Microbial transformation of diosgenin by Cunninghamella blakesleana AS 3.970 and potential inhibitory effects on P-glycoprotein of its metabolites†
Min Xu‡
a,
Xiao-Kui Huo‡a,
Xiang-Ge Tiana,
Pei-Pei Dongb,
Chao Wang*acd,
Shan-Shan Huanga,
Bao-Jing Zhanga,
Hou-Li Zhanga,
Sa Denga and
Xiao-Chi Ma*abcd
aCollege of Pharmacy, Dalian Medical University, Dalian 116044, P. R. China. E-mail: wach_edu@sina.com; maxc1978@163.com; Fax: +86-411-86110419; Tel: +86-411-86110419
bAcademy of Integrative Medicine, Dalian Medical University, Dalian 116044, P. R. China
cState Key Laboratory of Bioactive Substance and Function of Natural Medicines, Institute of MateriaMedica, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing 100050, P. R. China
dLiaoning Province Key Laboratory of Natural Products for Neurodegenerative Diseases, Dalian Medical University, Dalian, China
First published on 10th September 2015
Abstract
Microbial transformation of diosgenin ((25R)-spirost-5-en-3β-ol) using Cunninghamella blakesleana AS 3.970, afforded eleven polyhydroxylated derivatives, including seven previously unreported steroids, such as 25(R)-spirost-5-en-3β,7α,12β-triol (1), 25(R)-spirost-5-en-3β,7α,12β,15α,21-pentaol (3), 25(R)-spirost-5-en-3β,7α,12β,18-tetraol (4), 25(R)-spirost-5-en-3β,7α,12β,15α-tetraol (5), 25(R)-spirost-5-en-3β,7α,11α,21-tetraol (6), 25(R)-spirost-5-en-3β,7β,15α,21-tetraol (8), and 25(R)-spirost-5-en-3β,7β,12β,18-tetraol (10). The structures of metabolites 1–11 were elucidated by 1D-, 2D-NMR as well as HRESIMS techniques. Additionally, the biotransformation time-course of diosgenin by C. blakesleana AS 3.970 was presented. And the transformation pathway was also proposed on the basis of structural analyses and biotransformation time-courses. The P-glycoprotein (P-gp) inhibitory effects of these metabolites 1–11 were evaluated in a adriamycin resistant human breast adenocarcinoma cell line (MCF-7/ADR) at 20 μM. And compounds 4 and 6 could increase the accumulation of adriamycin in MCF-7/ADR cells by approximately four times that of the control group, which suggested the significant potential P-glycoprotein inhibitory activities of 4 and 6. In silico docking analysis suggested that compound 4 had a similar P-gp recognition mechanism with verapamil (a classical inhibitor).
Introduction
Biotransformation is mainly an enzymatic biochemical reaction to modify chemical substances by vegetal cellular or organs, animal cellular, microorganisms and organelles, and isolated enzymes.1 It is thought of as an efficient and important tool to diversify structures, especially for complex natural products, which are difficult to modify and prepare by chemical approaches. Compared to chemical synthesis, microbial transformations are usually used as an alternative method to modify complex structures with multiple enzymatic reactions, such as hydroxylation, oxidation, reduction, hydrolysis, epimerization, isomerization, and rearrangement.2,3 Microbial transformations are generally carried out under mild conditions, following the principles of green chemistry, and have the advantage of novel reactions, high chemo-, regio- and stereo-selectivity, less by-products, and are easy to operate under mild conditions.2–4Diosgenin [(25R)-spirost-5-en-3β-ol] is a steroidal sapogenin possessing seven rings with 27 carbons existing in a variety of plants species, especially Dioscerea plants. Traditionally, diosgenin was obtained from glycosidal steroidal saponins with high production. It is an important resource for the steroid drugs in the pharmaceutical industry.5 In recent years, several pharmacological attributes of diosgenin have been reported, such as apoptosis induction in erythroleukemia cell,6,7 inhibition of the migration of human breast cancer cells,8 inhibition of TNF-α-induced adhesion molecule expression,9 anti-inflammatory,10 and anti-hepatitis C virus.11 The structural modification studies of diosgenin have been conducted by microorganisms to provide some new derivatives, which improved the chemical diversity and gave some bioactive substances.12–17 In the previous investigations, we reported the biotransformation of some complex natural products and obtained a lot of transformed products with the new structures and potential bioactivities.18–21 Therefore, the biotransformation of diosgenin was carried out to provide some derivatives which could be utilized for screening for the new activities or better activity/side effects profile.
Results and discussion
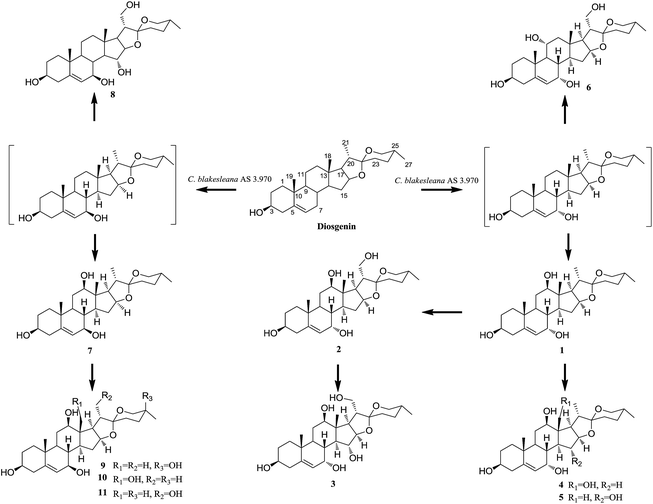
Identification of transformed products
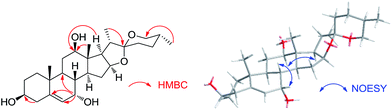
After screening 10 strains of filamentous fungi, Cunninghamella blakesleana AS 3.970 was chosen as a potent organism to be used for the biotransformation of diosgenin. The cultures of C. blakesleana AS 3.970 incubated with diosgenin were extracted with EtOAc and the EtOAc extract was collected and subjected to silica gel column, RP C18 to afford eleven metabolites 1–11. The structures of eleven metabolites were characterized by ESIMS, HRESIMS, 1D-, and 2D-NMR data (Fig. 1). Metabolites 2, 7 and 11 were characterized as (25R)-spirost-5-en-3β,7α,12β,21-tetraol,15 (25R)-spirost-5-en-3β,7β,12β-triol15 and 25(R)-spirost-5-en-3β,7β,12β,21-tetrol15 respectively, which could be transformed derivatives of diosgenin catalyzed by white-rot fungus Coriolus versicolor. Metabolite 9 was previously identified as (25S)-spirost-5-en-3β,7β,12β,25β-tetrol,22 a transformed product of diosgenin by Absidia coerulea.Metabolite 1, a white amorphous powder, gave the molecular formula C27H42O5, as deduced by its HRESIMS (m/z 447.3091, calcd for 447.3110 [M + H]+), suggesting a dihydroxylated derivative of diosgenin. Compared with diosgenin, two extra oxygenated methines (δC 65.7, 80.0) were observed in the 13C NMR spectrum. The corresponding proton signals were also observed in the 1H NMR spectrum at δH 3.27 (m), 3.80 (m). In the HMBC spectrum, the long range correlations between δH 3.80 and C-5, C-6, C-9 suggested that one hydroxyl group was located at C-7. The 12-OH moiety was established by the HMBC correlations of H-18 (δH 0.78)/δC 80.0, H-17 (δH 1.92)/δC 80.0 (Fig. 2). Therefore, the planar structure of metabolite 1 was same to the known compound 25(R)-spirost-5-en-3β,7β,12β-triol.15
However, 13C NMR data comparisons between two metabolites displayed the upfield shift (Δδ 7.8) of C-7 (δC 73.5 to δC 65.7) in metabolite 1, which indicated α orientation of 7-OH according to those 7α-oriented metabolites.15 7α-OH was further confirmed by the NOE effects of H-7 (δH 3.80)/H-8 (δH 1.57) in the NOESY spectrum (Fig. 2). Meantime, in the NOESY experiment, the correlation between H-12 (δH 3.27) and H-17 (δH 1.92) determined the 12β-OH (Fig. 2). Thus, the structure of 1 was elucidated as 25(R)-spirost-5-en-3β,7α,12β-triol. The 1D-NMR data (Table 1 and 2) were assigned accurately by 2D-NMR spectra.
| No. | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1.85 m | 1.85 m | 1.84 m | 1.83 m | 1.87 m | 1.60 m | 1.83 m | 1.88 m | 1.85 m | 1.84 m | 1.82 m |
| 1.17 m | 1.17 m | 1.14 m | 1.66 m | 1.13 m | 1.25 m | 1.06 m | 1.08 m | 1.08 m | 1.08 m | 1.06 m | |
| 2 | 2.17 m | 1.80 m | 1.80 m | 1.81 m | 1.82 m | 2.16 m | 1.81 m | 1.88 m | 1.81 m | 1.84 m | 1.80 m |
| 1.82 m | 1.68 m | 1.69 m | 1.15 m | 1.51 m | 1.29 m | 1.65 m | 1.69 m | 1.51 m | 1.64 m | 1.68 m | |
| 3 | 3.48 m | 3.48 m | 3.46 m | 3.46 m | 3.46 m | 3.49 m | 3.43 m | 3.43 m | 3.42 m | 3.42 m | 3.40 m |
| 4 | 2.31 m | 2.26 m | 2.34 m | 2.31 m | 2.35 m | 2.30 m | 2.29 m | 2.30 m | 2.27 m | 2.29 m | 2.29 m |
| 2.26 m | 2.27 m | 2.24 m | 2.26 m | 2.22 m | 2.24 m | 2.22 m | 2.23 m | 2.22 m | |||
| 6 | 5.55 dd (5.0, 11.5) | 5.56 dd (5.0, 1.5) | 5.52 dd (5.5, 2.0) | 5.56 m | 5.51 m | 5.60 d (5.5) | 5.26 brs | 5.30 m | 5.26 m | 5.28 m | 5.27 brs |
| 7 | 3.80 m | 3.81 brs | 4.19 m | 3.82 m | 4.19 m | 3.78 m | 3.69 dt (8.5, 2.0) | 3.85 dt (8.0, 2.5) | 3.69 dt (8.0, 2.0) | 3.70 d (8.0) | 3.71 m |
| 8 | 1.57 m | 1.62 m | 1.87 m | 1.59 m | 1.79 m | 1.60 m | 1.62 m | 1.74 m | 1.55 m | 1.55 m | 1.63 m |
| 9 | 1.47 m | 1.46 m | 1.41 m | 1.46 m | 1.41 m | 1.40 m | 1.22 m | 1.11 m | 1.19 m | 1.18 m | 1.22 m |
| 11 | 1.69 m | 1.66 m | 1.64 m | 1.67 m | 1.71 m | 4.00 m | 1.69 m | 1.59 m | 1.70 m | 1.69 m | 1.66 m |
| 1.47 m | 1.43 m | 1.33 m | 1.15 m | 1.41 m | 1.60 m | 1.48 m | 1.48 m | 1.15 m | |||
| 12 | 3.27 m | 3.43 m | 3.48 dd (11.5, 5.0) | 3.44 m | 3.32 m | 1.67 m | 3.26 dd (11.5, 5.0) | 1.75 m | 3.26 m | 3.43 m | 3.41 m |
| 1.19 m | |||||||||||
| 14 | 1.58 m | 1.62 m | 1.76 dd (22.5, 11.0) | 1.59 m | 1.68 m | 1.19 m | 1.23 m | 1.31 m | 1.24 m | 1.26 m | 1.29 m |
| 15 | 1.51 m | 2.22 m | 4.13 dd (10.5, 3.5) | 2.38 m | 4.08 dd (11.0, 3.5) | 1.76 m | 2.32 m | 3.99 dd (11.0, 4.0) | 2.31 m | 2.48 m | 2.37 m |
| 1.39 m | 1.51 m | 1.51 m | 1.53 m | 1.70 m | 1.62 m | 1.78 m | 1.64 m | ||||
| 16 | 4.43 m | 4.56 m | 4.30 dd (9.0, 3.5) | 4.62 m | 4.18 m | 4.51 m | 4.40 m | 4.28 dd (8.5, 4.0) | 4.23 dd (14.5, 7.5) | 4.59 m | 4.53 m |
| 17 | 1.92 m | 2.16 m | 2.36 m | 2.15 d (6.0) | 2.09 dd (9.0, 6.5) | 1.97 dd (8.0, 5.5) | 1.89 m | 2.09 m | 1.90 m | 2.09 d (6.0) | 2.11 m |
| 18 | 0.78 s | 0.83 s | 0.85 s | 3.85 d (12.0) | 0.80 s | 0.83 s | 0.79 s | 0.86 s | 0.80 s | 3.83 d (11.5) | 0.83 s |
| 3.63 d (12.0) | 3.61 d (11.5) | ||||||||||
| 19 | 1.037 s | 1.04 s | 1.03 s | 0.98 s | 1.03 s | 1.15 s | 1.10 s | 1.10 s | 1.10 s | 1.05 s | 1.10 s |
| 20 | 1.88 m | 2.12 m | 2.15 m | 2.49 m | 1.91 m | 2.05 m | 1.88 m | 2.07 m | 1.91 m | 2.49 m | 2.12 m |
| 21 | 1.035 d (6.5) | 3.70 dd (10.0, 4.0) | 3.68 dd (10.0, 4.0) | 0.91 d (5.5) | 1.02 d (6.0) | 3.68 dd (11.0, 6.0) | 1.03 d (6.5) | 3.68 dd (11.0, 6.0) | 1.09 d (6.0) | 0.911 d (6.5) | 3.69 m |
| 3.63 t (10.0) | 3.62 t (10.0) | 3.54 dd (11.0, 7.0) | 3.52 dd (11.0, 7.0) | 3.62 m | |||||||
| 23 | 1.77 m | 1.80 m | 1.83 m | 1.64 m | 1.71 m | 1.88 m | 1.65 m | 1.82 m | 2.08 m | 1.60 m | 1.64 m |
| 1.45 m | 1.41 m | 1.51 m | 1.60 m | 1.59 m | 1.66 m | 1.50 m | 1.51 m | 1.41 m | 1.50 m | 1.49 m | |
| 24 | 1.63 m | 1.65 m | 1.71 m | 1.60 m | 1.64 m | 1.62 m | 1.59 m | 1.63 m | 1.83 m | 1.59 m | 1.59 m |
| 1.43 m | 1.40 m | 1.45 m | 1.13 m | 1.45 m | 1.42 m | 1.45 m | 1.48 m | 1.58 m | 1.13 m | 1.41 m | |
| 25 | 1.71 m | 1.60 m | 1.61 m | 1.56 m | 1.62 m | 1.64 m | 1.58 m | 1.66 m | 1.56 m | 1.66 m | |
| 26 | 3.45 m | 3.40 m | 3.41 m | 3.40 m | 3.45 m | 3.45 m | 3.43 m | 3.46 m | 3.64 d (11.5) | 3.41 m | 3.44 m |
| 3.34 m | 3.32 m | 3.37 t (11.0) | 3.34 m | 3.40 m | 3.34 m | 3.34 m | 3.41 m | 3.29 d (11.5) | 3.34 m | 3.30 m | |
| 27 | 0.80 d (6.5) | 0.78 d (6.5) | 0.78 d (6.0) | 0.90 d (5.5) | 0.79 d (8.0) | 0.78 d (6.0) | 0.80 d (7.0) | 0.79 d (6.5) | 1.07 s | 0.90 d (6.5) | 0.78 d (6.5) |
| No. | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 38.1 | 38.1 | 38.28 | 38.2 | 38.3 | 39.7 | 38.2 | 38.2 | 38.2 | 38.3 | 38.3 |
| 2 | 32.10 | 33.2 | 33.3 | 32.1 | 32.1 | 32.4 | 32.5 | 33.7 | 32.2 | 32.3 | 33.3 |
| 3 | 72.0 | 72.0 | 71.9 | 72.0 | 71.9 | 72.1 | 72.1 | 72.1 | 72.1 | 72.1 | 72.1 |
| 4 | 42.8 | 42.8 | 42.8 | 42.8 | 42.8 | 43.5 | 42.5 | 42.5 | 42.5 | 42.5 | 42.5 |
| 5 | 146.6 | 146.6 | 147.1 | 146.5 | 147.1 | 147.3 | 144.0 | 144.6 | 144.0 | 144.0 | 144.1 |
| 6 | 124.9 | 124.9 | 123.8 | 125.0 | 123.8 | 124.6 | 127.5 | 125.8 | 127.5 | 127.6 | 127.5 |
| 7 | 65.7 | 65.6 | 65.8 | 65.6 | 65.8 | 65.7 | 73.5 | 73.5 | 73.5 | 73.6 | 73.5 |
| 8 | 37.6 | 37.8 | 38.25 | 38.6 | 37.8 | 38.9 | 40.0 | 41.6 | 40.0 | 40.2 | 40.3 |
| 9 | 43.3 | 43.1 | 43.7 | 43.1 | 43.8 | 49.7 | 49.3 | 48.6 | 49.3 | 48.9 | 49.3 |
| 10 | 38.7 | 38.6 | 38.0 | 37.9 | 38.2 | 40.1 | 37.8 | 38.0 | 37.8 | 37.8 | 37.9 |
| 11 | 31.2 | 29.7 | 29.9 | 31.1 | 31.4 | 69.2 | 31.5 | 21.4 | 31.5 | 31.4 | 29.8 |
| 12 | 80.0 | 80.3 | 80.3 | 78.3 | 79.9 | 50.5 | 80.0 | 40.5 | 80.0 | 78.3 | 80.4 |
| 13 | 46.6 | 46.6 | 46.5 | 51.3 | 46.4 | 41.8 | 47.3 | 42.0 | 47.3 | 51.8 | 47.3 |
| 14 | 49.3 | 49.6 | 54.8 | 47.9 | 54.7 | 50.8 | 55.8 | 61.8 | 55.8 | 54.1 | 55.8 |
| 15 | 32.08 | 32.2 | 78.7 | 37.3 | 78.6 | 32.3 | 34.7 | 79.5 | 34.7 | 39.7 | 34.8 |
| 16 | 82.2 | 83.1 | 92.0 | 81.9 | 91.0 | 83.1 | 82.5 | 90.1 | 82.8 | 82.2 | 83.5 |
| 17 | 63.1 | 61.1 | 59.0 | 56.7 | 61.1 | 60.1 | 62.7 | 57.9 | 62.8 | 56.3 | 60.6 |
| 18 | 10.9 | 11.0 | 12.0 | 60.2 | 12.0 | 17.4 | 11.0 | 17.7 | 11.0 | 60.1 | 11.1 |
| 19 | 18.6 | 18.6 | 18.8 | 18.5 | 18.7 | 18.3 | 19.4 | 19.6 | 19.4 | 19.4 | 19.5 |
| 20 | 43.7 | 51.0 | 51.1 | 38.6 | 43.9 | 51.0 | 43.8 | 51.0 | 43.7 | 38.7 | 51.1 |
| 21 | 14.0 | 64.2 | 64.0 | 16.9 | 13.9 | 63.0 | 14.0 | 62.9 | 14.1 | 14.6 | 64.3 |
| 22 | 110.8 | 110.2 | 109.9 | 112.1 | 110.4 | 110.2 | 110.7 | 110.0 | 110.7 | 111.8 | 110.2 |
| 23 | 31.2 | 32.1 | 32.1 | 32.2 | 32.5 | 33.8 | 32.2 | 32.3 | 28.2 | 32.3 | 32.3 |
| 24 | 29.9 | 29.8 | 29.7 | 28.6 | 30.0 | 29.8 | 29.9 | 29.9 | 33.7 | 28.7 | 29.9 |
| 25 | 31.5 | 31.3 | 31.4 | 37.2 | 31.2 | 31.3 | 31.5 | 31.3 | 67.4 | 37.2 | 31.4 |
| 26 | 67.9 | 67.8 | 67.9 | 68.5 | 67.9 | 67.9 | 67.9 | 67.9 | 69.8 | 68.5 | 67.8 |
| 27 | 17.5 | 17.4 | 17.5 | 14.6 | 17.5 | 17.5 | 17.5 | 17.5 | 26.4 | 16.9 | 17.5 |
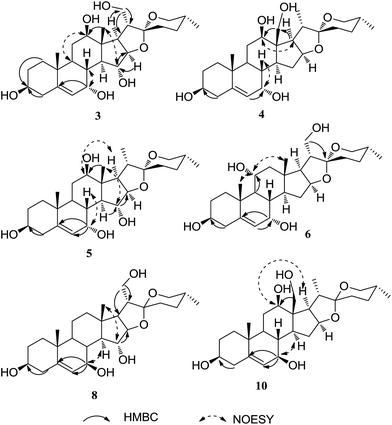
Metabolite 3 had the molecular formula C27H42O7 as determined by its HRESIMS at m/z 501.2823 [M + Na]+ (calcd for 501.2828). Analyses of the spectroscopic data suggested that 3 was a tetrahydroxylated derivative of diosgenin. Three extra oxygenated methines (δC 65.8, 78.7, 80.3) and one more oxygenated methene (δC 64.0) were deduced by the carbon signals observed in the 13C NMR spectrum. NMR data analyses suggested that metabolite 3 was similar to 2 (25R)-spirost-5-en-3β,7α,12β,21-tetraol, except for one extra hydroxyl. On the basis of HMBC experiment, 3,7,12,21-tetrahydroxyls were determined. Furthermore, in the HMBC spectrum, the long range correlations of H-14 (δH 1.57)/C-15 (δC 78.7), H-16 (δH 1.57)/C-15 (δC 78.7) established the 15-OH (Fig. 3). The relative configurations of 7,12,15-trihydroxyls were determined by the NOE correlations of H-7/H-8, H-12/H-9, H-15/H-18 in the NOESY spectrum. Therefore, 3 was determined as a high oxygenated derivative of diosgenin with four extra hydroxyl groups, named 25(R)-spirost-5-en-3β,7α,12β,15α,21-pentaol.
Analyses of the spectroscopic data of metabolite 4 indicated that it was a trihydroxylated derivative of diosgenin, with two extra oxygenated methines and one more oxygenated methene, which was similar to metabolite 2, except for the oxygenated methene. The 1H NMR spectrum displayed two methyl signals with doublets [δH 0.91 d (J = 5.5 Hz), 0.90 d (J = 5.5 Hz)], corresponding to 21-CH3, 27-CH3, respectively. The singlet of 19-CH3 was also deduced from the 1H NMR spectrum. Therefore, 4 was supposed to be 7,12,18-trihydroxylated diosgenin. On the basis of the HMBC spectrum, the planar structure of 4 was determined. The relative configuration of 4 was established by the NOE effects of H-7/H-8, H-12/H-17 (Fig. 3). Thus, metabolite 4 was elucidated as 25(R)-spirost-5-en-3β,7α,12β,18-tetraol. It is worth noting that 18-hydroxylated derivative of diosgenin was previously not reported in the modification of diosgenin.
Metabolites 5 and 6 were both trihydroxylated derivatives of diosgenin, deduced by the HRESIMS and NMR data. In the structure of 5, three hydroxyls were assigned at C-7, C-12, and C-15, respectively, on the basis of the observed long range correlations in its HMBC spectrum. For the structure of 6,7-OH, 11-OH, and 21-OH were established by the HMBC experiment. Furthermore, 7α-oriented hydroxyl groups were deduced for metabolites 5 and 6 by the carbon chemical shifts of C-7 (δC 65.8, δC 65.7), which were also supported by the NOESY experiments. The relative configurations of 5 and 6 were accurately determined by the NOESY spectra. On the basis of the spectroscopic data analyses, 5 and 6 were determined as 25(R)-spirost-5-en-3β,7α,12β,15α-tetraol, 25(R)-spirost-5-en-3β,7α,11α,21-tetraol, respectively.Metabolite 8 was obtained as an amorphous powder, with the molecular formula C27H42O6 established by HRESIMS (485.2857, calcd for 485.2879). Compared with diosgenin, the 13C NMR spectrum suggested the presences of two more oxygenated methines (δC 73.5, 79.5) and one extra oxygenated methene (δC 62.9). 1H NMR spectrum also displayed the corresponding protons at δH 3.85 dt (J = 8.0, 2.5 Hz), 3.99 dd (J = 11.0, 4.0 Hz), 3.68 dd (J = 11.0, 6.0 Hz), 3.52 dd (J = 11.0, 7.0 Hz). In the HMBC spectrum, the long range correlations of δH 3.85/C-5, C-6, C-14, δH 1.31/δC 79.5, δH 3.99/C-16, H-20/δC 62.9, and δH 3.52/C-22 assigned the 7-OH, 15-OH, and 21-OH, respectively (Fig. 3). The chemical shift of C-7 at δC 73.5 suggested the β orientation of 7-OH. Furthermore, the relative configuration of 8 was determined by the NOESY experiment. Therefore, metabolite 8 was elucidated as 25(R)-spirost-5-en-3β,7β,15α,21-tetraol.
Metabolite 10 was deduced as a trihydroxylated derivative of diosgenin on the basis of its HRESIMS and NMR data. Comparison of the spectroscopic data between 10 and 4 indicated that they have same planar structure. 2D-NMR data analysis also established the 7,12,18-trihydroxyl diosgenin planar structure for 10. Compared with the C-7 (δC 65.6) with 7α-OH, the δC 73.5 (C-7) suggested 7β-OH for 10. Additionally, the NOE effects of H-7/H-8, and H-12/H-14, H-17 accurately established the relative configuration of 10 (Fig. 3). Thus, metabolite 10 was determined as 25(R)-spirost-5-en-3β,7β,12β,18-tetraol.
Biotransformation time-course
The biotransformation time-course of diosgenin by C. blakesleana AS 3.970 was investigated in the present work (Fig. 4). The cultures of C. blakesleana AS 3.970 incubated with diosgenin were analyzed by LC-MS/MS each day in the incubation period of 7 days. Metabolites 1–11 and substrate diosgenin were used as standard substances. The concentrations of major transformed products 1–4, 7, 10, and 11 were detected by a HPLC-DAD instrument. Additionally, the variation of diosgenin as the substrate was also studied each day. The substrate diosgenin was almost exhausted within 24 h to produce metabolites with compound 1 as the major metabolite (yield 11.2%). 25(R)-Spirost-5-en-3β,7α,12β-triol (1) was a dihydroxylated derivative of diosgenin, which also was the lowest oxygenated derivative among metabolites 1–11. The amounts of metabolite 1 decreased progressively, which suggested that metabolite 1 was substituted by the more hydroxyl groups to yield the high oxygenated metabolites. Therefore, it was conceivable that metabolite 1 was an important intermediate in the metabolic progress of diosgenin by C. blakesleana AS 3.970. The time-courses of metabolites 2 and 7 indicated that the highest amounts were obtained at 4 and 5 days, respectively. And, the amounts were decreased significantly at 6–7 days, suggesting that they were further oxygenated to produce other metabolites such as 3, which may be a derivative of 2. Thus, the biotransformation time-course suggested that C. blakesleana AS 3.970 transformed diosgenin with high hydroxylation abilities.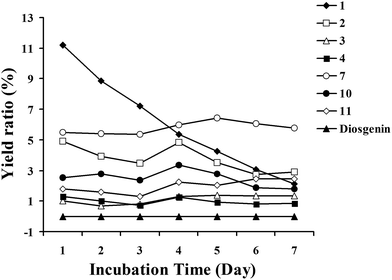 | ||
| Fig. 4 Biotransformation time-course of metabolites 1–11 by C. blakesleana AS 3.970 and the residual of diosgenin. | ||
On the basis of the structures analyses and time-course of metabolites 1–11, a possible biotransformed pathway of diosgenin by C. blakesleana AS 3.970 was proposed as shown in Fig. 1. The allylic moiety (C-5/C-6/C-7) of diosgenin played a crucial role in the metabolism of diosgenin by C. blakesleana AS 3.970, which gave two style metabolites possessing 7α- or 7β-OH. So, 25(R)-spirost-5-en-3β,7β-diol, and 25(R)-spirost-5-en-3β,7α-diol were proposed as the important intermediates in the transformation of diosgenin to form high oxygenated metabolites. To support the predicted pathway, the transformation of metabolites 1, 2, and 7 by C. blakesleana AS 3.970 was also investigated. In the qualitative analyses of the extracts of cultures, metabolites 4 and 5 were detected in the transformed culture of 1, metabolite 3 was detected in the transformed culture of 2, and metabolites 9–11 were detected in the transformed culture of 7. This transformation experiment could be used as an evidence for the predicted pathway, although metabolites 2 and 3 were not detected in the transformed culture of 1.
Inconsideration of the low recovery of metabolites, the mycelial mass was also extracted with EtOAc. And the extracts were analyzed by HPLC with substrate and metabolites as standard compounds. As a result, substrate and metabolites 1–11 were not detected. Therefore, it was deduced that some trace metabolites existed in the culture, which influenced the recovery rate.
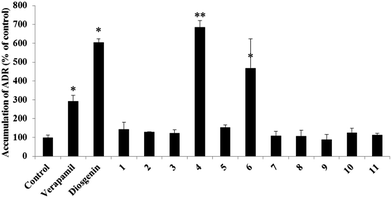
P-glycoprotein inhibitory effects in adriamycin (ADM)-resistant human breast adenocarcinoma cell line (MCF-7/ADR)
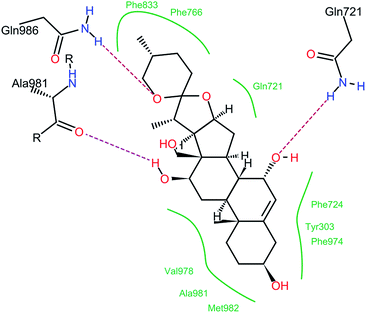
The P-glycoprotein (P-gp) inhibitory activities of compounds 1–11 were evaluated using adriamycin (ADM)-resistant human breast adenocarcinoma cell line (MCF-7/ADR). The accumulation of ADM in MCF-7/ADR cells was analyzed after cells were exposed to compounds 1–11, the substrate diosgenin or the positive control verapamil at 20 μM. In this bioassay, an increase of intracellular adriamycin accumulation was used to evaluate a P-gp inhibitory effect. Diosgenin and transformed products 4, 6 and verapamil increased the accumulation of ADM significantly when compared with the negative control (Fig. 5). The bioassay results suggested that diosgenin, metabolites 4 and 6 had significant inhibitory effects on P-gp, and especially that compound 4 had more potential inhibitory effect than diosgenin. The analyses of structures (1–11) and their corresponding P-gp inhibitory effects indicated that the 7α-OH group may play important role for the inhibitory effect.Molecular docking analyses predicted that compound 4 would bind at the drug binding site of P-gp. The preferable binding conformation is shown in Fig. 6, with the residues interacted with P-gp indicated. Compound 4 interacted with P-gp through several H-bonds interaction, such as 7-OH with Gln721, 12-OH with Gln981, O in the (six membered ring) with Ala 986. Hydrophobic interaction was formed between compound 4 and both aliphatic and aromatic amino acids (Tyr303, Gln721, Phe724, Phe766, Phe833, Phe974, Val978, Ala981 and Met982). The interaction between 12-OH and Ala981 of P-gp through H-bonds was similar with verapamil.23 This docking result suggests that P-gp recognizes compound 4 through a mechanism similar with verapamil.
Experimental
General experimental procedures
1H-, 13C- and 2D-NMR were recorded in methanol-d4 on a Bruker-500 spectrometer (500 MHz for 1H-NMR and 125 MHz for 13C-NMR). HRESIMS spectra were measured on an Agilent 1100 series LC/MSD ion trap mass spectrometer. ESIMS data were determined on API 3200 mass spectrometer (AB SCIEX, Framingham, MA). Analytical liquid chromatography was performed on a Ultimate 3000 HPLC instrument equipped with photodiode array detector (PAD). Preparative HPLC was performed on an Agel instrument with an UV detector and a YMC ODS A column (250 × 20 mm, 5 μm). Column chromatography was performed with silica gel (200–300 mesh, Qingdao Marine Chemical Inc., Qingdao, People's Republic of China). TLC was carried out on glass pre-coated silica gel GF254 plates. Spots were visualized under UV light or by spraying with 10% sulfuric acid in EtOH followed by heating. All solvents including ethyl acetate, methanol, CHCl3 and acetone are A.R. grade and were purchased from Tianjin Kemiou Chemical Reagent Company (Tianjing, China). Methanol and acetonitrile for HPLC analysis are chromatographic grade (Merck, Darmstadt, Germany).Microorganisms
Cunninghamella blakesleana AS 3.970, Mucor polymorphosporus AS 3.3443, Cunninghamella echinulata AS 3.3400, Aspergillus niger AS 3.1858, Penicillium janthinellum AS 3.510, Cunninghamella echinulata AS 3.2004, Cunninghamella elegans AS 3.1207, Syncephalastrum racemosum AS 3.264, Doratomyces stemonitis AS 3.1411 and Mucor spinosus AS 3.3450 were purchased from the Chinese General Microbiological Culture Collection Center, Beijing, P.R. China. These organisms were stored on potato-dextrose agar (PDA) medium slants at 4 °C.Culture medium
All biotransformation experiments were performed in liquid potato medium, which was consisted of water (1 L), potato (200 g) and glucose (20 g).Culture conditions and biotransformation procedures
Preliminary screening scale biotransformation of diosgenin was carried out in 250 mL Erlenmeyer flasks containing 100 mL of liquid medium. The flasks were placed on a rotary shaker (140 rpm, 30 °C). The substrates (diosgenin) gave a concentration of 10 mg mL−1 by acetone. After 24 h of pre-culture, 2 mg of diosgenin were added into each flask. The incubation was allowed to continue for 4 days. Culture controls consisted of fermentation blanks with microorganisms grown under identical non-substrate conditions. Substrate controls were composed of sterile medium with substrate, and they were incubated without microorganisms.Preparative scale biotransformation of diosgenin by C. blakesleana AS 3.970 was conducted in a 1 L Erlenmeyer flask. The substrate (10 mg) dissolved in acetone (0.4 mL) was added to pre-cultured medium (400 mL) and was incubated for an additional 4 days. Finally, 500 mg diosgenin was used for the preparative experiment.
Extraction and isolation of biotransformation products
The culture was filtered and the filtrate was extracted with the same volume of ethyl acetate (EtOAc). The organic phase was evaporated in vacuo at 38 °C. The EtOAc extracts were subjected to a silica gel column and eluted with CHCl3–CH3OH (100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 1
1 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to give 40 fractions. On the basis of TLC analysis, the metabolites of diosgenin were detected in Fr 24-30. Then, Fr 24-30 were successively purified by preparative HPLC (mobile phase: 40–70% acetonitrile in 0.03% aqueous acetic acid; flow rate: 8 mL min−1) to afford compounds 1 (22.51 mg, 4.5%), 2 (20.72 mg, 4.1%), 3 (5.25 mg, 1.05%), 4 (5.13 mg, 1.03%), 5 (8.85 mg, 1.77%), 6 (11.43 mg, 2.29%), 7 (25.6 mg, 5.12%), 8 (7.9 mg, 1.58%), 9 (10.5 mg, 2.1%), 10 (12.4 mg, 2.48%), and 11 (11 mg, 2.2%).
1) to give 40 fractions. On the basis of TLC analysis, the metabolites of diosgenin were detected in Fr 24-30. Then, Fr 24-30 were successively purified by preparative HPLC (mobile phase: 40–70% acetonitrile in 0.03% aqueous acetic acid; flow rate: 8 mL min−1) to afford compounds 1 (22.51 mg, 4.5%), 2 (20.72 mg, 4.1%), 3 (5.25 mg, 1.05%), 4 (5.13 mg, 1.03%), 5 (8.85 mg, 1.77%), 6 (11.43 mg, 2.29%), 7 (25.6 mg, 5.12%), 8 (7.9 mg, 1.58%), 9 (10.5 mg, 2.1%), 10 (12.4 mg, 2.48%), and 11 (11 mg, 2.2%).
Structure characterization of new compounds
Intracellular adriamycin accumulation assay24
Human breast adenocarcinoma cells (MCF-7) and adriamycin (ADM) resistant MCF-7 (MCF-7/ADR) cells were seeded in a 24-well plate at a density of 5 × 105 cells per well and incubated for 48 h at 37 °C in a 95% relative humidity atmosphere containing 5% CO2. After pre-incubation with fresh medium containing either the commonly used P-gp inhibitor verapamil (20 μM) or one of the test compounds (20 μM), 10 μM adriamycin was added to the medium. The plates were incubated for one hour at 37 °C with gentle shaking. The reaction was terminated by removal of the medium. Cells were then washed three times with 1 mL of ice-cold PBS. The cell monolayers were subsequently lysed with 0.3 mL of 0.1% Triton X-100®, and the concentration of adriamycin in the cell lysate was determined by LC-MS/MS. Protein concentrations served as the loading control and were measured using the bicinchoninic acid procedure with bovine serum albumin as the standard (BCA; Solarbio, China). MCF-7 cells were used as a positive control for maximum adriamycin accumulation.LC-MS/MS analysis
LC-MS/MS parameters were set as previously described with modification.25,26 An Agilent LC system (Agilent HP1200, Agilent Technology Inc., Palo Alto, CA, USA) and an API 3200 triple-quadrupole mass spectrometer (Applied Biosystems, Concord, Ont, Canada) were used for LC-MS/MS analysis. Chromatographic separation was performed on a Hypersil BDS-C18 column (150 mm × 4.6 i.d., 5 μm; Dalian Elite Analytical Instruments Co. Ltd, China) at ambient temperature.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 70, v/v) at a flow rate of 0.5 mL min−1. The run time was 3 min for each injection, and the column temperature was controlled at room temperature. An electrospray ionization (ESI) source was used, and the system was operated in positive mode. The optimized ionspray voltage and temperature were set at 4500 V and 500 °C, respectively. The curtain gas (CUR) was set at 10 psi; gas 1 and gas 2 (nitrogen) were set at 30 and 40 psi, respectively, and the dwell times were 200 ms. The quantification assay was performed using multiple reaction monitoring (MRM). The selected m/z transitions were 544.000 → 397.000 for adriamycin.
70, v/v) at a flow rate of 0.5 mL min−1. The run time was 3 min for each injection, and the column temperature was controlled at room temperature. An electrospray ionization (ESI) source was used, and the system was operated in positive mode. The optimized ionspray voltage and temperature were set at 4500 V and 500 °C, respectively. The curtain gas (CUR) was set at 10 psi; gas 1 and gas 2 (nitrogen) were set at 30 and 40 psi, respectively, and the dwell times were 200 ms. The quantification assay was performed using multiple reaction monitoring (MRM). The selected m/z transitions were 544.000 → 397.000 for adriamycin.Analyst 1.4.1 software (Applied Biosystems) was used to control the equipment and for data acquisition and analysis.
Molecular modeling
For better understanding the molecular interactions between ligand and protein, the docking was performed using Autodock Version 4.2.27 The ligand (compound 4) was docked into three-dimensional structure of P-gp (PDB Code:3G61).23 In the preparatory phase of docking process, the non-polar hydrogen atoms of P-gp were merged, and the Kollman and Gasteiger charges were assigned to protein and ligand 4 by Autodock Tools. The Autogrid Version 4.2 was used to calculate the grid maps for the proteins in the docking process. The grid dimensions covering the entire protein-binding site were 90 × 90 × 90 grid points with spacing of 0.375 Å in each dimension. Lamarckian genetic algorithm with default parameter settings was used to perform docking simulations. The best docking conformation for protein–ligand was selected for post-docking analysis. The two-dimensional diagrams of protein–ligand interactions were analyzed using the web-based online tool, PoseView (http://poseview.zbh.uni-hamburg.de/poseview).28Conclusions
In this paper, biotransformation was used to catalyze the modification of diosgenin. After screening 10 strains of filamentous fungi, C. blakesleana AS 3.970 was chosen to transform diosgenin due to high catalysis capability. Eleven polyhydroxylated derivatives, including seven previously unreported steroids were obtained from the culture of C. blakesleana AS 3.970 incubated with diosgenin. The transformed products suggested that C. blakesleana AS 3.970 preferred to catalyze hydroxylation reaction. Especially, hydroxylation of 7-OH was the key catalytic reaction. Additionally, hydroxylation of C-18 was obtained in the transformation, which was not reported about the biotransformation of diosgenin previously. Combined with in vitro bioassay, product 4 was obtained as a potential P-gp inhibitor. For the continuing investigation, it was designed to conduct screening of a series of microbial strains in order to find out a microorganism, which displayed selective catalytic capability. Thus, the bioactive derivative of diosgenin could be obtained with high yield.Acknowledgements
This research program is financially supported by the National Natural Science Foundation of China (No. 81274047, 81473334, 81303146, and 81503201), Education Department of Liaoning Province (LR2014025, L2014352), Outstanding Youth Science and Technology Talents of Dalian (2014J11JH132), Distinguished professor of Liaoning Province and Innovation Team of Dalian Medical University for financial supports.Notes and references
- B. P. Ma, B. Feng, H. Z. Huang and Y. W. Cong, Mode. Tradit. Chin. Med. Mater. Med., 2010, 12, 150–154 Search PubMed.
- P. Fernandes, A. Cruz, B. Angelova, H. M. Pinheiro and J. M. S. Cabral, Enzyme Microb. Technol., 2003, 32, 688–705 CrossRef CAS.
- D. A. Rathbone and N. C. Bruce, Curr. Opin. Microbiol., 2002, 5, 274–281 CrossRef CAS.
- K. B. Borges, W. S. Borges, D. P. Rosa, M. T. Pupo, P. S. Bonato and I. G. Collado, Tetrahedron: Asymmetry, 2009, 20, 385–397 CrossRef CAS PubMed.
- R. Saunders, P. S. J. Cheetham and R. Hardman, Enzyme Microb. Technol., 1986, 8, 549–555 CrossRef CAS.
- D. Y. Léger, B. Liagre, P. Cardot, J. L. Beneytout and S. Battu, Anal. Biochem., 2004, 335, 267–278 CrossRef PubMed.
- S. M. Mu, X. S. Tian, Y. W. Ruan, Y. T. Liu, D. Z. Bian, C. Y. Ma, C. X. Yu, M. Feng, F. R. Wang, L. Gao and J. J. Zhao, Biochem. Biophys. Res. Commun., 2012, 418, 347–352 CrossRef CAS PubMed.
- Z. M. He, H. Y. Chen, G. F. Li, H. Y. Zhu, Y. G. Gao, L. X. Zhang and J. M. Sun, Phytomedicine, 2014, 21, 871–876 CrossRef CAS PubMed.
- K. W. Choi, H. J. Park, D. H. Jung, T. W. Kim, Y. M. Park, B. O. Kim, E. H. Sohn, E. Y. Moon, S. H. Um, D. K. Rhee and S. Pyo, Vasc. Pharmacol., 2010, 53, 273–280 CrossRef CAS PubMed.
- D. H. Jung, H. J. Park, H. E. Byun, Y. M. Park, T. W. Kim, B. O. Kim, S. H. Um and S. Pyo, Int. Immunopharmacol., 2010, 10, 1047–1054 CrossRef CAS PubMed.
- Y. J. Wang, K. L. Pan, T. C. Hsieh, T. Y. Chang, W. H. Lin and J. T. A. Hsu, J. Nat. Prod., 2011, 74, 580–584 CrossRef CAS PubMed.
- T. Dong, G. W. Wu, X. N. Wang, J. M. Gao, J. G. Chen and S. S. Lee, J. Mol. Catal. B: Enzym., 2010, 67, 251–256 CrossRef CAS PubMed.
- F. Q. Wang, B. Li, W. Wang, C. G. Zhang and D. Z. Wei, Appl. Microbiol. Biotechnol., 2007, 77, 771–777 CrossRef CAS PubMed.
- G. W. Wu, X. J. Li, Y. Gao, Y. Jia and J. M. Gao, Chin. Chem. Lett., 2010, 21, 446–448 CrossRef CAS PubMed.
- G. W. Wu, J. M. Gao, X. W. Shi, Q. Zhang, S. P. Wei and K. Ding, J. Nat. Prod., 2011, 75, 2095–2101 CrossRef PubMed.
- X. Xiao, X. K. Liu, S. B. Fu and D. A. Sun, J. Asian. Nat. Prod. Res., 2011, 13, 270–275 CrossRef CAS PubMed.
- Y. Zhao, L. M. Sun, X. N. Wang, T. Shen, M. Ji, X. Li and H. X. Lou, Chin. Chem. Lett., 2010, 21, 76–80 CrossRef CAS PubMed.
- X. Lv, D. Liu, J. Hou, P. P. Dong, L. B. Zhan, L. Wang, S. Deng, C. Y. Wang, J. H. Yao, X. H. Shu, K. X. Liu and X. C. Ma, Food Chem., 2013, 138, 2260–2266 CrossRef CAS PubMed.
- Z. P. Mai, C. Wang, Y. Wang, H. L. Zhang, B. J. Zhang, W. Wang, X. K. Huo, S. S. Huang, C. Y. Wang, K. X. Liu, X. C. Ma and X. B. Wang, Fitoterapia, 2014, 99, 352–361 CrossRef CAS PubMed.
- Y. Wang, Y. Sun, C. Wang, X. K. Huo, P. D. Liu, C. Y. Wang, B. J. Zhang, L. B. Zhan, H. L. Zhang, S. Deng, Y. Y. Zhao and X. C. Ma, Phytochemistry, 2013, 96, 330–336 CrossRef PubMed.
- C. Wang, P. P. Dong, L. Y. Zhang, X. K. Huo, B. J. Zhang, C. Y. Wang, S. S. Huang, X. B. Wang, J. H. Yao, K. X. Liu and X. C. Ma, RSC Adv., 2015, 5, 12717–12725 RSC.
- Y. Zhao, L. M. Sun, X. N. Wang, T. Shen, M. Ji and X. H. Lou, Nat. Prod. Commun., 2010, 5, 373–376 CAS.
- S. G. Aller, J. Yu, A. Ward, Y. Weng, S. Chittaboina, R. Zhuo, P. M. Harrell, Y. T. Trinh, Q. H. Zhang, I. L. Urbatsch and G. Chang, Sci. Mag., 2009, 323, 1718–1722 CAS.
- X. K. Huo, Q. Liu, C. Y. Wang, Q. Meng, H. J. Sun, J. Y. Peng, X. X. Ma, P. Y. Sun and K. X. Liu, J. Pharm. Sci., 2014, 103, 719–729 CrossRef CAS PubMed.
- W. Z. Ma, J. L. Wang, Q. Guo and P. F. Tu, J. Pharm. Biomed. Anal., 2015, 111, 215–221 CrossRef CAS PubMed.
- K. Li, Y. W. Wang, J. K. Gu, X. Y. Chen and D. F. Zhong, J. Chromatogr. B: Analyt. Technol. Biomed. Life. Sci., 2005, 817, 271–275 CrossRef CAS PubMed.
- G. M. Morris, R. Huey, W. Lindstrom, M. F. Sanner, R. K. Belew, D. S. Goodsell and A. J. Olson, J. Comput. Chem., 2009, 30, 2785–2791 CrossRef CAS PubMed.
- K. Stierand and M. Rarey, ACS Med. Chem. Lett., 2010, 1, 540–545 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: The spectra including 1D, 2D-NMR, HRESIMS of compounds 1–11. See DOI: 10.1039/c5ra12253h |
| ‡ Min Xu and Xiaokui Huo contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2015 |