Open Access Article
Open Access ArticleDNA-based visual majority logic gate with one-vote veto function†
Daoqing
Fan
ab,
Kun
Wang
abc,
Jinbo
Zhu
ab,
Yong
Xia
ab,
Yanchao
Han
ab,
Yaqing
Liu
*ab and
Erkang
Wang
*ab
aState Key Laboratory of Electroanalytical Chemistry, Changchun Institute of Applied Chemistry, Chinese Academy of Sciences, Changchun, Jilin 130022, P. R. China. E-mail: yaqingliu@ciac.ac.cn; ekwang@ciac.ac.cn
bUniversity of Chinese Academy of Sciences, Beijing, 100039, China
cDepartment of Chemistry and Environmental Engineering, Changchun University of Science and Technology, Changchun, China
First published on 7th January 2015
Abstract
A molecular logic gate is a basic element and plays a key role in molecular computing. Herein, we have developed a label-free and enzyme-free three-input visual majority logic gate which is realized for the first time according to DNA hybridization only, without DNA replacement and enzyme catalysis. Furthermore, a one-vote veto function was integrated into the DNA-based majority logic gate, in which one input has priority over other inputs. The developed system can also implement multiple basic and cascade logic gates.
Introduction
Untraditional molecular computing is of scientific interest and has drawn increasing attention across extensive research fields.1–4 Due to its low-cost, feasible synthesis, well-ordered and predictable structure, DNA has been confirmed as an excellent building block for the engineering of biomolecular circuits.5–9 Various basic and advanced DNA-based logic gates have been fabricated by making use of biotechnologies.10–15 Though great achievements have been obtained, the research on DNA computing is still at an early stage. Herein, our investigation focused on developing a DNA-based majority logic gate. It can be used in fault-tolerant computing and can also serve as a versatile building block for constructing reversible logic circuits and advanced logic circuits such as a full adder.16 In 2006, Porod's group reported the first majority logic gate based on magnetic quantum-dot cellular automata technology.17 Up to now, only three molecular majority logic gates have been experimentally validated according to the delicate design. By taking advantage of biocatalytic reactions operating in parallel, an enzyme-based majority gate was realized by Katz's group.18 Based on a toehold-mediated DNA displacement strategy, Liu's group constructed a FAM-labeled majority logic gate.19 By combining four-way junction (Holliday junction) and toehold-mediated DNA displacement, an enzyme-free majority logic gate was successfully realized.20Considering the potential applications, a simple operation, low-cost logic gate would present great advantages. Inspired by the previous achievements, herein we developed a label-free three-input majority logic gate according to DNA hybridization only, without enzyme or (deoxy)ribozyme catalysis and DNA replacement. In Boolean logic, a TRUE decision can be made by a majority logic gate only when more than half of its inputs are present. Otherwise, the majority logic gate makes a FALSE decision.21 In such a majority logic gate, the three inputs have equal priority. However, some members can have priority over the others who can deny the proposal by utilizing the right of one-vote veto, which plays a critical role for the final decision. A well-known example is that the permanent members of the United Nation security council have a veto over any proposal. Herein, a visual three-input majority logic gate with one-vote veto function was conceptually developed for the first time. Interestingly, the decision made by the DNA strands can not only be directly distinguished by the naked eye but can also be read by a fluorescence signal which may be used as a remote output signal.
Results and discussion
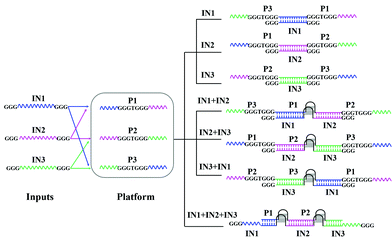
Scheme 1 outlines the operation mechanism of the developed majority logic gate, which is realized based only on hybridization among the input and platform DNA strands. The predictability of Watson–Crick base pairing can simplify the design and implementation of DNA devices.22 Herein, the DNA strands P1, P2 and P3, function as the platform and the DNA strands IN1, IN2 and IN3, function as the three inputs. To realize the majority logic function, a split G-quadruplex (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
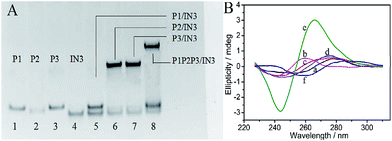
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) strategy is utilized. One G-riched DNA strand is split into three segments, GGGTGGG, GGG and GGG. The segment GGGTGGG is integrated into the middle of each platform DNA strand (see platform in Scheme 1). The two GGG segments are integrated into the 3′ and 5′ terminals of each input, respectively (see inputs in Scheme 1). The signal transduction is modulated by the formed G-quadruplex (G4) among the platform and input DNA strands. To perform the majority logic function, the right part of IN3 can hybridize with the left part of P3, and the left part of IN3 can hybridize with the right part of P2, while IN3 does not hybridize with P1. Hybridization among the DNA strands was validated by native polyacrylamide gel (PAGE) experiments. As shown in Fig. 1A, the DNA bands of P1, P2, P3 and IN3 appear at a similar position from Lane 1 to Lane 4. After adding IN3 to P2 or P3, new bands appear in Lane 6 and Lane 7, respectively, indicating the duplex formation of P2/IN3 and P3/IN3. After adding IN3 into P1, no new band appears in Lane 5, suggesting no hybridization between IN3 and P1. In coexisting P1, P2 and P3, the added IN3 can link P2 and P3 together to produce longer double stranded DNA, leading to a new band in Lane 8 whose position is quite different from the above bands. Another band appears at the position of P1, confirming that IN3 does not hybridize with P1. Similarly, IN1 can hybridize with P1 and P3 while not with P2 (see Fig. S1A in ESI†), and IN2 can hybridize with the platform DNA strands P1 and P2 while it does not hybridize with P3 (see Fig. S1B in ESI†). The sequence of oligonucleotides used in this work is given in Table S1.†
1) strategy is utilized. One G-riched DNA strand is split into three segments, GGGTGGG, GGG and GGG. The segment GGGTGGG is integrated into the middle of each platform DNA strand (see platform in Scheme 1). The two GGG segments are integrated into the 3′ and 5′ terminals of each input, respectively (see inputs in Scheme 1). The signal transduction is modulated by the formed G-quadruplex (G4) among the platform and input DNA strands. To perform the majority logic function, the right part of IN3 can hybridize with the left part of P3, and the left part of IN3 can hybridize with the right part of P2, while IN3 does not hybridize with P1. Hybridization among the DNA strands was validated by native polyacrylamide gel (PAGE) experiments. As shown in Fig. 1A, the DNA bands of P1, P2, P3 and IN3 appear at a similar position from Lane 1 to Lane 4. After adding IN3 to P2 or P3, new bands appear in Lane 6 and Lane 7, respectively, indicating the duplex formation of P2/IN3 and P3/IN3. After adding IN3 into P1, no new band appears in Lane 5, suggesting no hybridization between IN3 and P1. In coexisting P1, P2 and P3, the added IN3 can link P2 and P3 together to produce longer double stranded DNA, leading to a new band in Lane 8 whose position is quite different from the above bands. Another band appears at the position of P1, confirming that IN3 does not hybridize with P1. Similarly, IN1 can hybridize with P1 and P3 while not with P2 (see Fig. S1A in ESI†), and IN2 can hybridize with the platform DNA strands P1 and P2 while it does not hybridize with P3 (see Fig. S1B in ESI†). The sequence of oligonucleotides used in this work is given in Table S1.†
 | ||
| Scheme 1 The operation principle of the designed three-input majority gate based on DNA hybridization. | ||
As shown in Scheme 1, no matter which kind of hybridization, no G4 is formed in the presence of each input. When any two inputs are added, G4 is formed. For the addition of IN1 and IN2, IN1 can hybridize with P1 and P3, and IN2 can hybridize with P1 and P2. Thus, IN1 and IN2 can be linked together through their hybridization with P1. In other cases, the platform strand P2 can link IN2 and IN3 together and P3 can link IN1 and IN3 together. Thus, all three split parts, GGGTGGG in the platform DNA strands and GGG at the terminal of the inputs, are integrated together, resulting in the formation of G4. Then, it is easily understood that more G4 would be produced with the addition of all three inputs. The design was confirmed by circular dichroism (CD) spectrum measurements with the interaction among P1, IN1, and IN2 as a sample. From Fig. 1B, the CD spectra of P1 (curve (a)) and coexisting IN1 and IN2 (curve (b)) are relatively low amplitude, indicating that the DNA strands possess no obvious G4 configuration. After the addition of either IN1 (curve (c)) or IN2 (curve (d)) to P1, there was still no obvious G4 configuration found. However, a significant change is monitored once the two inputs, IN1 and IN2, are simultaneously added to P1 (curve (e)). A positive peak near 265 nm and a negative peak near 243 nm appear, indicating the formation of G4 with a parallel configuration.
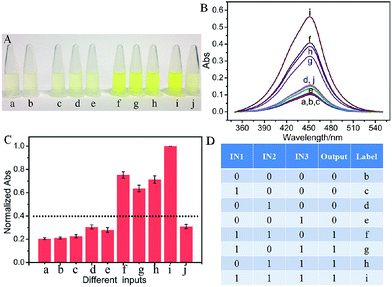
Colorimetric logic gates bring convenience for signal readout. According to the above DNA hybridization, a visual majority logic gate was conceptually realized for the first time. Hemin (Fe(III)-protoporphyrin IX) presents highly effective peroxidase-like properties once bound to G4, forming a G4/hemin complex. The complex can catalyze the oxidation reaction between TMB (3,3′,5,5′-tetramethylbenzidine) and H2O2.23–27 After stopping the reaction using H2SO4, the color of the solution is changed from blue to yellow, which can be monitored by the naked eye and is used as an output signal. As illustrated in Fig. 2A(a), in the absence of any input and platform DNA strands, the solution is a pale yellow color, which is contributed by the oxidation of TMB by H2O2. The solution color stays pale yellow in the presence of all the platform strands, P1, P2 and P3 (Fig. 2A(b)), indicating that the designed platform strands don't catalyze the colorimetric reaction. No obvious color change is monitored after adding any one of the inputs into the platform as shown in Fig. 2A(c)–(e). In the presence of any two inputs, however, the solution color became bright yellow, Fig. 2A(f)–(h), as a result of the high catalytic activity of the formed G4/hemin complex in the colorimetric reaction of TMB in the presence of H2O2. For the coexisting three inputs, more G4/hemin complexes are produced, leading to an even darker yellow output, Fig. 2A(i). Herein, the color shown in Fig. 2A(d) is defined as the controlled threshold, the system presents a TRUE output (brighter yellow) if more than half of the inputs exist. Otherwise, the system presents a FALSE output (paler yellow). The results fulfill the feature of a majority logic gate, whose decision can be conveniently distinguished by the naked eye. To establish the optimal conditions, optimized experiments were explored (see Fig. S2 in ESI†).
To further confirm the results, the absorbance values of the system in the presence of various input combinations are detected and shown in Fig. 2B. A low absorbance signal is detected in the absence or presence of only one input, Fig. 2B(a)–(e), while an enhanced absorbance signal is detected in the presence of any two or all three inputs, Fig. 2B(f)–(i), meeting the feature of a three-input majority logic gate. To make it clear, the inputs “0” and “1” are defined in the absence or presence of respective inputs. The normalized absorbance response at 450 nm is read as an output signal, Fig. 2C. The readout is defined as TRUE (1) when the normalized absorbance is higher than a threshold value of 0.40. Otherwise, it is defined as FALSE (0). The results generate the truth table as shown in Fig. 2D. It is obvious that the system makes a TRUE decision only if more than half of the inputs are introduced, meeting the feature of a majority logic gate. In the absence of the platform, no significant absorbance enhancement is found (see Fig. S3 in ESI†), indicating that the majority logic function is realized according to the interaction among the inputs and platform. The implementation of the logic function is only based on the interaction among the designed input and platform DNA strands and does not require strand displacement hybridization or enzymatic catalysis, which can reduce the time for signal transmission.8,12
In a majority logic gate, the inputs have the same priority as each other. One-vote veto plays an important role in various vote programs, which has never been realized on a molecular level. For the first time, herein, we extended a one-vote veto function to the developed majority logic gate. It has been reported that the cupric ion can unfold the G4 structure as shown in Fig. 1B(f).28 Once Cu2+ is added, the G4/hemin complex is correspondingly deformed, significantly reducing the catalysis activity of hemin in the colorimetric reaction. Here, Cu2+ is introduced as an input, which has priority over the other inputs and can deny the decision made by them. As shown in Fig. 2A(j), the system is a pale yellow color, which is much lighter than that in the presence of all three DNA inputs, validating its priority over the other inputs for the final decision. A low absorbance response further verified what the naked eye monitored, Fig. 2B(j). The operation of the three-input majority logic gate with one-vote veto function presents high repeatability, which can be found from Fig. 2C.
To fulfill the requirements of increased computational complexity, a molecular platform capable of performing multiple logic operations is in high demand.10,29,30 As discussed by Li and coworkers, a three-input majority logic gate can function as a two-input OR gate if any one of the three inputs exists in the initial platform. It also can function as a two-input AND gate if any one of the three inputs is absent in the initial platform.19 As well as those, the developed system here can also perform a cascade logic function. If one of the inputs, for example IN1, is defined as the initial platform and the coexistence of more than two input DNA strands as output “1”, the system would function as an OR logic gate. That is, the output of the OR logic gate presents “1” in the coexistence of IN1 and IN2, or IN1 and IN3, or IN1, IN2 and IN3. In a cascade logic gate, the output of the previous logic gate acts as the input of the next gate. If Cu2+ acts as another input, the system presents high output “1” (define the normalized Abs intensity of 0.4 as the threshold value) only when the output of the OR logic gate is “1”. Otherwise it is “0”, which is the feature of an INHIBIT logic gate. An OR–INHIBIT cascade logic gate is then realized, Fig. 3A–C. Additionally, by taking advantage of the strong coordination between EDTA (ethylenediaminetetraacetic acid) and Cu2+, the added EDTA results in the reforming of the G4/hemin complex and recovery of the absorbance signal, Fig. S4(k).† An IMPLICATION logic gate is then realized with Cu2+ and EDTA as inputs if all of the input and platform DNA strands work as the initial state, Fig. 3D–F. The system presents low output “0” when only Cu2+ is present (define the normalized Abs intensity of 0.4 as the threshold value). Otherwise it is “1”.
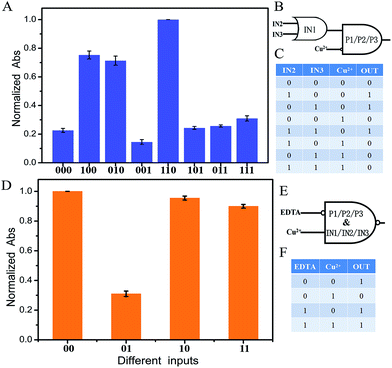 | ||
| Fig. 3 (A) Normalized Abs at 450 nm of the different input modes of the OR–INHIBIT cascade logic circuit with error bars. Abs at 450 nm of curve (i) in Fig. 2B was set as 1. The threshold value was set at 0.4 to judge the output signals. The input statuses of IN2, IN3 and Cu2+ were represented by three binary numbers. (B) Diagram of the OR–INHIBIT cascade logic circuit. (C) Truth table of the OR–INHIBIT cascade logic circuit. (D) Normalized Abs at 450 nm of the different input modes of the IMPLICATION logic circuit with error bars. Abs at 450 nm of curve (i) in Fig. 2B was set as 1. The threshold value was set at 0.4 to judge the output signals. The input statuses of EDTA and Cu2+ were represented by two binary numbers. (E) Diagram of the IMPLICATION logic circuit. (F) Truth table of the IMPLICATION logic circuit. The error bars were obtained according to three independent experimental results. | ||
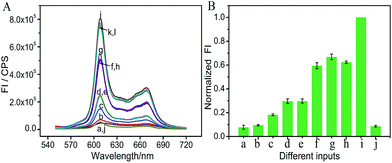
A fluorescence signal as an output is considered to have a multitude of practical applications since the signal enables easy remote reading with low cost.31–33 It is worth mentioning that the colorimetric majority logic gate can also be realized with fluorescence as an output signal, further confirming the operation repeatability. Here, NMM (N-methylmesoporphyrin IX) was selected as the signal reporter, which can bind to the G-quadruplex and produce a significantly enhanced fluorescence signal. As shown in Fig. 4A, a low fluorescence signal was detected in the absence of any input or in the presence of only one of the inputs since no G-quadruplex was formed in these cases. Once any two inputs were added, a complex of G-quadruplex/NMM was then formed, generating significantly enhanced fluorescence of NMM. For the coexisting three inputs and the platform, the system presented the highest fluorescence signal. The normalized fluorescence at 608 nm was read as the output signal, producing the column bar as a function of the various input combinations (Fig. 4B). The output was defined as “1” if the normalized fluorescence is higher than 0.40. Otherwise, it was defined as “0”. This produces the same truth table as given in Fig. 2D, indicating the successful implementation of the majority logic gate with the fluorescence signal as the output. Once Cu2+ was added, the low fluorescent response validates its priority over the other inputs for the final decision, Fig. 4A(j). That is, the system would produce a FALSE output once Cu2+ was added, confirming the one-vote veto function. The developed system with two kinds of optical signal as outputs would extend its potential application field.
Conclusions
In conclusion, a DNA-based majority logic gate with one-vote veto function has been successfully realized in a proof-of-principle study. The developed logic gate presents several advantages. First, one-vote veto function is integrated into the majority logic gate on the molecular level for the first time. In the system, one input has priority and can deny the decision made by the other inputs. Second, the logic implementation is simply based on DNA hybridization, reducing the reaction time and simplifying the design. The readout can be conveniently viewed by the naked eye for the first time, which can also be monitored with sensitive fluorescence as the output signal. Third, the developed logic gate is label-free and enzyme-free and realized with short oligonucleotides, reducing the cost. Last but not least, the developed system can also perform multiple basic and cascade logic gates. However, we should be aware that there is still a long road ahead to practical application. The investigation provides a conceptual prototype for constructing multicomponent devices on a single molecular platform.Experimental section
Materials
The sequence of oligonucleotides used in this work is given in Table S1.† The oligonucleotide was dissolved in water as a stock solution and quantified by UV-vis absorption spectroscopy with the following extinction coefficients (ε 260 nm, M−1 cm−1): A = 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400, G = 11
400, G = 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500, C = 7400, T = 8700. The stock solution of NMM (50 μM) was prepared in dimethyl sulfoxide (DMSO) and stored in darkness at −20 °C. The water used in the experiments was purified through a Millipore system. Other chemicals were reagent grade and were used without further purification.
500, C = 7400, T = 8700. The stock solution of NMM (50 μM) was prepared in dimethyl sulfoxide (DMSO) and stored in darkness at −20 °C. The water used in the experiments was purified through a Millipore system. Other chemicals were reagent grade and were used without further purification.
Native polyacrylamide gel electrophoresis (PAGE)
10 μM DNA stock solution was diluted to a concentration of 5 μM by an equal volume of 2× HEPES (4-(2-hydroxyethyl)piperazine-1-ethanesulfonic acid) buffer (50 mM HEPES, 40 mM KCl, 400 mM NaCl, pH 7.4). The solution was then heated at 90 °C for 10 min and slowly cooled down to room temperature. After that, the desired volume of the platform and input DNA solution was mixed and a suitable volume of 1× HEPES buffer (25 mM HEPES, 20 mM KCl, 200 mM NaCl, pH 7.4) was added into the mixture to give a final volume of 50 μL. After 30 min incubation, the DNA solution was analyzed in 15% native polyacrylamide gel. Electrophoresis was conducted in 1× TBE (17.80 mM Tris, 17.80 mM boric acid, 2 mM EDTA, pH 8.0) at a constant voltage of 120 V for 1.5 h. The gels were scanned by a UV transilluminator after staining with Gel-dye.Circular dichroism measurements
CD spectra were measured on a JASCO J-820 spectropolarimeter (Tokyo, Japan) at room temperature. Spectra were recorded from 220 to 320 nm in 1 mm path length cuvettes and averaged from three scans.Operation of the developed majority logic gate
10 μM DNA stock solution was diluted to a concentration of 5 μM by an equal volume of 2× HEPES buffer (50 mM HEPES, 40 mM KCl, 400 mM NaCl, pH 7.4), then the solution was heated at 90 °C for 10 min and slowly cooled down to room temperature. After that, the desired volume of platform and input DNA solution was mixed and a suitable volume of 1× HEPES buffer (25 mM HEPES, 20 mM KCl, 200 mM NaCl, pH 7.4) was added into the mixture to give a final volume of 100 μL. For the cupric ion reaction, the desired volume of CuSO4 was added into the mixed DNA solution. Then an equal volume of 2× HEPES buffer (50 mM HEPES, 40 mM KCl, 400 mM NaCl, 0.1% (w/v) Triton X-100, 2% (v/v) DMSO, pH 7.4) was added and the mixture was incubated for 40 min. Finally, an equal volume of the same concentration of hemin dissolved in the HEPES buffer (25 mM HEPES, 20 mM KCl, 200 mM NaCl, 0.05% (w/v) Triton X-100, 1% (v/v) DMSO, pH 7.4) was added to the solution. The mixture was incubated at room temperature for 1 h. The final volume of the sample is 500 μL with a final concentration of 300 nM of each platform DNA, 350 nM IN1, 300 nM IN2, 300 nM IN3, 1 μM NMM, 10 μM Cu2+.Colorimetric reaction
In the oxidation reaction, 10 μL of the final DNAzyme was mixed with 5 μL 0.5% (w/v) TMB, 5 μL 30% (w/v) H2O2, and 480 μL MES-Ac substrate buffer (25 mM MES-Ac, 20 mM KAc, pH 4.5), where MES is 2-(4-morpholino)ethanesulfonic acid. An equal volume of 2 M H2SO4 was added into the solution after 4 min to stop the reaction. UV-vis absorbance analysis was performed on a Cary 50 Scan UV/Vis/NIR Spectrophotometer (Varian, USA).Fluorescence spectra measurements
The fluorescence emission spectra of different samples were collected on a Fluoromax-4 Spectrofluorometer (HORIBA Jobin Yvon, Inc., NJ, USA) at room temperature from 550 to 750 nm with an excitation wavelength of 399 nm. Slit widths for the excitation and emission were 3 nm and 5 nm, respectively.Acknowledgements
This work was supported by the National Natural Science Foundation of China (no. 21105095 and 211900040), the State Key Instrument Developing Special Project of Ministry of Science and Technology of China (2012YQ170003), the Instrument Developing Project of the Chinese Academy of Sciences (YZ201203) and the Natural Science Foundation of Jilin Province, China (no. 20130101117JC).Notes and references
- P. A. de Silva, N. H. Q. Gunaratne and C. P. McCoy, Nature, 1993, 364, 42–44 CrossRef
.
- L. Adleman, Science, 1994, 266, 1021–1024 CAS
.
- Z. Xie, L. Wroblewska, L. Prochazka, R. Weiss and Y. Benenson, Science, 2011, 333, 1307–1311 CrossRef CAS PubMed
.
- D. C. Magri, G. J. Brown, G. D. McClean and A. P. de Silva, J. Am. Chem. Soc., 2006, 128, 4950–4951 CrossRef CAS PubMed
.
- A. J. Genot, J. Bath and A. J. Turberfield, J. Am. Chem. Soc., 2011, 133, 20080–20083 CrossRef CAS PubMed
.
- D. Han, Z. Zhu, C. Wu, L. Peng, L. Zhou, B. Gulbakan, G. Zhu, K. R. Williams and W. Tan, J. Am. Chem. Soc., 2012, 134, 20797–20804 CrossRef CAS PubMed
.
- H. Li, W. Hong, S. Dong, Y. Liu and E. Wang, ACS Nano, 2014, 8, 2796–2803 CrossRef CAS PubMed
.
- B. Shlyahovsky, Y. Li, O. Lioubashevski, J. Elbaz and I. Willner, ACS Nano, 2009, 3, 1831–1843 CrossRef CAS PubMed
.
- W. Hong, Y. Du, T. Wang, J. Liu, Y. Liu, J. Wang and E. Wang, Chem.–Eur. J., 2012, 18, 14939–14942 CrossRef CAS PubMed
.
- S. Xu, H. Li, Y. Miao, Y. Liu and E. Wang, NPG Asia Mater., 2013, 5, e76 CrossRef CAS
.
- R. Orbach, F. Remacle, R. D. Levine and I. Willner, Chem. Sci., 2014, 5, 1074–1081 RSC
.
- A. Lake, S. Shang and D. M. Kolpashchikov, Angew. Chem., Int. Ed., 2010, 49, 4459–4462 CrossRef CAS PubMed
.
- S. Bi, Y. Yan, S. Hao and S. Zhang, Angew. Chem., Int. Ed., 2010, 49, 4438–4442 CrossRef CAS PubMed
.
- T. Li, D. Ackermann, A. M. Hall and M. Famulok, J. Am. Chem. Soc., 2012, 134, 3508–3516 CrossRef CAS PubMed
.
- R. Orbach, S. Lilienthal, M. Klein, R. D. Levine, F. Remacle and I. Willner, Chem. Sci., 2015 10.1039/c4sc02930e
.
- X. Yang, L. Cai and Q. Kang, J. Comput. Theor. Nanosci., 2012, 9, 621–625 CrossRef CAS PubMed
.
- A. Imre, G. Csaba, L. Ji, A. Orlov, G. H. Bernstein and W. Porod, Science, 2006, 311, 205–208 CrossRef CAS PubMed
.
- S. Mailloux, N. Guz, A. Zakharchenko, S. Minko and E. Katz, J. Phys. Chem. B, 2014, 118, 6775–6784 CrossRef CAS PubMed
.
- W. Li, Y. Yang, H. Yan and Y. Liu, Nano Lett., 2013, 13, 2980–2988 CrossRef CAS PubMed
.
- J. Zhu, L. Zhang, S. Dong and E. Wang, ACS Nano, 2013, 7, 10211–10217 CrossRef CAS PubMed
.
- M. Goldmann and M. Karpinski, SIAM J. Comput., 1998, 27, 230–246 CrossRef
.
- G. Seelig, D. Soloveichik, D. Y. Zhang and E. Winfree, Science, 2006, 314, 1585–1588 CrossRef CAS PubMed
.
- Y. Xiao, V. Pavlov, T. Niazov, A. Dishon, M. Kotler and I. Willner, J. Am. Chem. Soc., 2004, 126, 7430–7431 CrossRef CAS PubMed
.
- Y. Li and D. Sen, Nat. Struct. Mol. Biol., 1996, 3, 743–747 CAS
.
- T. Li, L. Shi, E. Wang and S. Dong, Chem.–Eur. J., 2009, 15, 3347–3350 CrossRef CAS PubMed
.
- J. Zhu, T. Li, L. Zhang, S. Dong and E. Wang, Biomaterials, 2011, 32, 7318–7324 CrossRef CAS PubMed
.
- E. Golub, R. Freeman and I. Willner, Angew. Chem., Int. Ed., 2011, 50, 11710–11714 CrossRef CAS
.
- D. Monchaud, P. Yang, L. Lacroix, M. P. Teulade-Fichou and J. L. Mergny, Angew. Chem., Int. Ed., 2008, 47, 4858–4861 CrossRef CAS PubMed
.
- A. Okamoto, K. Tanaka and I. Saito, J. Am. Chem. Soc., 2004, 126, 9458–9463 CrossRef CAS PubMed
.
- R. Orbach, F. Wang, O. Lioubashevski, R. D. Levine, F. Remacle and I. Willner, Chem. Sci., 2014, 5, 3381–3387 RSC
.
- W. A. L. van Otterlo, E. L. Ngidi and C. B. de Koning, Tetrahedron Lett., 2003, 44, 6483–6486 CrossRef CAS
.
- Q.-C. Wang, D.-H. Qu, J. Ren, K. Chen and H. Tian, Angew. Chem., Int. Ed., 2004, 43, 2661–2665 CrossRef CAS
.
- D. H. Qu, Q. C. Wang, X. Ma and H. Tian, Chem.–Eur. J., 2005, 11, 5929–5937 CrossRef CAS PubMed
.
Footnote |
| † Electronic supplementary information (ESI) available: Table S1 and Fig. S1–S4. See DOI: 10.1039/c4sc03495c |
| This journal is © The Royal Society of Chemistry 2015 |