Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Enzyme repurposing of a hydrolase as an emergent peroxidase upon metal binding†
Nobutaka
Fujieda
*a,
Jonas
Schätti
a,
Edward
Stuttfeld
b,
Kei
Ohkubo
cd,
Timm
Maier
b,
Shunichi
Fukuzumi
cde and
Thomas R.
Ward
*a
aDepartment of Chemistry, University of Basel, Spitalstrasse 51, CH-4056 Basel, Switzerland. E-mail: fujieda@mls.eng.osaka-u.ac.jp; thomas.ward@unibas.ch
bBiozentrum, University of Basel, Klingelbergstr. 50/70, CH-4056 Basel, Switzerland
cDepartment of Material and Life Science, Graduate School of Engineering, Osaka University, ALCA and SENTAN, Japan Science and Technology Agency (JST), 2-1 Yamada-oka, Suita, Osaka 565-0871, Japan
dDepartment of Bioinspired Science, Ewha Womans University, Seoul 120–750, Korea
eFaculty of Science and Technology, Meijo University and ALCA and SENTAN, Japan Science and Technology Agency (JST), Tempaku, Nagoya, Aichi 468–8502, Japan
First published on 7th May 2015
Abstract
As an alternative to Darwinian evolution relying on catalytic promiscuity, a protein may acquire auxiliary function upon metal binding, thus providing it with a novel catalytic machinery. Here we show that addition of cupric ions to a 6-phosphogluconolactonase 6-PGLac bearing a putative metal binding site leads to the emergence of peroxidase activity (kcat 7.8 × 10−2 s−1, KM 1.1 × 10−5 M). Both X-ray crystallographic and EPR data of the copper-loaded enzyme Cu·6-PGLac reveal a bis-histidine coordination site, located within a shallow binding pocket capable of accommodating the o-dianisidine substrate.
Introduction
Metal ions are present in nearly half of the characterized proteome,1,2 and metalloenzymes catalyze some of nature's most challenging reactions.3 This versatility has inspired a number of different strategies to create artificial metalloenzymes in the past twenty years.4–6 Guided either by computation or intuition,4–7 artificial metalloenzyme design by-and-large relies on the de novo introduction of a metal-binding site within a protein scaffold or peptide.8–13 For this purpose, various strategies have been pursued including covalent-, supramolecular- and dative anchoring.4–6 The latter approach may rely on incorporating either natural- or non-natural aminoacids as ligands (i.e. bipyridinylalanine etc.).14–16 Alternatively, metal-substitution within a metalloenzyme can lead to novel catalytic activity.17–21 Eventually, the nascent catalytic activity may be further improved by directed evolution strategies.22–25To complement these efforts and in an “enzyme tinkering spirit”,26,27 we hypothesized that non-metal containing proteins may, through the course of random mutations, evolve a putative metal binding site. Upon acquisition of a metal, nascent catalytic activity may arise, Fig. 1. As with enzyme promiscuity,28–30 if the newly acquired asset provides a competitive advantage to the cell,31,32 it may evolve to a highly efficient metalloenzyme.
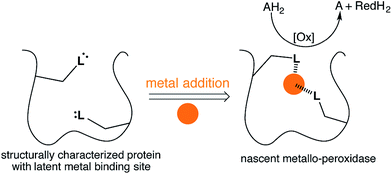 | ||
| Fig. 1 Emergent enzymatic activity resulting from metal acquisition by a protein bearing a latent metal binding motif. | ||
To test this hypothesis we employed the “Search for Three dimensional Atom Motifs in Protein Structure” (STAMPS) algorithm,33 which allows to identify proteins by a systematic search in the protein databank (PDB) for structures possessing a given motif in a topology similar to that adopted by the functional motif in a reference protein. Using this approach, we recently reported on the in silico identification of non-metallated two-histidine one-carboxylate metal binding motifs within the structurally characterized proteins of the PDB.34 Herein we report our efforts to valorise the in silico study by creating an artificial metallo-peroxidase upon addition of a metal cofactor to a protein harboring a latent mononuclear metal binding site, Fig. 1.
Results and discussion
The proteins bearing a latent non-metallated two-histidine one-carboxylate motif (HHD/E, hereafter) were identified previously using the STAMPS algorithm. For this study, we selected seven scaffolds bearing an HHD/E motif located within a binding pocket predisposed to bind metals.33,34 The search was expanded to include HHN/Q motifs, revealing six additional potential metal binders following a single point mutation. In total, six of the thirteen cloned proteins could be overexpressed in E. coli and purified using a Strep-tag II (Fig. S1–S4†).These wild-type proteins or their single mutant isoforms (pdb code: 3D53, 2F99, 1JSY, 1FHI (bearing a Q83E mutation), 1MEJ (bearing a N106D mutation), 3OC6 (bearing an N131D), see Table S1†) were tested for their peroxidase activity in the presence of various transition metal salts including VOSO4, MnCl2, FeCl2, CoSO4, NiCl2 and CuSO4. For this purpose, o-dianisidine (0.75 mM) and hydrogen peroxide (1.5 mM) were added to a buffered solution containing the protein (20 μM) and the metal salt (25 μM). The appearance of the quinone-diimine oxidation product was monitored at 460 nm.35 Gratifyingly, this screen revealed one hit: in the presence of CuSO4, 6-phosphogluconolactonase bearing an N131D mutation (Cu·6-PGLac hereafter) catalyzes the oxidation of o-dianisidine (Fig. 2A, S5 and S6†).
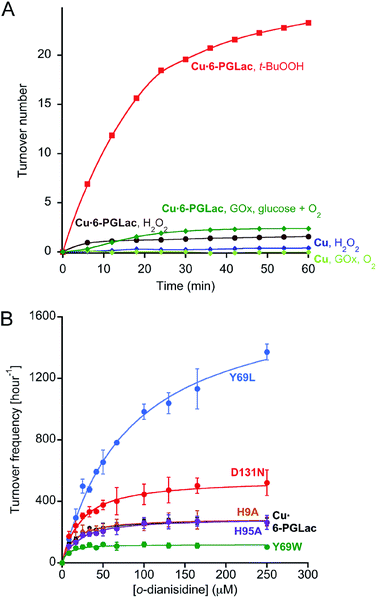 | ||
| Fig. 2 (A) Identification of nascent peroxidase activity resulting from copper addition to a lactonase using H2O2 (brown) and t-BuOOH (red) as oxidizing agent. In the presence of glucose oxidase (GOx) and glucose, Cu·6-PGLac displays peroxidase activity (green). None of the other metals tested (including Ni2+, Mn2+, Fe2+, Co2+, VO2+) displayed significant peroxidase activity in the presence of t-BuOOH and 6-PGLac (TON < 2 after 60 min, Fig. S6A†). (B) Michaelis–Menten saturation kinetics for the oxidation of o-dianisidine with t-BuOOH (6.6 mM) for selected mutants (1.25 μM) in the presence of copper (4.5 μM), [o-dianisidine] = 8–250 μM in MES-buffer (pH 6.5, 50 mM, 150 mM NaCl, 25 °C). Measured data (symbols); fitted data (solid lines). The minimal background reaction caused by free copper was subtracted from the raw data for fitting purposes (Fig. S12B†). | ||
Inspection of the X-ray structure of the putative 6-phosphogluconolactonase (6-PGLac, apo form) at 1.81 Å reveals a Rossmann fold annotated to the glucosamine-6-phosphate isomerases/6-phosphogluconolactonase family (PF01182) in PFAM database, with the hydrolytic Glu149–His151 dyad and the latent metal binding site 26 Å apart, (Fig. 3A and Table S4†). The alleged 6-phosphogluconolactonase activity36 was confirmed by 31P NMR, both in the absence and in the presence of cupric ions, emphasizing the moonlighting nature of this emergent peroxidase activity, Fig. S7.†29 Among the PF01182 family proteins, the triad residues (His67, His104, and Asn(Asp)131) are not conserved, thus suggesting that this potential metal binding motif is not functionally relevant (Fig. S8†).
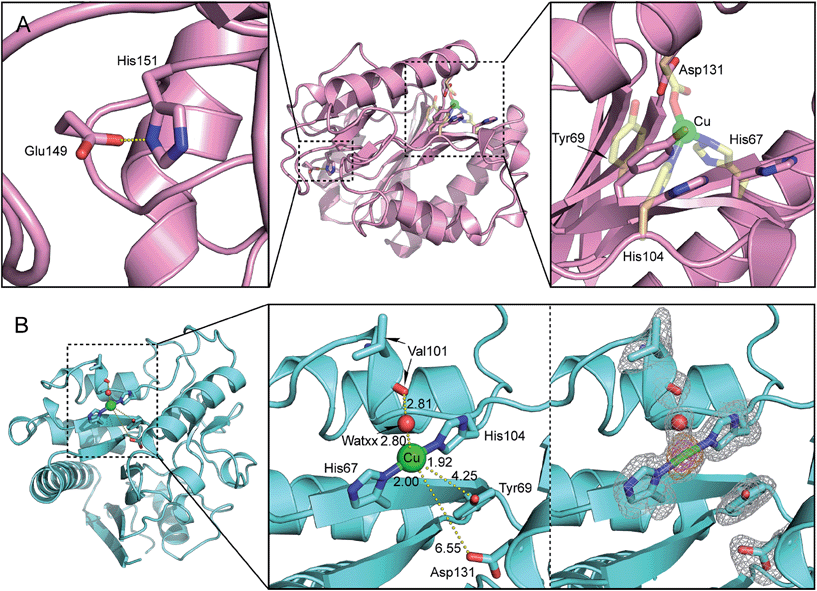 | ||
| Fig. 3 (A) Close-up view of the putative metal binding site of 6-phosphogluconolactonase 6-PGLac (PDB code 4TM8); the native hydrolytic site (left), located 26 Å away from the putative metal binding site (right). The protein main chain is displayed as ribbon and key amino acid residues are highlighted as sticks (the putative facial triad motif was metallated manually in silico and is displayed as transparent yellow sticks); (B) close-up view of the Cu1 binding site in Cu·6-PGLac (PDB code 4TM7, 2Fo − Fc (gray) and anomalous difference Fourier (orange and magenta) maps contoured at 1σ, 5σ, and 15σ, respectively): Cu1–Nδ(His67), 2.00 Å; Cu1–Nε(His104), 1.92 Å; Cu1–WatXX, 2.80 Å; Nδ(His67)–Cu1–WatXX, 94.2°; Nε(His104)–Cu1–WatXX, 95.8°; and (His67)–Cu1–Nε(His104), 170.0°. | ||
No peroxidase activity was observed upon substitution of hydrogen peroxide by the dioxygen/ascorbate couple. However, the enzyme cascade consisting of glucose oxidase and Cu·6-PGLac restores catalytic activity in the presence of glucose, dioxygen and o-dianisidine (Fig. 2A and S6B†). Significant rate enhancement was achieved using t-butyl hydroperoxide (t-BuOOH) instead of H2O2 as oxidant (Fig. 2A). The peroxidation of catechol and guaiacol by Cu·6-PGLac was investigated (Fig. S5†). No peroxidation product was detected for guaiacol with either H2O2 or t-BuOOH as oxidant. Cu·6-PGLac catalyzed the peroxidation of catechol. However, the background peroxidation with CuSO4 alone was significant, which contrasts to both o-dianisidine and guaiacol.
To gain structural insight into the metalloenzyme activity, 6-PGLac crystals (vide supra) were soaked with a mother liquor containing excess CuSO4. This procedure however did not yield suitable diffraction data. Thus, 6-PGLac was co-crystallized with 3 mM CuSO4 followed by soaking with 12 mM CuSO4 (Cu·6-PGLac). The X-ray structure was refined to a resolution of 1.39 Å (Fig. 3B, Table S4 and Fig. S9†). The overall structure is nearly identical to that of 6-PGLac, apo form (Root Mean Square Deviation (RMSD) = 0.32 over 243 Cα atoms, Fig. 3A and B and S9A†). The structure contains three fully occupied copper ion binding sites with strong anomalous signals (Table S6†): two copper ions are bound to surface histidines and aspartates crosslinking two protein monomers in the crystal (Cu2 is bound to His9 and Asp3′ and Cu3 is bound to His95 and Asp158′ and Asp220′ respectively, see Fig. S9B†). The third copper (Cu1) is located in the putative metal binding site and displays a T-shaped [2 + 1] coordination geometry. The copper is bound to His67 and His104 (at a distance of 2.00 Å and 1.92 Å, respectively) and displays a weak contact with a water molecule (WATxx 2.80 Å), which is held in place via a hydrogen bond to the carbonyl oxygen of Val101 (O⋯O 2.81 Å). This structure also contains further unspecifically bound copper ions in the lactonase active site as well as on the surface (Fig. S9C and Table S6†). However, these binding sites are characterized by low occupancy, as reflected by the height of anomalous difference density peaks, and thus presumably have a far lower binding affinity for copper ions.
The coordination geometry around Cu1 is T-shaped with two trans-histidines and a water ligand. Although reminiscent of the Cu-coordination recently reported for the lytic polysaccharide monooxygenase from Aspergillus oryzae, it lacks the terminal amine ligand, characteristic of the “histidine brace”.37 While two- and three coordinate copper geometries are frequently encountered in metalloenzymes and coordination complexes, earlier transition metals often prefer higher coordination numbers.3 We speculate that the lack of peroxidase activity observed with all other metal salts tested in the presence of 6-PGLac may be caused by the ill-suited low coordination geometry imposed by the putative active site.
The affinity of 6-PGLac for Cu(II) was determined using tryptophan-fluorescence quenching. Two dissociation constants could be extracted from the titration profile: Kd1 = 0.83 ± 0.11 μM, Kd2 = 130 ± 3.3 μM; confirming the presence of one tight and weaker copper binding sites (Fig. S13†).
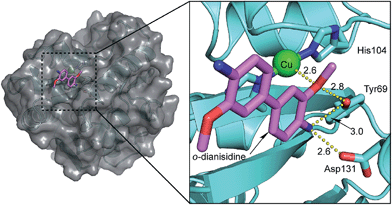
The potentially coordinating oxygen of Asp131 identified by the in silico search and of the Tyr69 are located 6.55 Å and 4.25 Å from Cu1 respectively. The low coordination number of Cu1, coupled with the possibility of photoreduction by the X-ray dose, led us to explore whether cupric state may have additional ligands.
In order to investigate the involvement of neighboring amino acids in the coordination of catalytically competent cupric ions, the kinetic saturation profiles of Cu·6-PGLac and mutants (Fig. S12†) thereof were determined. The results are summarized in Table 1, Fig. 2B and S12A.†Cu·6-PGLac bearing the HHD triad displays Michaelis–Menten behavior with an efficiency of kcat/KM = 6.9 × 103 M−1 s−1 (Table 1, entry 1). Interestingly, the KM value of Cu·6-PGLac is comparable to that of the naturally occurring peroxidase (For horseradish peroxidase (HRP), 13 μM).38 The kcat/KM however is lower than that of HRP (7.1 × 107 M−1 s−1),38 whereas it's equal to or greater than those of the artificial peroxidases (ferric porphyrin-binding antibody (3.4 × 104 M−1 s−1)39 and ferric porphyrin-binding xylanase (3.7 × 102 M−1 s−1)40). Deletion of the surface histidines confirms that both Cu2 and Cu3 (bound to His9 and His95 respectively) are not catalytically competent. Indeed both Cu·6-PGLac H9A and Cu·6-PGLac H95A display nearly identical kinetic behavior when compared to Cu·6-PGLac (Table 1, entries 2 and 3). Unfortunately, all attempts to mutate two surface histidine residues invariably led to inclusion bodies which could not be renatured. In stark contrast, mutation of either His67 or His104 binding residues into non-coordinating amino acids completely shuts off catalytic activity, (Table 1, entries 4 and 5), confirming that the catalytic metal is indeed located in the cavity flanked with residues His67, His104, Asp131 and Tyr69. Unexpectedly, mutation of Asp131 to either a hydrophobic, a coordinating or a polar amino acid does not affect the catalytic performance. This suggests that this residue does not bind either to cupric- or cuprous ions (Table 1, entries 6–9). Position 69 in contrast has a larger impact on the catalytic performance: Cu·6-PGLac Y69L displays a moderately improved kcat, albeit at the cost of KM (Table 1, entries 10–12). Based on this limited mutagenesis study, we are confident that the catalytic performance could be improved significantly with a larger mutagenesis campaign.
| Entry | Variant | K M (μM) | k cat × 103 (s−1) | k cat/KM (M−1 s−1) |
|---|---|---|---|---|
| a The kinetic measurements were determined in triplicate and fitted using the Michaelis–Menten equation. The minimal background reaction caused by free copper was subtracted from the raw data for fitting purposes. b The catalytic activity was too small to be determined (Fig. S12B). | ||||
| 1 | 6-PGLac | 11 ± 3 | 78 ± 4 | (6.9 ± 2) × 103 |
| 2 | H9A | 12 ± 5 | 80 ± 6 | (6.5 ± 3) × 103 |
| 3 | H95A | 17 ± 5 | 80 ± 6 | (4.6 ± 2) × 103 |
| 4 | H67F | N.D.b | N.D.b | N.D.b |
| 5 | H104F | N.D.b | N.D.b | N.D.b |
| 6 | D131A | 13 ± 5 | 100 ± 7 | (7.7 ± 3) × 103 |
| 7 | D131E | 10 ± 3 | 78 ± 4 | (7.7 ± 2) × 103 |
| 8 | D131H | 32 ± 6 | 150 ± 9 | (4.9 ± 1) × 103 |
| 9 | D131N | 21 ± 6 | 150 ± 10 | (7.2 ± 2) × 103 |
| 10 | Y69F | 36 ± 8 | 100 ± 7 | (2.9 ± 0.7) × 103 |
| 11 | Y69W | 4.8 ± 2 | 33 ± 2 | (6.9 ± 3) × 103 |
| 12 | Y69L | 82 ± 10 | 490 ± 30 | (5.9 ± 0.9) × 103 |
The involvement of Tyr69 was assessed by performing a docking simulation between Cu·6-PGLac and o-dianisidine. It revealed one major low-energy conformation with the polar edge of the biaryl-substrate pointing towards the copper active site, Fig. 4. One methoxy oxygen atom of o-dianisidine is hydrogen bonded to the oxygen atom of Tyr69 (H3CO⋯OY 2.8 Å) and interacts with Cu1 (H3CO⋯Cu 2.6 Å). The other hydrogen bonds were observed between nitrogen atom of o-dianisidine and the oxygen atom of Tyr69 at a distance of 3.0 Å, and the carboxyl oxygen atom of Asp131 at 2.6 Å.
The artificial peroxidase was scrutinized by X-band EPR analysis at 77 K (Fig. 5). The presence of multiple Cu2+ species in Cu·6-PGLac results in a complicated spectrum in the parallel region (Fig. 5 and S14A†), highlighting the presence of multiple Cu2+ binding sites as observed in the crystal structure (Cu1; His67 and His104, Cu2; His9, Cu3; His95). Analysis of histidine-deleted variants revealed that Cu·6-PGLac H9R exhibits an axial EPR spectrum, typical of a single Cu2+ species (Fig. 5 and S14A†), whereas Cu·6-PGLac H95F shows an almost identical spectrum to that of Cu·6-PGLac. This suggests that His9 binds to Cu2+ more tightly than His95. In contrast, the H67F and H104F single mutants resulted in markedly different spectra (Fig. 5), suggesting that both His67 and His104 are involved in copper coordination in Cu·6-PGLac. A reasonable simulation of the spectrum of Cu·6-PGLac H9R provided detailed EPR parameters (see Fig. S14C† for further details). Again here, a more detailed analysis was hampered by the formation of inclusion bodies upon mutation of multiple surface histidine residues. Given the parameters of the parallel region (gz = 2.230 and Az = 17.7 mT), Peisach–Blumberg analysis41 suggests that this Cu2+ species is a type 2, reminiscent of the type 2 copper in the multicopper oxidase (gz ∼ 2.24 and Az = 13–19 mT).42 This copper center bears two nitrogens and one or two water ligand(s) in the equatorial plane in the reported crystal structure.43,44 Superhyperfine coupling pattern due to the ligating nitrogen atoms is apparent in the perpendicular region (Fig. S14†). Thus, these results suggest that the 6-PGLac provides the Cu1 ion with a coordination environment suitable for coordination of both the peroxide and the substrate to yield a transient end-on (alkyl-)peroxide copper(II) species.45,46 Although, no known natural peroxidase contains a mononuclear copper active site to our knowledge, mononuclear cupric coordination complexes display peroxidase activity and have been shown to operate via concerted H atom abstraction with O–O bond scission and subsequent attack at a substrate by a Cu(II)-oxyl radical (i.e. LnCuII–O˙).45–47
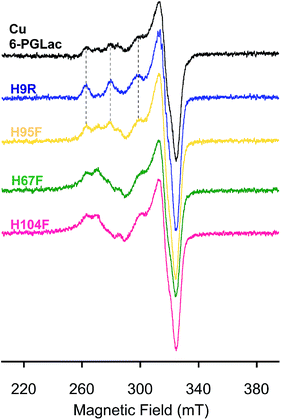 | ||
| Fig. 5 EPR spectra of Cu·6-PGLac (black), Cu·6-PGLac H9R (blue), Cu·6-PGLac H95F (yellow), Cu·6-PGLac H67F (green), and Cu·6-PGLac H104F (pink) at pH 6.5 (50 mM MES and 150 mM NaCl) at 77 K. | ||
Conclusions
The enzyme repurposing strategy contrasts with other artificial metalloenzyme approaches. Indeed, as we demonstrate herein, the latent metal binding site is present but not metallated in the wild-type 6-PGLac. Simple metal salt addition thus suffices to endow a protein with novel catalytic activity, very different from the native activity. Furthermore, this strategy relying upon metal acquisition can be viewed as complementary to enzyme promiscuity, offering a novel means to swiftly acquire enzymatic catalytic activity, vastly different from the native activity.Although the STAMPS algorithm suggested a possible facial triad coordination, a T-shaped Cu coordination was observed upon Cu(II) supplementation. We believe that this may be due to the size of the latent metal binding site which allows for significant side chain flexibility: a feature not taken into consideration in the STAMPS algorithm. Gratifyingly, this low-coordination bis-histidine copper geometry combined with the additional surrounding Tyr69 and Asp131 residues provides unique opportunities to fine-tune or alter the emergent peroxidase activity. The compatibility of the artificial metalloenzyme with other enzymes (e.g. glucose oxidase) as well as cell lysates suggests that directed evolution strategies may be used to further optimize its performance.
Acknowledgements
N.F. thanks MEXT, Japan (no. 20038784) as well as the Yoshida Foundation for Science and Technology. K.O. thanks MEXT, Japan for financial support (nos 26620154, and 26288037). T.R.W. thanks the Swiss National Science Foundation (SNF grant 200020_144354) as well as the NCCR Molecular Systems Engineering. T.M. acknowledges support by the R'Equip program of the Swiss National Science Foundation (SNF grant 316030_145023). We thank Dr D. Haeussinger, Ms L. Misson and Mr M. Barnet for their help with the 31P NMR, ESI-MS and docking simulation, respectively and the staff of beamlines PXI and PXIII at Swiss Light Source for support of crystallographic data collection.Notes and references
- K. J. Waldron and N. J. Robinson, Nat. Rev. Microbiol., 2009, 7, 25–35 CrossRef CAS PubMed
.
- A. Cvetkovic, A. L. Menon, M. P. Thorgersen, J. W. Scott, F. L. Poole, F. E. Jenney, W. A. Lancaster, J. L. Praissman, S. Shanmukh, B. J. Vaccaro, S. A. Trauger, E. Kalisiak, J. V. Apon, G. Siuzdak, S. M. Yannone, J. A. Tainer and M. W. W. Adams, Nature, 2010, 466, 779–782 CrossRef CAS PubMed
.
-
I. S. Bertini, A. Sigel and H. Sigel, Eds., Handbook of metalloproteins, Marcel Dekker Inc., New York, 2001 Search PubMed
.
- Y. Lu, N. Yeung, N. Sieracki and N. M. Marshall, Nature, 2009, 460, 855–862 CrossRef CAS PubMed
.
- J. C. Lewis, ACS Catal., 2013, 3, 2954–2975 CrossRef CAS
.
- F. T. Yu, V. M. Cangelosi, M. L. Zastrow, M. Tegoni, J. S. Plegaria, A. G. Tebo, C. S. Mocny, L. Ruckthong, H. Qayyum and V. L. Pecoraro, Chem. Rev., 2014, 114, 3495–3578 CrossRef CAS PubMed
.
- G. Kiss, N. Celebi-Ölcüm, R. Moretti, D. Baker and K. N. Houk, Angew. Chem., Int. Ed., 2013, 52, 5700–5725 CrossRef CAS PubMed
.
- M. Faiella, C. Andreozzi, R. T. M. de Rosales, V. Pavone, O. Maglio, F. Nastri, W. F. DeGrado and A. Lombardi, Nat. Chem. Biol., 2009, 5, 882–884 CrossRef CAS PubMed
.
- N. Yeung, Y. W. Lin, Y. G. Gao, X. Zhao, B. S. Russell, L. Y. Lei, K. D. Miner, H. Robinson and Y. Lu, Nature, 2009, 462, 1079–1082 CrossRef CAS PubMed
.
- J. Podtetenieff, A. Taglieber, E. Bill, E. J. Reijerse and M. T. Reetz, Angew. Chem., Int. Ed, 2010, 49, 5151–5155 CrossRef CAS PubMed
.
- B. S. Der, D. R. Edwards and B. Kuhlman, Biochemistry, 2012, 51, 3933–3940 CrossRef CAS PubMed
.
- M. L. Zastrow, A. F. A. Peacock, J. A. Stuckey and V. L. Pecoraro, Nat. Chem., 2012, 4, 118–123 CrossRef CAS PubMed
.
- T. A. Farid, G. Kodali, L. A. Solomon, B. R. Lichtenstein, M. M. Sheehan, B. A. Fry, C. Bialas, N. M. Ennist, J. A. Siedlecki, Z. Y. Zhao, M. A. Stetz, K. G. Valentine, J. L. R. Anderson, A. J. Wand, B. M. Discher, C. C. Moser and P. L. Dutton, Nat. Chem. Biol., 2013, 9, 826–833 CrossRef CAS PubMed
.
- H. Pedersen, S. Holder, D. P. Sutherlin, U. Schwitter, D. S. King and P. G. Schultz, Proc. Natl. Acad. Sci. U. S. A., 1998, 95, 10523–10528 CrossRef CAS
.
- H. S. Lee, G. Spraggon, P. G. Schultz and F. Wang, J. Am. Chem. Soc., 2009, 131, 2481–2483 CrossRef CAS PubMed
.
- I. Drienovskà, A. Rioz-Martinez, A. Draksharapu and G. Roelfes, Chem. Sci., 2015, 6, 770–776 RSC
.
- Z. P. Wu and D. Hilvert, J. Am. Chem. Soc., 1989, 111, 4513–4514 CrossRef CAS
.
- F. van de Velde, L. Könemann, F. van Rantwijk and R.
A. Sheldon, Chem. Commun., 1998, 1891–1892 RSC
.
- A. Fernández-Gacio, A. Codina, J. Fastrez, O. Riant and P. Soumillion, ChemBioChem, 2006, 7, 1013–1016 CrossRef PubMed
.
- Q. Jing, K. Okrasa and R. Kazlauskas, Top. Organomet. Chem., 2009, 25, 45–61 CrossRef CAS
.
- N. Fujieda, A. Hasegawa, K. Ishihama and S. Itoh, Chem.–Asian J., 2012, 7, 1203–1207 CrossRef CAS PubMed
.
- H. S. Park, S. H. Nam, J. K. Lee, C. N. Yoon, B. Mannervik, S. J. Benkovic and H. S. Kim, Science, 2006, 311, 535–538 CrossRef CAS PubMed
.
- B. Seelig and J. W. Szostak, Nature, 2007, 448, 828–831 CrossRef CAS PubMed
.
- S. D. Khare, Y. Kipnis, P. J. Greisen, R. Takeuchi, Y. Ashani, M. Goldsmith, Y. F. Song, J. L. Gallaher, I. Silman, H. Leader, J. L. Sussman, B. L. Stoddard, D. S. Tawfik and D. Baker, Nat. Chem. Biol., 2012, 8, 294–300 CrossRef CAS PubMed
.
- R. Blomberg, H. Kries, D. M. Pinkas, P. R. E. Mittl, M. G. Grutter, H. K. Privett, S. L. Mayo and D. Hilvert, Nature, 2013, 503, 418–421 CrossRef CAS PubMed
.
- F. Jacob, Science, 1977, 196, 1161–1166 CAS
.
- A. Bar-Even and D. S. Tawfik, Curr. Opin. Biotechnol., 2013, 24, 310–319 CrossRef CAS PubMed
.
- R. A. Jensen, Annu. Rev. Microbiol., 1976, 30, 409–425 CrossRef CAS PubMed
.
- S. D. Copley, Curr. Opin. Chem. Biol., 2003, 7, 265–272 CrossRef CAS PubMed
.
- O. Khersonsky and D. S. Tawfik, Annu. Rev. Biochem., 2010, 79, 471–505 CrossRef CAS PubMed
.
- U. T. Bornscheuer and R. J. Kazlauskas, Angew. Chem., Int. Ed., 2004, 43, 6032–6040 CrossRef CAS PubMed
.
- M. D. Toscano, K. J. Woycechowsky and D. Hilvert, Angew. Chem., Int. Ed., 2007, 46, 3212–3236 CrossRef CAS PubMed
.
- G. Debret, A. Martel and P. Cuniasse, Nucleic Acids Res., 2009, 37, W459–W464 CrossRef CAS PubMed
.
- B. Amrein, M. Schmid, G. Collet, P. Cuniasse, F. Gilardoni, F. P. Seebeck and T. R. Ward, Metallomics, 2012, 4, 379–388 RSC
.
- K. Okrasa and R. J. Kazlauskas, Chem.–Eur. J., 2006, 12, 1587–1596 CrossRef CAS PubMed
.
- E. Miclet, V. Stoven, P. A. M. Michels, F. R. Opperdoes, J. Y. Lallemand and F. Duffieux, J. Biol. Chem., 2001, 276, 34840–34846 CrossRef CAS PubMed
.
- G. R. Hemsworth, B. Henrissat, G. J. Davies and P. H. Walton, Nat. Chem. Biol., 2014, 10, 122–126 CrossRef CAS PubMed
.
- N. N. Ugarova, O. V. Lebedeva and I. V. Berezin, J. Mol. Catal., 1981, 13, 215–225 CrossRef CAS
.
- Y. Kawamura-Konishi, A. Asano, M. Yamazaki, H. Tashiro and H. Suzuki, J. Mol. Catal. B: Enzym., 1998, 4, 181–190 CrossRef CAS
.
- R. Ricoux, R. Dubuc, C. Dupont, J. D. Marechal, A. Martin, M. Sellier and J. P. Mahy, Bioconjugate Chem., 2008, 19, 899–910 CrossRef CAS PubMed
.
- J. Peisach and W. E. Blumberg, Arch. Biochem. Biophys., 1974, 165, 691–708 CrossRef CAS PubMed
.
- E. I. Solomon, D. E. Heppner, E. M. Johnston, J. W. Ginsbach, J. Cirera, M. Qayyum, M. T. Kieber-Emmons, C. H. Kjaergaard, R. G. Hadt and L. Tian, Chem. Rev., 2014, 114, 3659–3853 CrossRef CAS PubMed
.
- N. Hakulinen, L. L. Kiiskinen, K. Kruus, M. Saloheimo, A. Paananen, A. Koivula and J. Rouvinen, Nat. Struct. Biol., 2002, 9, 601–605 CAS
.
- M. Ferraroni, I. Matera, A. Chernykh, M. Kolomytseva, L. A. Golovleva, A. Scozzafava and F. Briganti, J. Inorg. Biochem., 2012, 111, 203–209 CrossRef CAS PubMed
.
- A. Kunishita, H. Ishimaru, S. Nakashima, T. Ogura and S. Itoh, J. Am. Chem. Soc., 2008, 130, 4244–4245 CrossRef CAS PubMed
.
- T. Tano, M. Z. Ertem, S. Yamaguchi, A. Kunishita, H. Sugimoto, N. Fujieda, T. Ogura, C. J. Cramer and S. Itoh, Dalton Trans., 2011, 10326–10336 RSC
.
- L. Que and W. B. Tolman, Nature, 2008, 455, 333–340 CrossRef CAS PubMed
.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental details, additional characterization and catalytic data. See DOI: 10.1039/c5sc01065a |
| This journal is © The Royal Society of Chemistry 2015 |