removed from the NF-H fractions. NF-L and NF-M were separated by HT column with a gradient of 0.1 to 0.4 M potassium phosphate pH 7.0. Purity and separation were verified by SDS-PAGE (Fig. S1, ESI†).
2.2 Recombinant protein purification
Bovine NF-L and α-Inx were purified using BL21 E. coli strains transformed with pET30a vectors. Overnight cultures were grown in 5 ml LB medium supplemented with 50 μg ml−1 kanamycin. The overnight cultures were diluted into fresh 0.5 L LB medium supplemented with 550 μg ml−1 kanamycin, grown to 0.6 OD, at 600 nm. Protein expression was induced by the addition of 0.4 mM isopropyl-b-thiogalactoside (IPTG), and allowed to grow for additional 4 h. The bacteria suspension was centrifuged at 6000 RPM with a Fiberlite F14 rotor (Thermo Scientific) for 20 min and resuspended in Tris buffer (50 ml of 25 mM Tris-HCl pH 7.5, 150 mM NaCl and protease inhibitors) and stored overnight at −80 C. Frozen cells were thawed and treated with 1% (w/v) of Triton X-100 and were then sonicated on ice-cold water with a 500 watt tip at 20% amplitude. We sonicated for 2 min using short 5 second pulses followed by 5 second pauses. Clearing of the lysates was performed by two rounds of centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 900 RPM with a Fiberlite F14 rotor for 60 min at 4 °C and resuspended in Tris buffer. The last pellet was then resuspended in 8 M urea buffer (100 mM sodium phosphate for NF-L and 10 mM sodium phosphate for α-Inx, pH 7). The debris was pelleted by centrifugation at 8100 RPM with a Fiberlite F14 rotor for 10 min, and the NF containing supernatant was applied to a DEAE Sepharose column. The α-Inx was eluted with a sodium phosphate gradient of 10 to 100 mM, while NF-L was eluted by a NaCl gradient of 0 to 200 mM. NF-L required additional purification with a size exclusion HiLoad 16/600 Superdex 200 (GE Healthcare) equilibrated in 8 M urea buffer (100 mM sodium phosphate, 150 mM NaCl, pH 7). Purity was verified by SDS-PAGE (Fig. S1, ESI†).
900 RPM with a Fiberlite F14 rotor for 60 min at 4 °C and resuspended in Tris buffer. The last pellet was then resuspended in 8 M urea buffer (100 mM sodium phosphate for NF-L and 10 mM sodium phosphate for α-Inx, pH 7). The debris was pelleted by centrifugation at 8100 RPM with a Fiberlite F14 rotor for 10 min, and the NF containing supernatant was applied to a DEAE Sepharose column. The α-Inx was eluted with a sodium phosphate gradient of 10 to 100 mM, while NF-L was eluted by a NaCl gradient of 0 to 200 mM. NF-L required additional purification with a size exclusion HiLoad 16/600 Superdex 200 (GE Healthcare) equilibrated in 8 M urea buffer (100 mM sodium phosphate, 150 mM NaCl, pH 7). Purity was verified by SDS-PAGE (Fig. S1, ESI†).
2.3 Filament and hydrogel assembly
Purified fractions of recombinant α-Inx and NF-L as well as native NF triplet proteins were mixed to reach the desired stoichiometric ratios. The solution was dialyzed at 37 °C against assembly buffer A with added NaCl to form filaments at the desired different monovalent ion concentrations. Filaments for atomic force and electron microscopy experiments were dialyzed for 4 h and prepared for imaging. Filaments for X-ray scattering and crossed-polarized light microscopy were dialyzed for 48 h and centrifuged for 1 h at 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 RPM using a TLA 120.1 rotor in a Beckman Coulter Optima TLXl ultracentrifuge. The supernatant was immediately removed from the pellet. The NF pellet was then transferred to 1.5 mm quartz capillaries, overlaid with ∼100 μl assembly buffer and sealed with epoxy glue to prevent dehydration. The protein concentration of the supernatant was determined by a Bradford assay (Sigma-Aldrich) and the NF subunit composition of the pelleted assembled filaments was determined by SDS-PAGE followed by image analysis using ImageJ software (National Institutes of Health). Subtracting the amount of each subunit protein remaining in the supernatant from the initial amount gave the composition of the pelleted hydrogel. Any protein remaining in the supernatant was considered unassembled NF precursors.24
000 RPM using a TLA 120.1 rotor in a Beckman Coulter Optima TLXl ultracentrifuge. The supernatant was immediately removed from the pellet. The NF pellet was then transferred to 1.5 mm quartz capillaries, overlaid with ∼100 μl assembly buffer and sealed with epoxy glue to prevent dehydration. The protein concentration of the supernatant was determined by a Bradford assay (Sigma-Aldrich) and the NF subunit composition of the pelleted assembled filaments was determined by SDS-PAGE followed by image analysis using ImageJ software (National Institutes of Health). Subtracting the amount of each subunit protein remaining in the supernatant from the initial amount gave the composition of the pelleted hydrogel. Any protein remaining in the supernatant was considered unassembled NF precursors.24
2.4 Osmotic stress technique
To osmotically pressure the NF network we added polyethylene glycol with molecular weight of 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g mol−1 (PEG20K). The osmotic pressure, Π, was determined by documented calibration25 to its weight percentage in the solution and following the formula log10
000 g mol−1 (PEG20K). The osmotic pressure, Π, was determined by documented calibration25 to its weight percentage in the solution and following the formula log10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Π = 1.57 + 2.75 (wt%)0.21. To overrule osmolyte interference with the NF network, we produce several control hydrogels with the smaller PEG6K.26
Π = 1.57 + 2.75 (wt%)0.21. To overrule osmolyte interference with the NF network, we produce several control hydrogels with the smaller PEG6K.26
2.5 Imaging
Crossed-polarized light microscopy was employed to characterize of the hydrogel structure (isotropic or birefringent nematic). Sedimented NFs were observed in 1.5 mm quartz capillaries using a Nikon Eclipse LV 100 POL microscope fitted with 5-20X objectives. Micrographs were taken with a Nikon D90 camera. Atomic force microscopy (AFM, JPK NanoWizard III) was preformed on diluted NF protein solutions (1–100 μg ml−1).27 Filaments were deposited onto a clean poly(ethyleneimine)-coated silicon wafer. After dehydration, the samples were imaged under ambient pressure in tapping mode and at 1 Hz per line speed using a silicon probe of k = 42 N m−1 and 300 KHz frequency. For transmission electron microscopy (TEM), we either fixed and negatively stained the samples28 or glycerol sprayed them for low angle rotary metal shadowing.29 Grids were imaged with a Zeiss EM 910 or a Philips Tecnai F20 microscope.2.6 Small angle X-ray scattering
The powder diffraction scattering data from NF hydrogels contained in quartz capillaries was integrated azimuthally and the intensity was plotted versus reciprocal distance q. The intensity, in arbitrary units, showed a broad peak with a maximum in the range of q = 0.1–0.2 nm−1 (see ref. 24 and 30). The peak location relates to the inter-filament spacing (d = 2π/q). Broadening of this peak is observed due to density fluctuations and the semi-flexible nature of the individual filaments. Baseline background of the form A·q−B + C with B = 2–3 is subtracted (Fig. S2, ESI†), and the resultant peak is fitted with a Lorentzian function using Matlab routines.13,30Preliminary experiments were performed at our home-lab using a Pilatus 300K detector and a Xenocs GeniX Low Divergence CuKα radiation source setup with scatterless slits.31 Subsequent measurements were performed at synchrotron facilities: P12 beamline in DESY, Hamburg; SWING beamline in SOLEIL, Paris; and I911 SAXS beamline in MAX-lab, Lund with 10 keV.
3 Results
3.1 α-Inx self-assembles with either NF-M or NF-H into filaments which form nematic hydrogels
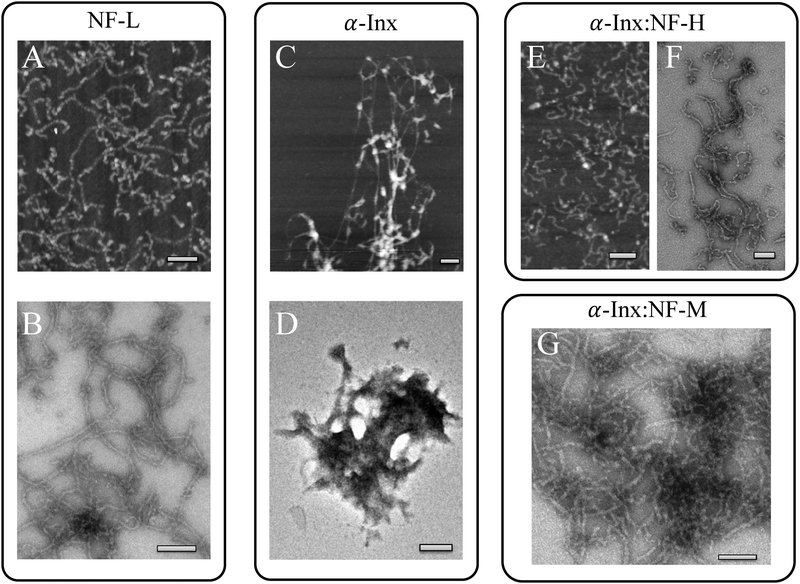
We purify NF proteins from bovine spinal cord and from transformed E. coli hosts, both under denaturing conditions. Using assembly buffer, subunit proteins self-assemble into heteropolymer filaments at near-physiological conditions (buffer A with added NaCl at 150 mM final monovalent ion concentration, see Materials and methods section). Filament formation is verified by AFM and TEM. We find that recombinant and native NF-L forms filaments with a diameter in the range of 10 nm similar to those reported previously.16 In the wide-scan AFM image (Fig. 2A), many long and short filaments are imaged. The negatively stained samples give more insight into the beaded appearance of these short filaments, which tend to associate laterally with the long filaments (Fig. 2B), as previously reported.16We also find that mixtures of α-Inx with NF-H (Fig. 2E and F) or α-Inx with NF-M (Fig. 2G) self-assemble into filaments that are similar to composite heteropolymer filaments of NF-L with NF-M or NF-H.27 In contrast, on its own α-Inx assembles into filaments and irregular aggregates (Fig. 2C and D) indicating that the presence of NF-M or NF-H prevents irregular aggregation. The formation of such irregular structures is consistent with previous observations for α-Inx.21,32 Notably, the recombinant NF-L proteins, purified similarly to α-Inx (see Materials methods) did form filaments (Fig. 2B).
Following filament formation, samples are centrifuged to produce hydrogels. The pelleted filaments generate a stable nematic liquid crystalline hydrogel, which is phase-separated from the supernatant (Fig. S4, ESI†). We note that filament assembly and centrifugation steps were performed under reducing conditions, with 0.1% (v/v) 2-mercaptoethanol to prevent covalent bonding. We also examined the possible role of hydrophobic interactions in hydrogel stability; we self-assembled an NF-L sample with 0.01% (w/v) Triton,33 which resulted in the formation of similar filaments and a stable nematic hydrogel. It appears that neither covalent Cys–Cys bonds nor hydrophobic interactions can account for the observed NF network stability. Instead, ionic cross-links are responsible for the attraction between filaments and the resultant stable physical hydrogel.11,13,30
3.2 α-Inx and NF-L based filaments include up to one third of long-tailed subunit proteins
Both NF-M and NF-H require either NF-L or α-Inx to form filaments. For each subunit pair we compare the assembled pellet to that of the denaturing solution, i.e., before assembly24 (Fig. S5, ESI†). We find a linear relation between initial long tail proteins mole fraction to their final assembled mole fraction, i.e., in the assembled pellet. The linear slope is approximately one, which indicates that as long as a maximal ratio is not reached, incorporating a long tail protein is almost as likely as incorporating another short tail protein into a filament. However, above roughly one third of initial long-tail mole fraction, we find that additional long-tail subunits are not incorporated into the filaments. The maximal assembled long-tailed protein fraction is not strongly regulated by the identity of the long- or short tail subunit proteins (Fig. S5, ESI†).3.3 The NF-L:NF-M heteropolymer filaments form significantly more condensed networks than α-Inx based filaments
To study the translational organization of the hydrogel ultrastructure, we determined the inter-filament spacing, d, using small angle X-ray scattering SAXS.24,30 This allows probing the structural and mechanical properties of NF networks in solution at near physiological salt concentrations and varying osmotic pressures, Π. We examine below networks formed by filaments with increasing number of subunit proteins. Filaments are self-assembled from either one (homopolymer), two (bipolymer), three (triplet) or four (quartet) different subunit proteins. All hydrogels are assembled by equilibration against assembly buffer, unless stated otherwise.Networks consisting of two NF components were designed to reveal the specific roles of each protein, by comparing α-Inx with NF-L, and NF-M with NF-H. Since only the short-tail proteins (NF-L and α-Inx) can serve as a backbone for filament formation, we assemble NF-L and α-Inx with either NF-M or NF-H. For osmotic pressure measurements, we assemble networks with a maximal long-tail fraction (dashed lines in Fig. S5, ESI†).
The osmotic pressure Π–d diagrams for the four bipolymer filament networks are presented in Fig. 3A–C. At low osmotic pressures (Π ≲ 104 Pa), both NF-H containing bipolymer filament networks are in expanded state with d ≈ 80 nm. In contrast, NF-M bipolymer filament networks strongly depend on their short-tailed partners. Here, NF-L:NF-M co-assembly is in a collapsed state with d ≈ 40 nm, whereas α-Inx:NF-M is in expanded state (Fig. 3B). These results are intriguing as the α-Inx tail is both shorter and less charged than the NF-L tail. Furthermore, the expanded state of the α-Inx:NF-M network compared to the NF-L:NF-M network does not agree with the mean field calculated trend.22 There, replacing NF-L with α-Inx in the NF triplet network was predicted to reduce the brush height. The disagreement suggests that the trend observed in the NF-L:NF-M network at low osmotic pressures is related to specific interactions between the two proteins. Unfortunately, a comparison between the expanded states of NF-L:NF-H, α-Inx:NF-M and α-Inx:NF-H is limited by the experimental error of our measurements which is larger than the differences previously predicted at comparable ionic strengths.34,35
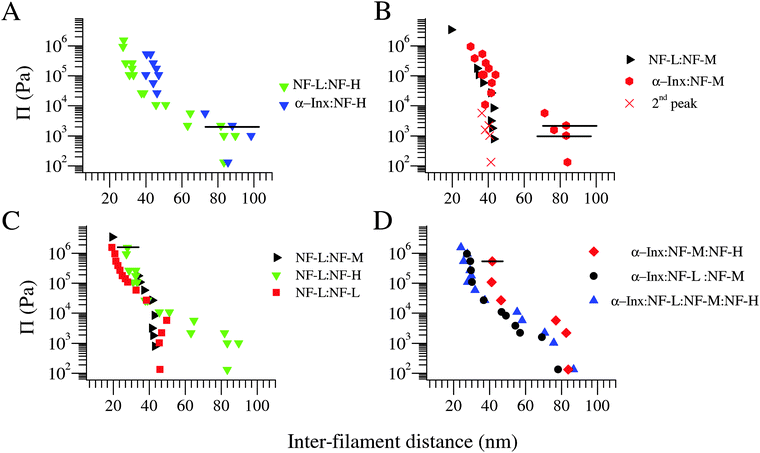 | ||
Fig. 3 Osmotic pressure vs. inter-filament distance curves for filaments of different protein compositions. (A) Networks of filaments comprising NF-H with different short tails, either α-Inx or NF-L, at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 mol 4 mol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mol subunit molar ratio respectively. A secondary low intensity peak at 35–45 nm is also fitted to SAXS data for both networks at the expanded state, as previously observed30 (not shown). (B) Networks of filaments comprising NF-M with either α-Inx or NF-L at 1 mol subunit molar ratio respectively. A secondary low intensity peak at 35–45 nm is also fitted to SAXS data for both networks at the expanded state, as previously observed30 (not shown). (B) Networks of filaments comprising NF-M with either α-Inx or NF-L at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 mol 3 mol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mol subunit ratio respectively. Here, the secondary correlation peak is shown and denoted by X symbol. (C) Comparison of NF-L based networks: NF-L homopolymer filaments, composite filament comprising NF-L:NF-M (taken from Beck et al.30) and NF-L:NF-H. (D) Comparison of two triplet filament networks to the quartet filament network. The latter comprises α-Inx mol subunit ratio respectively. Here, the secondary correlation peak is shown and denoted by X symbol. (C) Comparison of NF-L based networks: NF-L homopolymer filaments, composite filament comprising NF-L:NF-M (taken from Beck et al.30) and NF-L:NF-H. (D) Comparison of two triplet filament networks to the quartet filament network. The latter comprises α-Inx![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) NF-L NF-L![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) NF-M NF-M![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) NF-H at biologically relevant 4 NF-H at biologically relevant 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 subunit molar ratio. Typical horizontal error bars for distances larger than 50 nm, (i.e., in the expanded state) are shown in (A) and (B) only. Typical error bars for the collapsed state obtained for Π ≈ 104 Pa and higher are shown in (C) and (D) only. Measurements conducted at 150 mM monovalent salt. 1 subunit molar ratio. Typical horizontal error bars for distances larger than 50 nm, (i.e., in the expanded state) are shown in (A) and (B) only. Typical error bars for the collapsed state obtained for Π ≈ 104 Pa and higher are shown in (C) and (D) only. Measurements conducted at 150 mM monovalent salt. | ||
At high osmotic pressures (Π ≥ 104 Pa) we observe an irreversible collapse of the inter-filament distance down to ≈40 nm in all bipolymer filament networks except for NF-L:NF-M. Upon further compression, all bipolymer filament networks exhibit a similar trend, which follows the curve of the NF-L homopolymer filament network (Fig. 3C). Only the highly compressed α-Inx:NF-H curve appears slightly different, where a more extended inter-filament distance is observed (Fig. 3A). Notably, a comparison of recombinant and native NF-L networks has shown that the NF-L post-translational modifications, in particular the three potential tail phosphorylation sites,36 do not have a significant effect on the network response (Fig. S3, ESI†).
3.4 The expansion of the α-Inx:NF-M bipolymer filament network resembles that of three and four component networks
We examine the inter-filament distance in α-Inx based bipolymer filament networks at various monovalent salt concentrations ranging from 40 mM to 240 mM (Fig. 4). At low pressures, networks are found in the expanded state, with all inter-filament distances above 70 nm, regardless of salt concentrations. Therefore, unlike the NF-L:NF-M network which collapses at monovalent salt concentrations exceeding 70 mM,13,30 α-Inx based filament networks remain expanded with increasing ionic strength.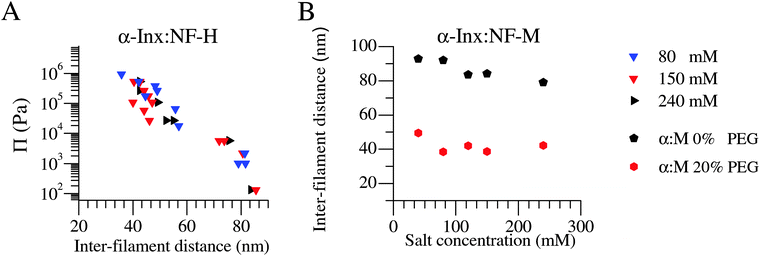 | ||
| Fig. 4 α-Inx based bipolymer filament networks do not condense with increasing monovalent salt concentrations. The compression response of (A) α-Inx:NF-H is salt independent in a near-physiological range. Similarly, (B) α-Inx:NF-M remains at an expanded state with increasing salt concentrations. This is in contrast to the reported salt dependent transition of NF-L:NF-M into a collapsed conformation at monovalent salt concentrations exceeding 70 mM.30 Experimental error for the inter-filament distance is ≈10 nm. | ||
The mature NF network is composed of all four subunit proteins. In Fig. 3D, we examine hetero-filament networks composed of either four (quartet) or three components (triplets) subject to osmotic pressure. The quartet is formed by assembling all four proteins at the biologically relevant stoichiometric ratio of 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (NF-L
1 (NF-L![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) α-Inx
α-Inx![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) NF-M
NF-M![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) NF-H).5 The two triplet networks measured are composed of α-Inx
NF-H).5 The two triplet networks measured are composed of α-Inx![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) NF-M
NF-M![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) NF-H filaments (with corresponding 7
NF-H filaments (with corresponding 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3
3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 ratios) and NF-L
2 ratios) and NF-L![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) α-Inx
α-Inx![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) NF-M filaments (3
NF-M filaments (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4
4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 ratios). The Π–d diagrams show that the omission of one component does not have a dramatic effect on the hydrogel response. Their response is also comparable to the NF-L:NF-M:NF-H network measured before.30 A possible exception is found for the α-Inx:NF-M:NF-H triplet which was slightly more expanded in comparison other triplet and quartet networks at pressures exceeding the pressure required for network irreversible collapse (Π ≥ 104 Pa). Interestingly, similar stability with regards to NF composition was recently predicted by simulations of several NF triplet compositions, which did not, however, include α-Inx.37 Unfortunately, the experimental error of d measured at low osmotic pressure is too large in order to allow us to examine the minute changes predicted by exchanging NF-L and α-Inx.22
3 ratios). The Π–d diagrams show that the omission of one component does not have a dramatic effect on the hydrogel response. Their response is also comparable to the NF-L:NF-M:NF-H network measured before.30 A possible exception is found for the α-Inx:NF-M:NF-H triplet which was slightly more expanded in comparison other triplet and quartet networks at pressures exceeding the pressure required for network irreversible collapse (Π ≥ 104 Pa). Interestingly, similar stability with regards to NF composition was recently predicted by simulations of several NF triplet compositions, which did not, however, include α-Inx.37 Unfortunately, the experimental error of d measured at low osmotic pressure is too large in order to allow us to examine the minute changes predicted by exchanging NF-L and α-Inx.22
3.5 Electrostatic analysis reveals multiple ionic cross-linking sites on NF tails
The structural role of physical, non-covalent, ionic cross-linkers in NF networks was previously demonstrated.14,38 Such cross-linkers should also affect the inter-filament distance.13,30,39,40 In order to find potential cross-linking sites we undertake an electrostatic “handshake” analysis of the NF tails.30 We evaluate short-range electrostatic attractive interactions, between tails, using a coarse-grained model; where segments engage in a zipper-like electrostatic interaction. Interacting segments may belong to a pair of tails protruding from the same filament or from two opposing filaments. We fix the distance between neighboring amino acids on the same tail to be 0.35 nm, while the opposing segments are separated by 0.28 nm (the span of anionic–cationic bonds in ionic crystals). The energy landscape between residues from two tails is represented as a two-dimensional matrix and estimated from Coulomb's law:30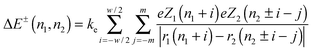 | (1) |
The generated matrices suggest energetically favorable sites for ionic bridging, which are responsible for tail attractions. Therefore, it is reasonable that the specific amino-acid sequence, and charge distribution in particular, is key in regulating the interactions between the tails and the inter-filament distance (Fig. 3).
We find that the anti-parallel (Fig. 5) and parallel (Fig. S7, ESI†) ionic handshake matrices are similar. The similarities between the two matrices are the result of the coarse-grained calculation. Significant differences can only be observed when examining segments which are comparable to the coarse-grained widow (≈10 amino acids), as can be seen by comparing the α-Inx matrices in Fig. 5 with Fig. S7 (ESI†). The locations of large attractive and repulsive areas strongly depend on the specific sequence, but not on the parallel or anti-parallel configuration. This is verified by handshakes calculated between randomly permuted tail sequences and will be discussed later.
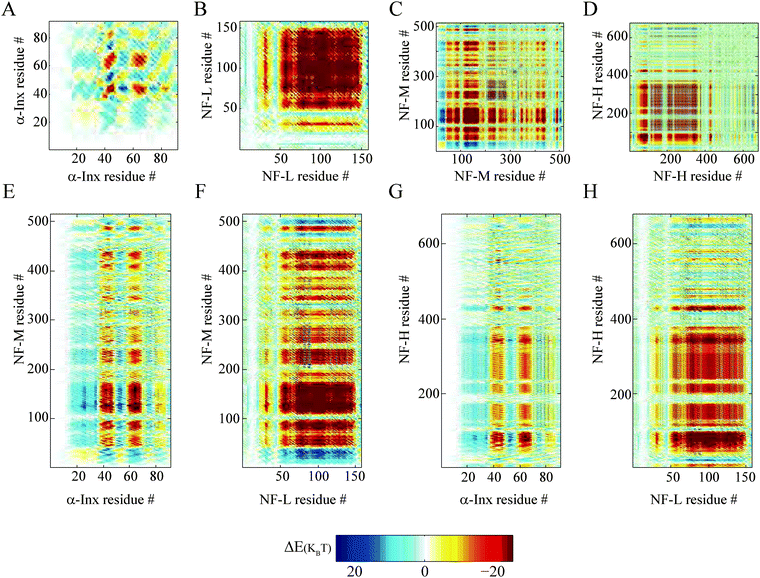 | ||
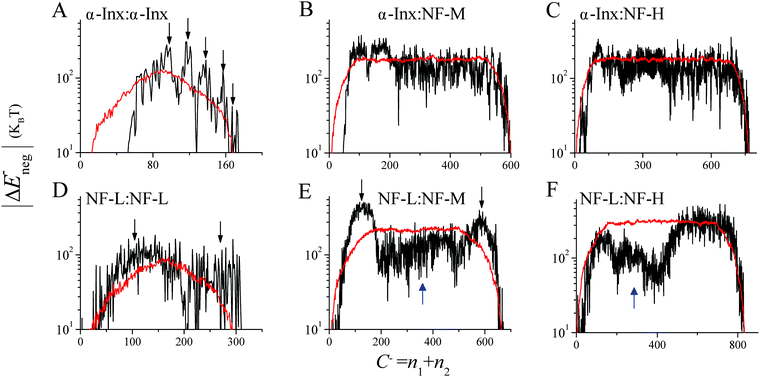
| Fig. 5 Handshake analysis of tail-to-tail interactions. Two tails aligned in an anti-parallel configuration, showing tail-to-tail interaction of ionic cross-linking sites on two opposite tails. The colors in the ΔE− (n1, n2, w = 10, m = 5) handshake matrices are given by eqn (1). Homopolymer filament handshakes of (A) α-Inx, (B) NF-L, (C) NF-M and (D) NF-H are on the first row. Below are bipolymer filament handshakes of NF-M with either (E) NF-M or (F) NF-H and NF-H with either (G) α-Inx or (H) NF-L. The phosphorylation of NF-L, NF-M and NF-H but not α-Inx was taken into account in these calculations as previously described.30 | ||
The numerous attractive sites observed in the matrices suggest multiple possible tail conformations. We evaluate the cumulative effect of attractive sites and correlate it to the inter-filament distance. For an anti-parallel configuration, the distance is correlated to the value of constant C− = n1 + n2, where residue n1 interacts with residue n2. For a parallel configuration, the distance is correlated to the constant C+ = N2 − (n2 − n1) for n2 > n1 and C+ = N1 − (n1 − n2) otherwise. Here, N1 and N2 are the total amino acid lengths and we set N2 ≥ N1 for clarity. For each given value of C−or C+ we sum over the attractive residue pairs only, obtaining ΔE−neg(C−) or ΔE±neg(C+) respectively (Fig. 6, Fig. S8–S10 and detailed calculations in ESI†).
To identify the role of a specific tail sequence we also calculate an average |ΔE±neg(C±)| for 100 permuted tail sequences in both parallel and anti-parallel configurations (Fig. 6 and Fig. S8–S10, ESI†). For each network, the permuted values are compared to the original, non-permuted |ΔE±neg(C±)|. The comparison emphasizes the significance of a specific sequence within each of the tails. Deviations from the average permuted sequences indicate more probable cross-linking sites and inter-filament spacing.
3.6 α-Inx forms a dense corona close to the filament backbone
α-Inx aggregation can be explained by the multiple attractive sites along the tail (Fig. 5A and 6A). In contrast, the NF-L:NF-L matrix shows a large (100 × 100 amino acids) repulsive domain and an attraction due to the positively charged C-terminal tip (Fig. 5B and 6D). Hydrophobic interactions may account for additional attraction between amino-acid sites in NF-L and α-Inx homopolymer filaments. We find that α-Inx is the least hydrophilic, with a hydrophobicity score17 of −0.6, whereas the NF triplet proteins are more hydrophilic with −1.4 to −1.6 scores (Fig. 1G and H and Table S1, ESI†). This finding further supports that in comparison to the NF-L tail, the α-Inx tail is more susceptible to collapse on the filament backbone.3.7 α-Inx prevents cross-linking interactions near the filament backbone
Both NF-L:NF-H and α-Inx:NF-H networks show a similar compression response (Fig. 3A). The corresponding handshake matrices show similar multiple weak attractive sites at NF-H's last 200 amino acids (Fig. 5G, H and 6C, F and Fig. S8C and F, ESI†). In fact, our analysis agrees with previous studies of NF-H mutants, where a minimal truncation of the last 191 amino acids resulted in network and cross-bridging disruption.39At low osmotic pressure, the inter-filament distance measured in the α-Inx:NF-M network is surprisingly larger than the inter-filament distance measured in the NF-L:NF-M network. An examination of NF-M handshake matrices reveals an attractive interaction of the NF-M tip with NF-M and NF-L segments close to the filament backbone (Fig. 5B, F and 6E and Fig. S10, ESI†). The analysis suggests two potential cross-linking interactions that promote the reduced inter-filament distance we observe. First, opposing NF-L tails can interact, as discussed before in the homopolymer case. The existence of such NF-L interactions can also explain why the measured inter-filament distance in the NF-L:NF-M and NF-L homopolymer networks is similar (Fig. 3C). Second, NF-M tail tips can interact with segments close to the filament backbone. This can occur either through penetration of the NF-M tail into opposite filament brushes, or by looping of the NF-M tail back to its filament backbone. Such loops were recently suggested by simulation34,41 and AFM pulling experiments.42 As for tail interpenetration, Monte-Carlo simulations predicted increased inter-penetration with simultaneous decrease in tail expansion, as the inter-filament distance was reduced from 60 to 40 nm.43 Nonetheless, a microscopic experimental validation of the internal organization of the tails would be needed in order to confirm our analysis and its correspondence to simulations and previous experiments.44
A comparison of α-Inx:NF-M and NF-L:NF-M matrices shows how α-Inx affects the conformation of the NF-M tails (Fig. 5E, F and 6B, E). The α-Inx:NF-M matrix indicates that multiple weak attractive sites exist along the NF-M tail. It resembles the matrices of α-Inx:NF-H and NF-L:NF-H networks which are also in an expanded state. Specifically, we find that |ΔE±neg(C±)| values of NF-L:NF-M deviate significantly, in favour of attractive interactions, from the averaged permuted sequences. The deviations favor cross-linking sites which result in shorter inter-filament spacing. In contrast, the nearly neutral α-Inx seems to flatten the energy landscape to be similar to the permuted sequences (Fig. 6B, C, E and F). In the absence of specific attraction, the most probable configuration will be an expanded one, as measured in our experiment.
The collapsed nature of α-Inx mentioned earlier can seemingly reduce the inter-filament spacing in α-Inx-based heteropolymer filament networks.22 However, it appears that the specific structural properties generated by the α-Inx collapsed tails prevent the attractive interactions close to the filament backbone, which accounted for the condensed state of NF-L:NF-M. Further support is given by comparing the inter-filament distance of α-Inx:NF-H and NF-L:NF-H bipolymer filament networks at high osmotic pressure (Fig. 3A). There, the distance is larger for bipolymer filaments networks containing the shorter α-Inx tail, which suggests that α-Inx tails effectively repel the longer tails from cross-linking nearby the backbone.
4 Discussion
We measured the inter-filament distance response to osmotic stress in multi-component networks of neuronal intermediate filaments. The NF network structural and mechanical properties are determined by synergistic interactions between the short and long protein tails. This effect is less pronounced in the three and four-component networks, where the omission of a single component does not change the network compression significantly.The coarse-grain handshake analysis shows preferable sites that induce attractions between different tails. Although our analysis lacks many molecular details including hydrophobicity, steric and entropic factors, it captures the key experimental findings. Further support for extensive tail interactions is given by available simulations on NF proteins.34,41,43
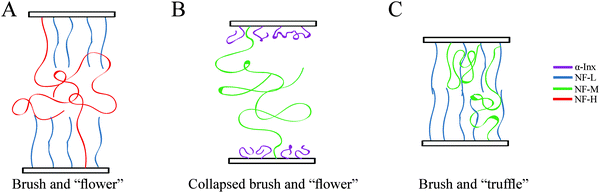
The experimental results and their analyses allow us to schematically illustrate the possible conformations that correspond to the dominant cross-linking sites (Fig. 7). The short-tailed NF-L and α-Inx form an inner layer corona close to the filament core.22 Due to the dissimilar properties of the α-Inx and NF-L tails with respect to charge, amino-acid length (N) and hydropathy, they are expected to form different coronas. The α-Inx tail is nearly neutral, with a −0.03 e per amino acid linear charge density (ϕ), which is much lower than the −0.29 e per amino acid calculated for NF-L tail (Table S1, ESI†). The α-Inx tail is also shorter and less hydrophilic (Fig. 1G and H). For physiological salt concentrations (cs), the concentration of tail counter-ions is lower than the concentration of the bulk ion solution. Therefore, such charged brushes are always in the “salted brush” regime, where the brush height holds H ∼ N3/4ϕ1/2cs−14 (ref. 45). Hence, NF-L is expected to stretch from the filament backbone due to its higher negative charge.46 On the other hand, α-Inx, which is almost neutral, is organized in a denser, collapsed brush surrounding the filament backbone (Fig. 7B). We note that the estimated radius of gyration (Rg) of α-Inx in solution is expected to be 40% smaller than that of NF-L (Table S2, ESI†), but we do not expect the distance between adjacent tails on the corona (2 nm) to be larger than Rg.
Heteropolymer filaments that contain long tails are thus predicted to form two distinctive layers.22 The inner layer corona is composed of the short tails while the longer tails are repelled farther away from the corona, into the outer layer. Since the tails within the outer layer are less dense, they are expected to form a “flower-like” conformation that can cross-link with opposite filaments (Fig. 7A and B). The “flower” conformation agrees with our results for heteropolymer filament networks containing NF-H. There, the inter-filament distance at low pressure is almost unaffected by the identity of the remaining tails, as the distance is determined by the NF-H “flowers” decorating the “collapsed brush” corona.
However, bipolymer filaments containing NF-M are organized differently. The condensed state of NF-L:NF-M networks implies that NF-M interpenetrates the opposite brush to form cross-links with tail segments close to the filament backbone. This may allow cross-linking between the apposing NF-L inner coronas. Consequently, the NF-M tail favors conformations in the vicinity of the filament backbone. As an analogue to the “mushroom” regime, we refer to these hidden tails as the “truffle” regime (Fig. 7C). A comparison of the expanded α-Inx:NF-M network to the condensed NF-L:NF-M network is indicative of the different structural roles of α-Inx and NF-L in multi-component assemblies. We suggest that the neutral charge and the less hydrophilic residues of the α-Inx tail cause the formation of a collapsed corona that hinders cross-linking interactions close to the filament backbone. Condensation of α-Inx results in an effective repulsion of long-tails from the backbone.
Notably, we show that the inter-filament distance response of the α-Inx:NF-M network resembles that of the quartet network, in contrast to the NF-L:NF-M network. Since most studies indicate that NF-M and α-Inx form a transitory network in early postnatal stages,6,7 the measured differences between NF-M bipolymer filament networks suggest a new structural rationale for the sequential expression of NF proteins during embryonic development. These findings also correspond well with in vivo measurements demonstrating that NF-M is a dominant factor in controlling axonal diameters.47,48 We show that the addition of α-Inx does not significantly alter the inter-filament spacing of the three component network (Fig. 3), in agreement with reports of transgenic mice, where the deletion of α-Inx had no apparent effect on axon caliber.49 However, our results show that careful attention must be taken to the early development expression level of α-Inx within neuronal IF networks due to its synergistic interactions with the NF triplet proteins.
On top of the complex interactions mediated by the primary amino acids of the various NF protein tails, a reversible regulatory mechanism for NF network stabilization is introduced by post-translation modifications.6 In particular, phosphorylation of the NF long tails significantly increases their negative charge fraction.50 We expect that the phosphorylation level will alter the electrostatic interaction between the tails. Such effects were indeed observed and will be addressed in future communications.
A recent study of the peripheral nervous system (PNS) identified peripherin in composite filaments along with the NF triplet proteins.51 Like α-Inx, peripherin expression in the PNS decreases postnatally until stabilizing at a non-negligible fixed stoichiometry with the NF triplet proteins. Both α-Inx and peripherin also act as the short tailed partner in these assemblies, as peripherin's tail is only 65 amino-acid long. Given these similarities in expression pattern and size, the role of peripherin in composite filament networks should be of physical and biological interest.
Acknowledgements
We are grateful to Dr Geraisy Wassim of Beit Shean abattoirs Tnuva for kindly providing us with the spinal cords. We thank the following beamlines for SAXS measurements: I911-SAXS at MAX IV Laboratory, Lund, Sweden; SWING at SOLEIL synchrotron, Paris, France; and P12 EMBL BioSAXS beamline at Hamburg, Germany. We thank Rona Shaharabani, Dr Stive Pregent, Dr Yuval Reiss and Guy Jacoby for their assistance in purification and measurements and to Ram Avinery also for his invaluable programming work. We are grateful to Dr Michael Wyrsta for his comments, Dr Yevgeny Berdichevsky for his advice on expression and purification, Dr Artium Khatchtouriants for the AFM guidance and support and Dr Yossi Lereah for TEM imaging. We acknowledge the help of Dorothee Möller and Dr Karsten Richter (both DKFZ) for TEM sample preparation and imaging. The work was supported by the Israeli Science Foundation (grant 571/11), the European Community's 7th Framework Programme (CIG-293402), the Tel Aviv University Center for Nanoscience and Nanotechnology and the Scakler Institute for Biophysics at Tel Aviv University. Travel grants to synchrotron facilities were provided by BioStruct-X. MK acknowledges a “Short term scientific mission” grant to DKFZ in Heidelberg by COST action BM1002.References
- B. Alberts, A. Johnson, J. Lewis, M. Raff, K. Roberts and P. Walter, Molecular biology of the cell, Garland, 2002 Search PubMed.
- H. Herrmann and U. Aebi, Annu. Rev. Biochem., 2004, 73, 749–789 CrossRef CAS PubMed.
- S. Sinha, Intermed. Filam. Cytoskelet., Academic Press, 2004, vol. 78, pp. 267–296 Search PubMed.
- C. Lépinoux-Chambaud and J. Eyer, Histochem. Cell Biol., 2013, 140, 13–22 CrossRef PubMed.
- A. Yuan, M. V. Rao, T. Sasaki, Y. Chen, A. Kumar, Veeranna, R. K. H. Liem, J. Eyer, A. C. Peterson, J.-P. Julien and R. A. Nixon, J. Neurosci., 2006, 26, 10006–10019 CrossRef CAS PubMed.
- A. Laser-Azogui, M. Kornreich, E. Malka-Gibor and R. Beck, Curr. Opin. Cell Biol., 2015, 32, 92–101 CrossRef CAS PubMed.
- R. A. Nixon and A. Yuan, Cytoskelet. Nerv. Syst., Springer New York, New York, NY, 2011, ch. 8, vol. 3, pp. 189–199 Search PubMed.
- G. M. Cooper and R. E. Hausman, The cell: a molecular approach, ASM Press, 2007 Search PubMed.
- A. A. Chernyatina, S. Nicolet, U. Aebi, H. Herrmann and S. V. Strelkov, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 13620–13625 CrossRef CAS PubMed.
- D. A. Parry, S. V. Strelkov, P. Burkhard, U. Aebi and H. Herrmann, Exp. Cell Res., 2007, 313, 2204–2216 CrossRef CAS PubMed.
- R. Beck, J. Deek and C. R. Safinya, Biochem. Soc. Trans., 2012, 40, 1027–1031 CrossRef CAS PubMed.
- H. C. Hesse, R. Beck, C. Ding, J. B. Jones, J. Deek, N. C. MacDonald, Y. Li and C. R. Safinya, Langmuir, 2008, 24, 8397–8401 CrossRef CAS PubMed.
- J. Deek, P. J. Chung, J. Kayser, A. R. Bausch and C. R. Safinya, Nat. Commun., 2013, 4, 2224 Search PubMed.
- Y.-C. Lin, N. Y. Yao, C. P. Broedersz, H. Herrmann, F. C. MacKintosh and D. A. Weitz, Phys. Rev. Lett., 2010, 104, 058101 CrossRef.
- J. P. Gou, T. Gotow, P. A. Janmey and J. F. Leterrier, Med. Biol. Eng. Comput., 1998, 36, 371–387 CAS.
- S. Heins, P. C. Wong, S. Müller, K. Goldie, D. W. Cleveland and U. Aebi, J. Cell Biol., 1993, 123, 1517–1533 CrossRef CAS.
- J. Kyte and R. F. Doolittle, J. Mol. Biol., 1982, 157, 105–132 CrossRef CAS.
- H. Hajarian, J. Biol. Chem., 1995, 270, 9334–9339 CrossRef PubMed.
- H. Herrmann, M. Häner, M. Brettel, N. O. Ku and U. Aebi, J. Mol. Biol., 1999, 286, 1403–1420 CrossRef CAS PubMed.
- G. Y. Ching and R. K. Liem, J. Cell Biol., 1993, 122, 1323–1335 CrossRef CAS.
- I. A. Abumuhor, P. H. Spencer and J. A. Cohlberg, J. Struct. Biol., 1998, 123, 187–198 CrossRef CAS PubMed.
- F. A. M. Leermakers and E. B. Zhulina, Eur. Biophys. J., 2010, 39, 1323–1334 CrossRef CAS PubMed.
- J. Kas, J. Biol. Chem., 1996, 271, 15687–15694 CrossRef PubMed.
- J. B. Jones and C. R. Safinya, Biophys. J., 2008, 95, 823–835 CrossRef CAS PubMed.
- V. A. Parsegian, R. P. Rand, N. L. Fuller and D. C. Rau, Methods Enzymol., 1986, 127, 400–416 CAS.
- D. C. Rau, B. Lee and V. A. Parsegian, Proc. Natl. Acad. Sci. U. S. A., 1984, 81, 2621–2625 CrossRef CAS.
- R. Beck, J. Deek, M. C. Choi, T. Ikawa, O. Watanabe, E. Frey, P. Pincus and C. R. Safinya, Langmuir, 2010, 26, 18595–18599 CrossRef CAS PubMed.
- N. Mücke, L. Kreplak, R. Kirmse, T. Wedig, H. Herrmann, U. Aebi and J. Langowski, Am. J. Mol. Biol., 2004, 335, 1241–1250 CrossRef PubMed.
- U. Aebi and W. Baschong, Cell Biology, Elsevier, 2006, vol. 3, pp. 241–246 Search PubMed.
- R. Beck, J. Deek, J. B. Jones and C. R. Safinya, Nat. Mater., 2010, 9, 40–46 CrossRef CAS PubMed.
- Y. Li, R. Beck, T. Huang, M. C. Choi and M. Divinagracia, J. Appl. Crystallogr., 2008, 41, 1134–1139 CrossRef CAS.
- P. M. Steinert, L. N. Marekov and D. A. D. Parry, J. Biol. Chem., 1999, 274, 1657–1666 CrossRef CAS PubMed.
- P. Pawelzyk, N. Mücke, H. Herrmann and N. Willenbacher, PLoS One, 2014, 9, e93194 Search PubMed.
- S. Jeong, X. Zhou, E. B. Zhulina and Y. Jho, Isr. J. Chem., 2014 DOI:10.1002/ijch.201400085.
- J. Lee, S. Kim, R. Chang, L. Jayanthi and Y. Gebremichael, J. Chem. Phys., 2013, 138, 015103 CrossRef PubMed.
- The UniProt Consortium, Nucleic Acids Res., 2014, 42, D191–D198 CrossRef PubMed.
- S. Kim, R. Chang, C. Teunissen, Y. Gebremichael and A. Petzold, J. Neurol. Sci., 2011, 307, 132–138 CrossRef CAS PubMed.
- J.-F. Leterrier and J. Eyer, Biochem. J., 1987, 245, 93–101 CAS.
- J. Chen, T. Nakata, Z. Zhang and N. Hirokawa, J. Cell Sci., 2000, 113(Pt 21), 3861–3869 CAS.
- E. B. Zhulina and F. A. M. Leermakers, Biophys. J., 2010, 98, 462–469 CrossRef CAS PubMed.
- R. Chang, Y. Kwak and Y. Gebremichael, J. Mol. Biol., 2009, 391, 648–660 CrossRef CAS PubMed.
- H. Aranda-Espinoza, P. Carl, J. F. Leterrier, P. Janmey and D. E. Discher, FEBS Lett., 2002, 531, 397–401 CrossRef CAS.
- L. Jayanthi, W. Stevenson, Y. Kwak, R. Chang and Y. Gebremichael, J. Biol. Phys., 2013, 39, 343–362 CrossRef PubMed.
- J. F. Hess, M. S. Budamagunta, A. Aziz, P. G. FitzGerald and J. C. Voss, Protein Sci., 2013, 22, 47–55 CrossRef CAS PubMed.
- L.-J. Qu, Commun. Theor. Phys., 2012, 57, 1091–1094 CrossRef.
- N. Srinivasan, M. Bhagawati, B. Ananthanarayanan and S. Kumar, Nat. Commun., 2014, 5, 5145 CrossRef CAS PubMed.
- M. L. Garcia, C. S. Lobsiger, S. B. Shah, T. J. Deerinck, J. Crum, D. Young, C. M. Ward, T. O. Crawford, T. Gotow, Y. Uchiyama, M. H. Ellisman, N. A. Calcutt and D. W. Cleveland, J. Cell Biol., 2003, 163, 1011–1020 CrossRef CAS PubMed.
- M. V. Rao, M. L. Garcia, Y. Miyazaki, T. Gotow, A. Yuan, S. Mattina, C. M. Ward, N. A. Calcutt, Y. Uchiyama, R. A. Nixon and D. W. Cleveland, J. Cell Biol., 2002, 158, 681–693 CrossRef CAS PubMed.
- F. Levavasseur, Q. Zhu and J. P. Julien, Brain Res. Mol. Brain Res., 1999, 69, 104–112 CrossRef CAS.
- J. F. Leterrier, J. Käs, J. Hartwig, R. Vegners and P. A. Janmey, J. Biol. Chem., 1996, 271, 15687–15694 CrossRef CAS PubMed.
- A. Yuan, T. Sasaki, A. Kumar, C. M. Peterhoff, M. V. Rao, R. K. Liem, J.-P. Julien and R. A. Nixon, J. Neurosci., 2012, 32, 8501–8508 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5sm00662g |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2015 |