Enhancement of performance in porous bead-based microchip sensors: effects of chip geometry on bio-agent capture†
Eliona Kulla‡
a,
Jie Chou‡b,
Glennon W. Simmonsab,
Jorge Wongab,
Michael P. McRaeb,
Rushi Patelb,
Pierre N. Florianoc,
Nicolaos Christodoulidesab,
Robin J. Leachd,
Ian M. Thompsond and
John T. McDevitt*abe
aDepartment of Chemistry, Rice University, Houston, Texas 77005, USA
bDepartment of Bioengineering, Rice University, Houston, Texas 77005, USA
cThe University of Texas MD Anderson Cancer Center, Houston, Texas 77030, USA
dUrology, University of Texas Health Science Center at San Antonio, Texas 78229, USA
eDepartment of Biomaterials, New York University, New York, 10010-4086, USA. E-mail: mcdevitt@nyu.edu; Tel: +1 212 998 9204
First published on 21st May 2015
Abstract
Measuring low concentrations of clinically-important biomarkers using porous bead-based lab-on-a-chip (LOC) platforms is critical for the successful implementation of point-of-care (POC) devices. One way to meet this objective is to optimize the geometry of the bead holder, referred to here as a micro-container. In this work, two geometric micro-containers were explored, the inverted pyramid frustum (PF) and the inverted clipped pyramid frustum (CPF). Finite element models of this bead array assay system were developed to optimize the micro-container and bead geometries for increased pressure, to increase analyte capture in porous bead-based fluorescence immunoassays. Custom micro-milled micro-container structures containing an inverted CPF geometry resulted in a 28% reduction in flow-through regions from traditional anisotropically-etched pyramidal geometry derived from Si-111 termination layers. This novel “reduced flow-through” design resulted in a 33% increase in analyte penetration into the bead and twofold increase in fluorescence signal intensity as demonstrated with C-reactive protein (CRP) antigen, an important biomarker of inflammation. A consequent twofold decrease in the limit of detection (LOD) and the limit of quantification (LOQ) of a proof-of-concept assay for the free isoform of prostate-specific antigen (free PSA), an important biomarker for prostate cancer detection, is also presented. Furthermore, a 53% decrease in the bead diameter is shown to result in a 160% increase in pressure and 2.5-fold increase in signal, as estimated by COMSOL models and confirmed experimentally by epi-fluorescence microscopy. Such optimizations of the bead micro-container and bead geometries have the potential to significantly reduce the LODs and reagent costs for spatially programmed bead-based assay systems of this type.
Introduction
Microfluidic LOC devices have received significant attention in the past two decades with a range of potential applications for detection of environmental, biological, and chemical agents.1–4 Through the miniaturization of existing components (i.e., chromatography paper and microspheres, mixers, micropumps, etc.), LOC devices offer a variety of benefits including high surface-to-volume ratios, rapid diffusion times, low sample and reagent volumes, low cost, reduced analysis times, portability, and on-site analysis of patient samples collected with minimal discomfort from body fluids, such as finger-stick blood or saliva.5–13 In addition to rapid turnaround of results, some devices with multiplex capability can simultaneously measure a panel of biomarkers from a single sample for disease diagnosis, prognosis, and monitoring.14–16A great deal of effort has been directed towards development of lateral flow assays (LFAs) (i.e., dipstick assays) for POC devices due to their low cost and simplicity.17 However, these simple dipstick assays typically suffer from lack of quantitation as well as poor performance at low concentrations.18 On the other hand, enzyme-linked immunosorbent assays (ELISAs) and immunoassays completed with the Luminex platform deliver quantitative results, but require long incubation times (on the order of hours) making them impractical for use at the POC.19 The ability to detect low concentrations of protein biomarkers using noninvasive sampling on a rapid time frame remains a critically important priority for the LOC community.20
Bead-based microfluidic platforms are attractive for these functions due to their enhanced reaction kinetics, reduced background noise, and potential for decreased system costs.21–25 A variety of bead-based solid support types including gel, silica, polystyrene, magnetic, and agarose beads have been used to capture target biomolecules in both benchtop and microfluidic systems.8,21,22,26–29 Porous forms of these beads demonstrate improved performance due to their large surface-to-volume ratios.30,31 The three-dimensional structure of porous beads can be more densely packed with immobilized capturing antibodies than their non-porous counterparts. The microporous geometry allows for greater capture efficiency and has potential to offer lower limits of detection.32
Methods to increase binding capacity in porous beads have focused on tuning the pore size or modifying the chemical functionalization of the bead surface.33,34 Recent, alternative methods demonstrate that the capture rate of analytes in porous beads may be tailored by simple physical methods such as bead compression. Thompson and Bau reported a twofold increase in mass transfer rates for binding of biotin-functionalized quantum dots to streptavidin-coated agarose beads by periodically compressing and expanding the beads as compared to stationary beads.35 Similarly, Ouyang and Liang found a twofold increase in adsorption rates when compressing chitosan beads in solutions containing dye molecules or nanoparticles.36
Porous bead sensors may also be positioned into microfluidic components while free flowing analytes are transported by diffusion- and convection-driven flow to the porous medium.37 Our group has developed a modular micro-bead array platform known as the programmable Bio-Nano-Chip (p-BNC), designed for the capture and quantitation of soluble biomarkers.38,39 The p-BNC system contains an array of individually addressable agarose bead sensors, each confined in flow-through micro-cavities resembling inverted, square pyramid frustums (i.e., inverted PF microchip). In contrast to other stationary bead-based array platforms, the bead-based p-BNC system is unique as it utilizes some of the three-dimensional interior of agarose through pressure-driven flow that is induced by the confinement of beads in the microchip environment. Studying and optimizing integrated LOC systems using computational fluid dynamics is both cost- and time-effective. Previous computational fluid dynamic model studies characterized bio-agent capture and fluid flow dynamics for pressure-driven flow in the inverted PF microchip of the p-BNC system.37 These results suggested that pressure-driven flow increases analyte capture within porous sensors.
In this paper we use computational simulations and experiments to examine the effect of microchip geometry and bead size in optimizing pressure-driven flow rates within the bead-based p-BNC system to enhance analyte binding in the context of the essential and well-characterized biomarkers: CRP and PSA. Because pressure build-up is generally proportional to flow-through area around the beads sitting in the micro-container, it is hypothesized that by using a new micro-container with smaller bypasses, increased pressure would be generated inside the bead chamber which would, in turn, force more analyte into the bead interior thereby producing a higher analyte-specific signal. While other groups have demonstrated the importance of optimizing geometries of microfluidic channels, biochips, and mixers to enhance performance of various LOC applications,40–47 to our knowledge, no other studies have examined the effects of bead and bead/micro-container geometries on the capture dynamics in porous stationary bead arrays.
Results and discussion
For the experimental portion of these studies, two key biomarkers are chosen as clinically relevant targets. First, CRP is selected as a model antigen here, due to the fact that it serves as an acute phase reactant that is involved in multiple diagnostic settings. Both low and high sensitivity CRP tests are used for various cardiac indications. Recently, CRP has been found to be useful in profiling malnourished children who are most at risk of dying from bacterial infections.48 In the case of trauma and radiation damage, CRP serum concentrations are found to increase dramatically following the traumatic events as well as from acute radiation exposure.49 The same biomarker is useful in diagnosis of lower respiratory tract infection, potentially reducing unnecessary use of antibiotics.50The second biomarker utilized in this study relates to prostate cancer (PC) diagnostics. Current methods of screening for PC, the most common non-skin malignancy in American men, include measurements of serum levels of prostate-specific antigen (PSA) produced primarily by the prostate. The biomarker PSA exists in the blood in four kallikrein forms: free PSA, intact PSA, complexed PSA and pro-PSA combined with kallikrein-related peptidase 2 (hK2).51 Measurement of PSA and its isoforms is widely used for PC early detection and recent efforts have been made to move PSA testing, primarily performed using ELISA, to testing at the POC.52
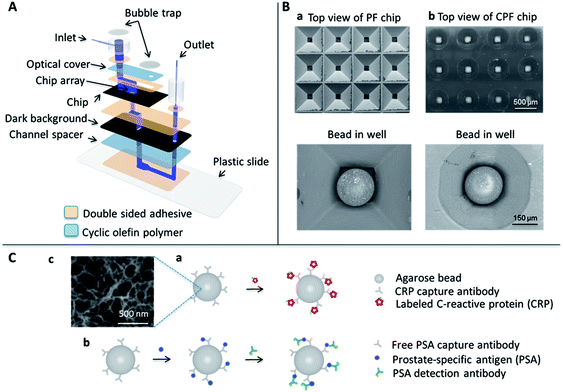
The components of the p-BNC system used in this study are shown in Fig. 1A. For the analyte penetration depth and flow rate study, a prototype device constructed out of several layers of double-sided adhesive and polymer films was used. Individual layers were fabricated using a rapid prototyping method called xurography which utilizes a cutting plotter to create micro-channels that are aligned to form the fluid flow path.54 The xurography approach allowed for the rapid development of reaction vessels and the associated fluidic network to test multiple geometries as required for these studies.
The biochip is at the heart of the detection system and is composed of a 3 × 4 array of micro-containers filled with agarose beads of 280 μm diameter. For these studies, the agarose beads were manually placed into the wells of the micro-container using tweezers under a dissecting microscope. In parallel, efforts have been completed to develop automated approaches suitable for mass production of these sensors (see ESI, Fig. S1†) as well as the long term storage of the agarose beads in the microfluidic cards (see ESI, Fig. S2†). Once the beads are placed in the micro-container, the fluid flows over the agarose beads and out through the bottom openings of each micro-container.
Fig. 1B shows the two different micro-container designs that contain the beads. For the detection of CRP and PSA two different immunoassay platforms were utilized. The cartoon in Fig. 1C(a) represents agarose beads labeled with capturing antibodies that bind fluorescently-labeled CRP in a one-step immunoassay format, while Fig. 1C(b) shows a cartoon of a sandwich immunoassay utilizing agarose beads with capturing antibodies that bind free PSA followed by a fluorescently-labeled detection antibody. The agarose beads have a porous structure with pore size ranging from 100–400 nm, as shown in Fig. 1C(c), which provides a dense framework for immobilizing capturing antibodies.55
In this report, the assay performances using the two micro-container geometries shown in Fig. 1B were compared. The first is a previously reported design constructed with tapered walls with angles of 54.7°, dictated by anisotropic etching of the crystalline lattice of silicon in the 111 direction plane. Using this process an inverted PF structure with micro-container dimensions of 623 ± 7.1 μm × 620 ± 1.9 μm at the top and 132 ± 4.5 μm × 130 ± 2.1 μm at the bottom with a height of 280 μm, was created, as shown in Fig. 1B(a). To generate the second design, computer numerical control (CNC) was used to create a cone-like geometry that we refer to as an inverted CPF design with micro-container radii dimensions of 268 ± 4.6 μm at the top, 60 ± 2.5 μm at the bottom with a height of 280 μm, was created, as depicted in Fig. 1B(b). The CPF structure was designed as the geometric intersection of the PF structure with an inverted conical frustum with a radius of 53 μm at the base. The image of the beads sitting in the wells in Fig. 1B shows large open regions at the corners of the micro-container, which we refer to as bypasses. However, the new custom-form micro-containers have a more rounded shape tailored to the bead geometry that significantly reduces the bypass area as visualized in the right column of Fig. 1B. Because fluid must flow through the bead-filled micro-containers during an assay, fluid must flow either through the open bypasses or be forced into the porous bead structure. Because pressure build-up is generally proportional to flow-through area, we hypothesized that by using the inverted CPF micro-container with smaller bypasses, more pressure would be generated inside the bead chamber that would, in turn, force more analyte into the bead interior.
Optimization of the inverted CPF geometry
The optimized cone-like geometry was inspired from a former geometry, the inverted PF design, which was developed with several key design considerations, including uniform capture signal, maximum analyte capture, and the physical fit of the bead inside the micro-container. Furthermore, it should be appreciated that this prior design structure was created using an anisotropic etching process with use of Si 100 wafers. The geometry of the resulting etched pit was dictated by the silicon 111 etch plane geometry. However, as these devices move closer to clinical practice, there has been a push away from the use of these expensive silicon wafers. From the challenge of switching the materials and processing bases comes the opportunity to refine the geometry to be more consistent with enhanced performance.Initial designs included monolithic straight through, triangular prism-based bead holders with rounded corners, fabricated through SU-8 photolithographic methods. These design features provided some promising results, however, due to the variability in the fit between the bead and well, bead signals in the array exhibited both high inter-bead and intra-bead variation. As such, it was useful to adapt to the tapered wall feature similar to that of inverted pyramidal pits from anisotropically etched silicon (Fig. 1B). This tapered design results in a more uniform flow around the medial slice of the beads where the highest signal occurs.
Initial computational modeling revealed that a true conical geometry micro-container provides the best performance. However, a conical design leads to high pressure buildups which create high potentials for device mechanical failures including the formation of leaks. Therefore, a cone-like geometry was chosen, which started with the former inverted PF design, but where the empty edges of the pyramid were clipped off, hence the term “clipped”. This situation creates a more rounded shape than the inverted PF design, but contains more bypass area than a true conical design. The clipping radius of the micro-container and the bypass area around the bead are the most important parameters for optimizing the performance of the structure. The extent of clipping is expressed by the clipping radius, defined as the distance from the center of the micro-container to the clipped edge at a depth corresponding to the medial slice of a 280 μm bead diameter sitting in the well. Next, modeling of analyte capture to agarose beads, under several clipping radii, revealed increased analyte capture as the clipping radius decreased. As such, several clipping radii were tested experimentally to determine the optimal geometry that does not result in leaks at typical flow rates used in the micro-bead array test ensembles. These clipping radii include 160, 170, and 180 μm. For the 160 μm clipping radius, flow rates ≥400 μL min−1 result in immediate leaks, and flow rates of 300 μL min−1 result in leaks after 45 s; no leaks are observed for flow rates ≤200 μL min−1 for up to 1 h. Subsequent experiments were performed using the micro-containers with a 160 μm clipping radius as it provided the best results (Fig. 1B).
Pressure gain from custom micro-containers
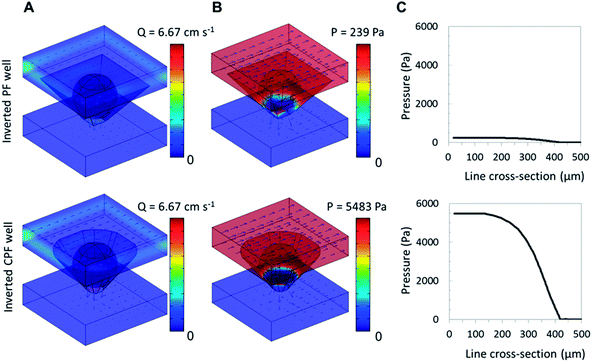
Using a computational fluid dynamics model, we have previously shown that some pressure develops when fluid flows over agarose beads sitting in the wells of the inverted PF micro-container. This pressure was an important factor in increasing capture of analyte within porous agarose beads. As the fluid delivered to the array of beads was forced to flow through each bead sensor, a bypass area-dependent pressure gradient was observed at and around the beads while the pressure at the bottom of the wells dropped to zero.37Fig. 2 shows results from computational fluid dynamics models and provides a comparison of the fluid velocity and pressure gain between the two designs as a result of the difference in bypass area. The bypass-to-bead area ratio at the medial x–y plane was estimated for 280 μm beads in both inverted PF and CPF wells at 83% and 60%, respectively. This 28% decrease in bypass for the inverted CPF geometry corresponds to a 23 times higher pressure and leads to an increase in exiting flow rate from 5.7 cm s−1 to 8.5 cm s−1. While the flow rate prior to the bead array remains constant, the nonlinear pressure drop across the well (accentuated by the small bypass area) increases the linear flow rate (or volumetric flow rate per cross sectional area) around the bead-well interface and drain exit. As a result, beads sitting in the inverted CPF micro-container exhibit higher pressure-driven internal flow due to the reduced bypass and higher flow rates in the container. This result is similar to the findings of Nischang and colleagues whereby geometry and size studies of the cross section of monolithic columns, used for chromatography separations, resulted in a significant increase in pressure as an effect of confinement of polymeric materials in very narrow conduits.56 Note that other tapered micro-container designs with similar bypass area may produce similar results.
Analyte capture and penetration depth in micro-container designs
While some polymer beads such as those made from silica and polystyrene rely primarily on binding of analyte at the bead's periphery, porous beads such as agarose have the capacity to bind more analyte due to their extended three-dimensional internal porous structure that offers a large density of binding sites. Achieving high binding capacity using a single bead sensor has been shown to be advantageous and allows for a reduction in reagent costs. We found that the use of microporous beads leads to a lowering of the limit of detection observed for the porous beads relative to standard solid spheres.57 However, mass transfer of analytes to the interior of these three-dimensional structures with small pore sizes is limited to diffusive and convective transport.58 One effective way to circumvent this apparent shortcoming is the placement of beads in micro-containers and subjecting them to high pressures and high internal flow rates that are regulated by the size of the bypass around the bead as discussed above. The new clipped design achieved 23 times higher pressure compared to the inverted PF chip design in the computational model. Further, computational fluid dynamics models predict higher quantities of analyte are driven into the porous bead by convective flow as it decreases the size of the analyte depletion region around the bead.Analyte depletion around porous materials is generally difficult to measure experimentally. However, by using theoretical simulations it is possible to gather important information (such as size) about the depletion region (area where the free analyte concentration is less than that of the bulk solution) surrounding the bead. Larger beads have larger depletion regions, assuming all other variables remain constant. For the theoretical simulations, the cutoff for the depletion width was defined as 90% to that of the bulk solution.59
Using CRP as a model analyte, a depletion width of 30 μm was simulated using the inverted PF chip, which was decreased to 19 μm when the inverted CPF chip was used. Higher signal also means that a greater fraction of the analyte is captured as opposed to flowing through the bypasses. Therefore, the fractional capture efficiency, as estimated through simulations, was ∼25% in the inverted CPF micro-container, a 4× increase from the inverted PF micro-structure. This supports our hypothesis that reducing the bypass increases the rate of replenishment to the depletion region, the region local to the bead surface, thereby forcing more analyte into the bead interior.
In these studies, the dimensionless Péclet number (Pe), defined as the ratio of the rate of convection to the rate of diffusion, was also investigated so as to understand the convection-driven flow within the agarose bead interior. Generally, if Pe ≫ 1 the fluid flow is governed by convection, and if Pe ≪ 1 the fluid flow is dominated by diffusion. Based on simulation data, the average Pe within the bead increases 26-fold, from 36 to 936, when the beads are sitting in the inverted CPF chip design. This increase in internal convection leads to the decrease in the depletion region and an increase in analyte capture within the bead.
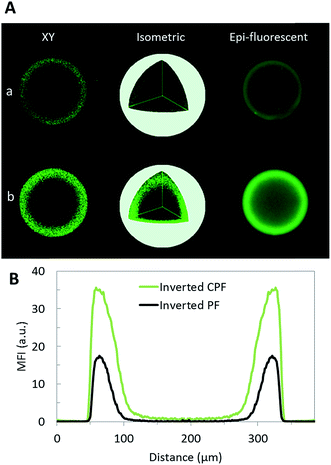
Experimental validation of the enhancement in analyte capture in the inverted PF and CPF designs using CRP as a model analyte was further obtained. CRP is an important biomarker of inflammation, and, although the relevant clinical concentration cutoff is relatively high,60 it was primarily chosen as a model to test the capture efficiency using the two different biochip geometries. Fig. 3 shows fluorescence images resulting from 50 ng mL−1 fluorescently-labeled CRP binding to agarose beads resting in inverted PF and CPF chip designs, using confocal and epifluorescense microscopy. The increased CRP capture observed in the inverted CPF design was seen in all axial planes of the bead as shown in the isometric view. Based on the full width at half maximum, the higher convection resultant of the reduced flow-through regions in the inverted CPF design, drives more analytes deeper into the bead. This leads to an analyte penetration depth of 40 μm using the inverted CPF chip as compared to 30 μm for the inverted PF chip, a 33% increase. In addition, there is a twofold increase in maximum fluorescence signal intensity for the inverted CPF design which should improve sensitivity.
Analyte flow rate dependence study
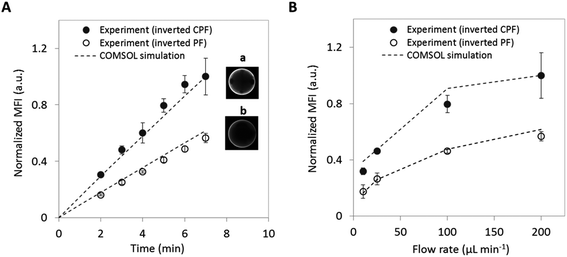
Further experiments confirm the increased analyte binding when using the inverted CPF chip design. Fig. 4A shows the increase in mean fluorescence intensity during a CRP assay for both the inverted PF and CPF chip designs. Both chips show a linear increase in signal with assay time as the bead captures more analyte from the sample; however, it is clearly evident that the capture rate in the inverted CPF design is significantly higher. Based on the slope of the linear regression shown, the binding rate for the CPF geometry is nearly doubled compared to the PF geometry. In addition, assays with varying flow rates for a fixed amount of time (7 min) were performed with derived results as shown in Fig. 4B. A consistent difference in performance is observed between the two designs, as evidenced by a doubling in signal for the inverted CPF geometrywith relatively low intra- and inter-assay measurements (see ESI†, Table S1). However, the increase in signal for higher flow rates is not linear and it further plateaus at higher flow rates, perhaps because the analyte capture moves from a transport-limited to a reaction-limited regime in which the agarose beads cannot bind the analyte fast enough before flowing out of the micro-container. Also important to note is the sample volume requirements for each chip. To reach a normalized mean fluorescence intensity of 0.4, the inverted PF design requires 700 μL of CRP analyte, while the CPF design requires only 175 μL. Thus, optimizing the geometry of the chip can lead to a significant decrease in required sample volume and overall reagent consumption.Standard curves for free PSA analyte
The experiments discussed above supported the theoretical modeling of analyte transport into porous agarose beads and revealed a twofold increase in the capture of CRP using the new inverted CPF biochip design. Here, a more practical application for employing the new biochip geometry is demonstrated. Given the previous results, it was hypothesized that the new biochip would lead to a decrease in the LOD and the LOQ of a clinically-relevant biomarker such as free PSA.Fig. 5A shows proof-of-concept standard curves derived from the measurement of different concentrations (0, 0.1, 0.2, 0.5, 1, 2, and 5 ng mL−1) of free PSA in phosphate buffered saline (PBS) buffer on a sandwich immunoassay platform using agarose beads in the two different biochip geometries. For these assays, a more integrated p-BNC platform was employed that allowed for the use of a lower (100 μL vs. 1000 μL) sample volume.39 Linear regression was used to characterize assay performance (sensitivity) of the two designs: inverted CPF design vs. inverted PF design. Results show that the inverted CPF design provides about 2.5× greater sensitivity (slope derived from the analyte captured/concentration) than the inverted PF design. Fig. 5B shows a visual representation of fluorescence intensity signal on the beads as a function of concentration and biochip design. The fluorescence intensity signal indicates that the inverted CPF chip captures more free PSA than the inverted PF design. In addition, both chip designs exhibit a high degree of signal uniformity suggesting that the tapered design provides adequate mechanical stabilization for the beads. However, because of the smaller bypasses in the inverted CPF design, it is important to properly filter reagents before use to minimize the amount of debris that could deposit on the bead's surface.
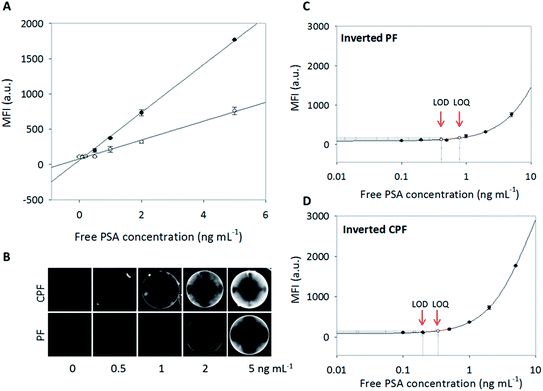 | ||
| Fig. 5 (A) Concentration dependence of free PSA sandwich-type immunoassay format in the p-BNC system39 with utilization of the inverted PF (open circles) and CPF (solid circles) geometric chips. MFI from the medial slice of the beads (intra-assay measurements) is fit to a linear curve for the inverted PF (y = 133x + 77.4; R2 = 0.991) and CPF (y = 339x + 58.4; R2 = 0.998) geometries. (B) Epi-fluorescent images show the capture of free PSA on an agarose bead (about 250 μm in diameter) in the chip array for both geometric chip designs at 1 s exposure time. (C) Standard curve of the capture of free PSA in the inverted PF chip fit to a four parameter logistic curve (R2 = 0.997) with LOD and LOQ corresponding to concentrations of free PSA antigen equal to 0.41 ng mL−1 and 0.79 ng mL−1, respectively. (D) Standard curve of the capture of free PSA in the inverted CPF chip is fit to a four parameter logistic curve (R2 = 0.999) with LOD and LOQ corresponding to concentrations of free PSA antigen equal to 0.20 ng mL−1 and 0.34 ng mL−1, respectively. | ||
Next, the influence of the change of geometry on the measured LOD and LOQ was explored. To determine the LOD and LOQ of both standard curves, data were plotted using four parameter logistic curves in SigmaPlot (Fig. 5C and D). The LODs were calculated according to the fluorescence intensity of the zero concentration plus three times its standard deviation, and LOQs were calculated according to the fluorescence intensity of the zero concentration plus ten times its standard deviation.61 A summary of these values is provided in Table 1. Importantly, the LOD obtained from the inverted CPF biochip was about twofold lower than that obtained from the inverted PF design. Similarly, the LOQ is about twofold lower in the CPF design with significant implication towards the real-world application of the assay. It should be noted that for real world testing, the free PSA assay must have an LOQ of 0.4 ng mL−1 or lower to be most useful.53 For the assay time and sample volume used, the inverted PF design did not meet this criteria with an LOQ of about 0.8 ng mL−1, while the LOQ using the inverted CPF design drops to 0.34 ng mL−1, below the desired value. This key increase in performance is attributed to the optimized bead micro-container which expands the utility of this approach.
| Chip geometry | Biomarker | Total time (min) | Total volume (μL) | Intra-assay CV% | LOD ng mL−1 | LOQ ng mL−1 |
|---|---|---|---|---|---|---|
| PF | Free PSA | 34 | 100 | 5.1–19.2 | 0.41 | 0.79 |
| CPF | Free PSA | 34 | 100 | 0.1–13.9 | 0.20 | 0.34 |
Another important benefit of the pressure-driven flow design is reduction of assay time. Immunoassays that use porous beads, but lack a flow-through design must rely on diffusion and typically report assay times in excess of 2 h.62 Given that the new inverted CPF flow-through geometry described here allows a further reduction in assay time compared to previous designs, it is possible to complete full protein-based bioassays in less than 30 min, a key factor during translation of these devices into POC testing settings.
Pressure dependence on bead size
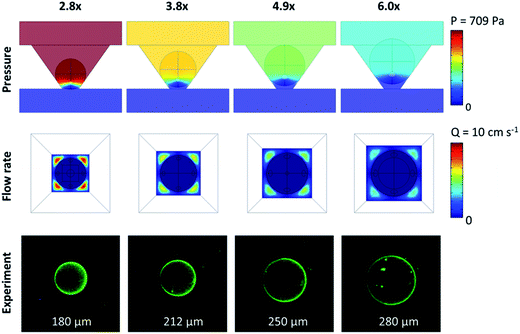
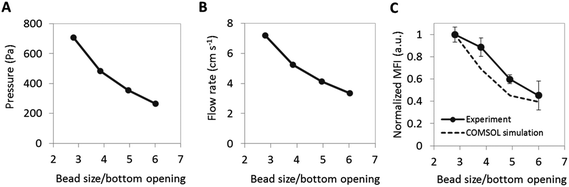
The aforementioned results demonstrated that the size of the bypass is important for increasing pressure driven flow into the agarose beads by modifying the micro-container geometry. Next, a different method was explored for achieving similar results by reducing the bead size using the inverted PF micro-container only. Smaller agarose beads are attractive because they have the potential to reduce reagent costs and localize capture in a smaller cross-sectional area that could potentially increase sensitivity.When looking at the geometry of a spherical bead in the inverted PF micro-container, the amount of bypass relative to the bead area at the medial slice of the bead remains constant at 45% of the cross-sectional area at the bead medial plane, mostly independent of the diameter of the bead (unless the diameter of the bead is smaller or equal to the diameter of the bottom opening of the micro-container, in which case the bead would fall through). However, the absolute value for total area of bypass (this is the area from the medial plane of the bead to the bottom opening of the micro-container) decreases with decreasing bead size since the bead sits deeper inside the well. Fig. 6 (top panel) shows that a decrease in bead size from 280 μm to 180 μm corresponds to a 55% decrease in bypass area and a pressure increase by 160% as determined from computational modeling. Similar to the effect of micro-container geometry, while the flow rate prior to the bead array remains constant, the nonlinear pressure drop across the well increases the linear flow rate, or volumetric flow rate per cross sectional area, around the bead-well interface and drain exit. As a result, smaller beads situated at the bottom of the well exhibit higher pressure-driven internal flow due to the reduced bypass and higher flow rates at the base of the container. While further gains are obtainable with much smaller beads, the high pressures resultant from the reduced bypasses and the diameter of the well opening of about 100 μm will lead to mechanical displacement of beads into the drain area instead of the desired pressure-driven confinement of the beads in the micro-wells.
Effect of bead size on analyte capture
Next, simulations and experiments were completed in an effort to measure the dependence of analyte capture on bead size and correlate the same to the pressures and exiting flow rates. Fig. 7A and B show the pressure increase corresponding to a maximum flow rate increase of 114% resultant from the reduced bypass. Based on simulation data, the Pe increased 3.5-fold with the reduction in bead size suggesting increased convective flow in the bead, which, as stated above is desirable for increasing capture efficiency in agarose beads. Next, to obtain experimental validation for the computational results, CRP assays were completed as shown in Fig. 1C(a). Here a 250% increase in the mean fluorescence intensity was achieved as the bead size decreased by 53%. Additionally, experimental results matched up very closely to the simulation data (Fig. 7C).Similar to the micro-container study, the depletion region with different sized beads was examined. For example, at 10 min the depletion width derived from simulations for a bead with bead size to bottom opening ratio of 6.0× exhibits a depletion width of 30 μm while a smaller bead of 2.8× exhibits a depletion width of 20 μm. This situation matches closely to the micro-container results which saw a decrease in depletion width from 30 μm to 19 μm and correlates with a similar increase in capture signal of about 2.5-fold. This behavior suggests that the effect on bypass area of the two geometric changes is similar although the signal intensity observed is slightly higher which may be attributed to higher analyte concentration per capture area given that the total area of the smaller beads is significantly less than that of the larger beads. Combining these two geometric changes into one chip design may further increase capture efficiency.
Scalability and practical considerations
In addition to increased sensitivity, another advantage offered by the smaller beads is the reduced cost associated with the reduction of capture agents coupled to the bead. From past efforts it has been shown that high concentrations of immobilized antibodies exhibit higher sensitivities, with diminishing benefits to signal intensities as higher amounts of antibody are loaded onto the beads.63 However, higher costs of reagents and sub-utilized core interiors by capture agents lead to underutilization of antibodies in the core of the sensors. As such, the percent of sub-utilized core decreases with decreasing bead size. For example, a shell containing a penetration depth of 30 μm equates to a larger available proportion of the sensor volume for smaller beads than for larger beads. From computational modeling, it is observed that a reduction in bead size from 280 μm to 180 μm leads to an increase in bead-localized capture ligand utilization from 12.3% to 28.4%. At the same time, with a cubic relationship between volume and bead diameter, a 36% reduction in bead diameter leads to a projected 73% decrease in costs of reagents per conjugation of capture probes. Further, while the research expenditure on agarose is currently $13.70 per g because of the ability to mass produce small beads in batches, the cost of the agarose material per bead becomes practically negligible. For example, a batch obtained from 1.002 g of agarose contains up to 10 mL of beads with diameter of 280 μm. Using Kepler's conjecture with a random packing efficiency yields a total of 662![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 beads.55 Further, the ability to reduce costs through less reagent consumption per bead allows for the capability to functionalize higher densities of antibodies to increase capture effectiveness.
500 beads.55 Further, the ability to reduce costs through less reagent consumption per bead allows for the capability to functionalize higher densities of antibodies to increase capture effectiveness.
Additionally, mechanical and practical constraints depend not only on the geometry and material of the micro-container, but also on the percent agarose, capture antibody loading and the fluid environment. Lower percent agarose beads may be susceptible to deformation under pressure (due to their soft mechanical properties) resulting in loss of beads through the bottom openings, while higher percent agarose beads (i.e., rigid beads), well capable of bearing higher backpressure, may be prone to clogging (due to their rigid mechanical properties and reduced porosity), thus stopping fluid flow and causing leaks. It should also be noted that for the purpose of this paper, there has been a focus on immunoassays in buffered solutions such as PBS with relatively low viscosities. Using more viscous fluids, such as saliva and whole blood, may displace rigid beads from the micro-container necessitating micro-containers with larger bypasses. The choice of container and the mechanical properties of the bead reactors are expected to influence the performance of the particular bioassay in the context of real-world clinical testing. Geometry considerations for serum, whole blood, and oral fluid samples as well as tuning the pore size of the agarose will be discussed in future publications.
Conclusions
While significant advancements in the field of POC diagnostics have been made recently, very few devices have found broad use in laboratories, clinics, and ambulances. Bead-based POC devices have the potential to serve as platform technologies with “plug and play” sensing elements that can be quickly reprogrammed to detect multiple biomarkers simultaneously for a wide panel of diseases. When these sensors are combined in a high pressure-driven array of flow-through micro-containers, a combination of both high analyte capture and high internal convection allow for the sensitive detection of low concentrations of analytes in fluidic samples.As demonstrated here, with the optimization of the micro-container geometry of the p-BNC systems through the reduction of inefficient flow-through regions, an improvement of the sensitivity in porous agarose beads is possible using only direct immunochemistry without optimizing reagents. Further, the use of smaller sensor sizes has the potential to reduce significantly the costs of both immobilized antibodies and detecting reagents. These activities may foster opportunities for low cost, single use, disposable lab cards, all consistent with POC applications.
Moreover, it has been demonstrated that the enhanced sensitivity afforded by these geometry-based optimizations improved the LOD and LOQ of an important biomarker, free PSA, without increasing analysis time or sample volume. Also, the possibility of shorter analysis times needed to achieve the same signal intensity using a CRP assay on the analytical platform, has been discussed. Thus, reduced-bypass structures with higher fractional capture may reduce the assay time and the required sample or reagent volume. With the aim of developing low cost, rapid decision devices, these initial geometric optimization findings can be incorporated into fully-integrated, injection-molded microfluidic cards to facilitate practical measurements at the POC on timeframes consistent with a typical doctor's visit that lasts 15–30 minutes.
Materials and methods
Device fabrication
Layers of double sided adhesive with fluidic channels and polyethylene terephthalate tube were sandwiched together to form the drain section of the test structure. The flow channel bonded on top of the polyethylene microchip measured 3 mm × 23 mm. The drain reservoir under the array measured 4 mm × 6.6 mm and led to a waste channel with dimensions 12 mm × 1.5 mm. The flow chamber was sealed with a polycarbonate cover slip. Modular, fluidic buses used to connect the device to external fluid delivery systems were cut using a laser cutter (Universal Laser System; Scottsdale, AZ) and fabricated in-house (Fig. 1A).Preparation of micro-containers
Inverted CPF bead arrays containing positive features of 3 × 4 inverted PF on a base plate with dimensions 150 mm × 150 mm × 12.7 mm were designed in Solidworks 2010 (Waltham, MA). Measured from SEM images, each micro-container structure measured 623 ± 7.1 μm × 620 ± 1.9 μm at the top and 132 ± 4.5 μm × 130 ± 2.1 μm at the bottom with a height of 280 μm. Clipping radius, centered about the axis of each structure, measured 268 ± 4.6 μm at the top, 60 ± 2.5 μm at the bottom with a height of 280 μm. The height of the microstructure is the thickness of the plastic which can be measured using a Micrometer graduated in the hundredths of a millimeter (0.01 mm) from Starrett (Athol, MA). The part was micro-milled using a high level CNC mill onto a brass slab by Jauvtis Microprecision (Brighton, MA). The tolerance on features was 10 μm and the precision ± 5 μm.Inverted PF structures were anisotropically etched onto a 400 μm thick 10 mm diameter silicon wafer using standard anisotropic etching procedures. Briefly, a p-type 〈100〉 silicon wafer with a protective nitride coating, purchased from Nova Wafers (Flower Mound, TX), was cleaned using acetone and isopropyl alcohol. S1813 was spun onto the wafer for 3 s at a rate of 1000 rpm with an acceleration of 500 rpm s−1 followed by a secondary spin at 3000 rpm for 60 s with an acceleration of 500 rpm s−1. The wafer was soft baked on a hot plate at 115 °C for 60 s. The process was repeated for the reverse side. A 100 mm mylar photomask film, designed in AutoCAD (San Rafael, CA) with arrays containing 3 × 4 squares with dimensions 550 μm × 550 μm was purchased from Fineline (Colorado Springs, Co). The wafer was exposed through the mask with a MJB4 mask aligner (SUSS MicroTec; Garching, Germany) for 17 s followed by development using MF-319 (Rohm and Haas Electronic Materials; Marlborough, MA) for approximately 10 s. The patterned wafer was etched by reactive ion etching (Oxford Plasma Lab 80 Plus; Concord, MA) with a mixture of 45 standard cubic centimeters (sccm) CF4 and 5 sccm O2, at an ICP power of 60 W and RIE forward power of 100 W for 80 s at a pressure of 50 mT. The wafer was cleaned with acetone and then anisotropically etched in a double bath setup containing KOH overnight until inverted wells completely etched through the wafer.
Casting of epoxy stamp
A mixture of 100 g of polydimethylsiloxane (PDMS) was prepared on a weigh boat at a 100/20 ratio of prepolymer to casting agent. The mixture was centrifuged for 2.5 min at a rate of 2500 rpm and poured over the brass mold with micro-milled positive features and heated to 70 °C in front of a heating fan (Stanley; #675945) for 1 h. The cured PDMS containing negative features of the brass mold was peeled off from the mold.Negative PDMS mold containing inverted CPF structures and silicon mold with negative features of unclipped structures were placed on top of a 150 mm × 150 mm scotch tape surrounded by plastic slabs to shape the aluminum-based epoxy mold. Ease Release 200 (Smooth-On; Easton, PA) was sprayed on the silicon surface to facilitate removal from the mold. A mixture containing 75 g of the epoxy, PT4925 (PTM&W Industries; Santa Fe Springs, CA), was prepared on a weigh boat at a 100/9.5 ratio of PT4925A/PT4925B. The mixture was centrifuged at 2500 rpm for 2.5 min and then heated in front of a heating fan at 70 °C for 30 s to improve its viscosity. After epoxy was poured over the wafer, manual confirmation using a wooden stick ensured the epoxy filled all the negative features. The epoxy was then degassed under vacuum for 2 min and later cured under continuous heating at 70 °C for 3 h. The epoxy mold was released and then hard baked at incrementally increasing temperatures each hour at 66, 121, and 177 °C.
Hot embossing
A layer of polyethylene thermoplastic measuring 75 mm × 75 mm × 375 μm, purchased from McMaster-Carr (#86255K61; Santa Fe Springs, CA) was cut and placed over the epoxy. A layer of elastomer rubber (#86075K22) was placed on top of the polymer to serve as a buffer layer. These three layers were then sandwiched between pairs of aluminum plate (#86075K22), elastomer rubber (#5787T31), and Kapton film (#2271K1), all purchased from McMaster-Carr (Santa Fe Springs, CA). The stack was then hot embossed inside an Autoseries AutoFour 15-NE Press (Carver; Wabash, IN) for 10 min under a pressure of 680 kg and temperature of 160 °C. Water was delivered to the platens to cool the part for 5 min. After the part cooled to 100 °C, the mold and part were removed from the press.Computational modeling
Computational simulations were performed with COMSOL 3.5a (Burlington, MA) with the Chemical Engineering add-on, using methods previously described.37 Briefly, a three-dimensional model consisting of incompressible Navier–Stokes and convection–diffusion equations allowed for the simulation of analyte transport and modeling in a single well. The well, modeled and imported from AutoCAD, consisted either of a PF or CPF micro-well. For the PF micro-well, the diameter at the top was 550 × 550 μm, 100 × 100 μm at the bottom opening and a 280 μm height. For the CPF micro-well the radius at the top was 275 μm, 53 μm at the bottom opening and a 280 μm height. The pore size of the bead was 243 nm with a porosity value of 0.98, based on the average pore size of SEM images of a 2%weight agarose bead. The flow rate at the outlet boundary at the top of the well was set to 75% of the entrance with a set pressure at the drain at 1 atm. The tubing was set to 7.94 × 10−2 cm and the velocity at the inlet at 0.67 cm s−1. The binding association rate for CRP was 105 L mol−1 s−1 and dissociation rate was 10−5 s−1. The solution to the time-independent Navier–Stokes equation was obtained prior to the time-dependent convection–diffusion and reaction equations.For the flow rate dependence study, 50 ng mL−1 of CRP antigen (corresponding to 3.7 × 10−13 mol cm−3) was delivered to anti-CRP capturing antibody sites of 6.0 mg mL−1 (corresponding to 2.0 × 10−7 mol cm−3) at flow rates of 10, 25, 100 and 200 μL min−1 (1.67 × 10−4, 4.17 × 10−4, 1.67 × 10−3, 3.33 × 10−3 cm3 s−1) for 7 min at room temperature. In the cross-section plot parameters, the concentration of the antibody–antigen complex was plotted at the medial slice of the bead and the maximum bound concentration in mol cm−3 was normalized and compared to normalized experimental fluorescent values at the medial slice.
Reagents and buffers
All materials were purchased from Sigma-Aldrich, Inc. (St. Louis, MO) unless otherwise noted. Reagents and proteins were used without further purification unless further stated. To achieve working concentrations, all antibodies and proteins were diluted in PBS buffer at pH 7.2 (Thermo Fisher Scientific; Rockford, IL). The PBS buffer was prepared from powder packets, filtered using a Corning® bottle-top vacuum filter equipped with a nylon membrane of pore size 0.2 μm (Corning Incorporated; Corning, NY) and used up to one week when stored at room temperature. Note that the justification for using buffers rather that serum samples for this study is that this matrix could be reproduced easily throughout the entire time course of these measurements. Many prior studies have been completed using clinical samples derived from patients recruited from multiple trials.Preparation of agarose bead sensors
Homogeneous, 2%weight agarose beads were prepared using micro-emulsion chemistry. Briefly, a 2%weight agarose solution was prepared by dissolving 1.001 g of agarose in 49 mL of nanopure water and heated to 87 °C until fully dissolved. Simultaneously, a suspending solution consisting of 9 mL sorbitan trioleate in 91 mL of heptane was heated to 58 °C. Once dissolved, the agarose solution was allowed to cool to 62 °C and poured into the suspending solution while stirring at 550 rotations per minute (rpm) with an overhead stirrer with rod size 33 mm diameter × 230 mm height. The resulting reaction was allowed to cool to room temperature while continuously stirred. Beads were washed with a 50% ethanol–water solution, diluted down from 190-proof ethanol reagent (DLI; King of Prussia, PA). Beads with estimated diameters of 180, 212, 250 and 280 μm were collected using size selective sieves (Cole-Parmer; Vernon Hills, IL) and cross-linked with divinyl sulfone and glyoxal-activated using glycidol (Acros Organics; Fair Lawn, NJ).Coupling of CRP antibody to agarose beads
A volume of 500 μL of settled glyoxylated agarose beads of 280 μm diameter were conjugated overnight with 1000 μL of 2.0 mg mL−1 rabbit anti-human CRP polyclonal antibody (Fitzgerald; Concord, MA) and 60 μL of 0.43 M NaBH3CN. For the bead size dependency study, glyoxylated beads of 180, 212, 250 and 280 μm were conjugated in four separate reactions overnight. Each reaction consisted of 400 μL of 6.0 mg mL−1 mouse anti-human CRP monoclonal antibodies (Fitzgerald; Concord, MA) per 200 μL of beads plus 20 μL of 0.43 M NaBH3CN. Unreacted aldehyde groups on the beads were blocked with 50 mM Tris buffer at pH 7.0 for 1 h (Thermo Fisher Scientific; Rockford, IL).Coupling of free PSA antibody to agarose beads
A volume of 100 μL of settled glyoxylated agarose beads of 250 μm diameter were conjugated for 8 h with 102.6 μL of 7.8 mg mL−1 free PSA monoclonal antibody (Cal Bioreagents; San Mateo, CA) plus 113.4 μL of PBS and 12 μL of 0.43 M NaBH3CN. Unreacted aldehyde groups on the beads were blocked with 215 μL of 50 mM Tris buffer at pH 7.0 for 1 h (Thermo Fisher Scientific; Rockford, IL).Fluorescence-labeling of CRP antigen
The effects of bypass on capture between bead micro-containers were studied using 50 ng mL−1 of fluorescently-labeled CRP antigen labeled with AlexaFluor®488 (Invitrogen; Carlsbad, CA). Briefly, purified native human CRP (Fitzgerald; Concord, MA) was diluted to 2 mg mL−1, 50 μL of 1 M NaHCO3 was added, and the solution was reacted with 1 vial of bis (triethylammonium salt) (TFP ester) for 1 h, as per manufacturer's instructions. A 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 MW size-exclusion resin was used to purify the labeled antigen from unreacted dye and the concentrations of labeled antigen and the dye were determined from absorbance values at 280 and 494 nm, respectively. A concentration of 50 ng mL−1 of conjugated CRP with AlexaFluor®488 resulted in 4.51 moles of AlexaFluor®488 dye per mole of antigen and was aliquoted and stored in 0.1% bovine serum albumin (BSA) at −20 °C in the dark. The antigen was centrifuged prior to use at 1.5 × 1000 rpm for 5 min and the supernatant was used.
000 MW size-exclusion resin was used to purify the labeled antigen from unreacted dye and the concentrations of labeled antigen and the dye were determined from absorbance values at 280 and 494 nm, respectively. A concentration of 50 ng mL−1 of conjugated CRP with AlexaFluor®488 resulted in 4.51 moles of AlexaFluor®488 dye per mole of antigen and was aliquoted and stored in 0.1% bovine serum albumin (BSA) at −20 °C in the dark. The antigen was centrifuged prior to use at 1.5 × 1000 rpm for 5 min and the supernatant was used.
Fluorescence-labeling of detection antibody for detection of free PSA
The detection antibody to detect free PSA was purchased from Fitzgerald (Concord, MA) and was labeled with AlexaFluor®488 (Invitrogen; Carlsbad, CA) as per manufacturer's instructions and briefly described in the section above. The resultant stock concentration was diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 25 before use.
25 before use.
Assay delivery
For the flow rate dependence study, 50 ng mL−1 of fluorescently-labeled CRP antigen was delivered using a programmable NE-1000 syringe pump from New Era Pump systems Inc. (Farmingdale, NY) at flow rates of 10, 25, 100 and 200 μL min−1 for 7 min. Sensor beads labeled with 6 mg mL−1 of anti-CRP monoclonal antibodies were used.For the penetration depth study, 36 mL of 50 ng mL−1 fluorescently-labeled CRP was re-circulated for 2 h using a peristaltic pump (FiaLAB, Bellevue, WA) at 200 μL min−1. Sensor beads labeled with 2 mg mL−1 of anti-CRP polyclonal antibodies were used.
For the sandwich immunoassay standard curves, PSA antigen was purchased from Fitzgerald (Concord, MA) and concentrations of 5, 2, 1, 0.5, 0.2, and 0.1 ng mL−1 were prepared by serial dilutions. A volume of 100 μL of sample and 8 μL of the detection antibody was loaded into the fully integrated p-BNC card as previously described.39 The sample was delivered at 5 μL min−1 for 20 min. A PBS buffer wash was delivered at 100 μL min−1 for 1 min. The detection antibody was delivered at 20 μL min−1 for 3 min. A final PBS buffer wash was delivered at 100 μL min−1 for 4 min. The total assay time was 28 min and the total assay volume used was 660 μL. Sensor beads labeled with 2.53 mg mL−1 of anti-free PSA monoclonal antibodies were used.
Bead loading
For loading the beads onto the chip, a drop of PBS buffer, pH 7.2, containing 0.1% BSA and 20% isopropyl alcohol (VWR, West Chester, PA) was placed on the chip. Adding 20% isopropyl alcohol in PBS buffer helped to seal the beads in the micro-container tightly as the material of the micro-container is hydrophobic. For all assays, beads were loaded onto the chip arrays using tweezers under the microscope and a cover slip was placed over the loaded beads before starting the assay. To pre-wet the chips, block, and wash away nonspecific binding, PBS buffer, pH 7.2, containing 0.1% BSA was used and delivered at a flow rate of 100 μL min−1 for 1 min; prior and after the antigen delivery.Image analysis and data collection
A DVC 1312 camera (DVC; Austin, TX) mounted to an Olympus BX-2 microscope (Center Valley, PA) with 4× objectives, saved 8-bit TIFF at 0.1 s exposure for the flow rate study and 3 s exposure for the penetration depth study. Images were split into respective red, green and blue (RGB) components with the green channel used for analysis. Beads free of bubbles and debris were manually located and analyzed using a custom Java-based macro in ImageJ (NIH; Bethesda, MD). A collection of mean values consisting of the maximum fluorescent intensity of horizontal line profiles, scanned from the medial slice of the bead, provided the resultant signal intensity value on the bead. SEM images were acquired using a Hitachi TM3000 and SU6600 (Pleasanton, CA) for the imaging of the micro-structures and agarose pore size, respectively. Confocal images were acquired using a LSM 510 META laser scanning microscope system equipped with 10×/0.45 Plen-apochromat objective lens from Carl Zeiss MicroImaging (Gena, Germany) equipped with an argon laser at 50% output. Configuration controls to accommodate excitation and emission maxima of AlexaFluor®488 were applied at 495 and 519 nm, respectively. A laser power of 100% was used for fluorescence excitation at 488 nm and Z-stacks of 1 μm height were acquired using a pinhole of 396 and detector gain at 700.Conflict of interest
Principal Investigator, John T. McDevitt, has an equity interest in SensoDX, LLC. Inc, and also serves on Scientific Advisory Board.Acknowledgements
Funding was provided by through the National Institute of Dental and Craniofacial Research (NIH Grant no. 3 U01 DE017793-02S1 and 5 U01 DE017793-2), the Cancer Prevention Research Institute of Texas (RP140024), and the National Cancer Institute (P30 CA054174 and U01 CA086402). The content is solely the responsibility of the authors and does not necessarily represent or reflect views of the NIH, state of Texas or the Federal Government. The authors also would like to thank scientists from Scienion AG (Berlin, Germany) for providing images presented in the ESI.†References
- S. K. Sia, IEEE Pulse, 2011, 2, 35–39 CrossRef PubMed.
- S. K. Kim, D. R. Burris, J. Bryant-Genevier, K. A. Gorder, E. M. Dettenmaier and E. T. Zellers, Environ. Sci. Technol., 2012, 46, 6073–6080 CrossRef CAS PubMed.
- B. R. Schudel, B. Harmon, V. V. Abhyankar, B. W. Pruitt, O. A. Negrete and A. K. Singh, Lab Chip, 2013, 13, 811–817 RSC.
- P. S. Dittrich and A. Manz, Nat. Rev. Drug Discovery, 2006, 5, 210–218 CrossRef CAS PubMed.
- T. Vilkner, D. Janasek and A. Manz, Anal. Chem., 2004, 76, 3373–3385 CrossRef CAS PubMed.
- S. Haeberle and R. Zengerle, Lab Chip, 2007, 7, 1094–1110 RSC.
- A. W. Martinez, S. T. Phillips, G. M. Whitesides and E. Carrilho, Anal. Chem., 2010, 82, 3–10 CrossRef CAS PubMed.
- B. L. Schmidt, J. Clin. Microbiol., 2004, 42, 2833–2835 CrossRef CAS PubMed.
- C. Liu, M. G. Mauk, R. Hart, M. Bonizzoni, G. Yan and H. H. Bau, PLoS One, 2012, 7, e42222 CAS.
- S. Wang, S. Tasoglu, P. Z. Chen, M. C. Chen, R. Akbas, S. Wach, C. I. Ozdemir, U. A. Gurkan, F. F. Giguel, D. R. Kuritzkes and U. Demirci, Sci. Rep., 2014, 4, 1–9 Search PubMed.
- F. B. Mayer, R. H. Henrikson, J. Bone and L. P. Lee, PLoS One, 2013, 8, e60159 Search PubMed.
- V. Gau and D. Wong, Ann. N. Y. Acad. Sci., 2007, 1098, 401–410 CrossRef CAS PubMed.
- N. Zhang, Z. Zhang, S. Feng, Q. Wang, D. Malamud and H. Deng, Anal. Chim. Acta, 2013, 774, 61–66 CrossRef CAS PubMed.
- J. Verbarg, W. D. Plath, L. C. Shriver-Lake, P. B. Howell, J. S. Erickson, J. P. Golden and F. S. Ligler, Anal. Chem., 2013, 85, 4944–4950 CrossRef CAS PubMed.
- C. D. Chin, V. Linder and S. K. Sia, Lab Chip, 2007, 7, 41–57 RSC.
- A. Groisman, M. Enzelberger and S. R. Quake, Science, 2003, 300, 955–958 CrossRef CAS PubMed.
- J. Hu, L. Wang, F. Li, Y. L. Han, M. Lin, T. J. Lu and F. Xu, Lab Chip, 2013, 13, 4352–4357 RSC.
- C. D. Chin, V. Linder and S. K. Sia, Lab Chip, 2012, 12, 2118–2134 RSC.
- K. S. Anderson, Methods Mol. Biol., 2011, 723, 227–238 CAS.
- C. L. Hansen, E. Skordalakes, J. M. Berger and S. R. Quake, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 16531–16536 CrossRef CAS PubMed.
- E. Verpoorte, Lab Chip, 2003, 3, 60N–68N RSC.
- C. T. Lim and Y. Zhang, Biosens. Bioelectron., 2007, 22, 1197–1204 CrossRef CAS PubMed.
- W. H. Tan and S. Takeuchi, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 1146–1151 CrossRef CAS PubMed.
- J. Nilsson and M. Evander, Anal. Chim. Acta, 2009, 649, 141–157 CrossRef CAS PubMed.
- T. F. Leary, S. Manafirasi and C. Maldarelli, Lab Chip, 2015, 15, 459–477 RSC.
- Y. Zhao, X. Zhao, C. Sun, J. Li, R. Zhu and Z. Gu, Anal. Chem., 2008, 80, 1598–1605 CrossRef CAS PubMed.
- S. Nahavandi, S. Baratchi, R. Soffe, S. Tang, S. Nahavandi, A. Mitchell and K. Khoshmanesh, Lab Chip, 2014, 14, 1496–1514 RSC.
- H. C. Tekin and M. A. M. Gijs, Lab Chip, 2013, 13, 4711–4739 RSC.
- J. A. Thompson and H. H. Bau, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2010, 878, 228–236 CrossRef CAS PubMed.
- Z. Yang, Z. Fu, F. Yan, H. Liu and H. Ju, Biosens. Bioelectron., 2008, 24, 35–40 CrossRef CAS PubMed.
- T. Song, Q. Zhang, C. Lu, X. Gong, Q. Yang, Y. Li, J. Liu and J. Chang, J. Mater. Chem., 2011, 21, 2169–2177 RSC.
- D. Sung, S. Yang, J. W. Park and S. Jon, Biomed. Microdevices, 2013, 15, 691–698 CrossRef CAS PubMed.
- P. E. Gustavsson, A. Axelsson and P. O. Larsson, J. Chromatogr. A, 1999, 830, 275–284 CrossRef CAS PubMed.
- Y. Xiao and X. Zhou, React. Funct. Polym., 2008, 68, 1281–1289 CrossRef CAS.
- J. A. Thompson and H. H. Bau, Anal. Chem., 2011, 83, 2858–2861 CrossRef CAS PubMed.
- A. Ouyang and J. Liang, RSC Adv., 2014, 4, 25835 RSC.
- J. Chou, A. Lennart, J. Wong, M. F. Ali, P. N. Floriano, N. Christodoulides, J. Camp and J. T. McDevitt, Anal. Chem., 2012, 84, 2569–2575 CrossRef CAS PubMed.
- N. Christodoulides, P. N. Floriano, C. S. Miller, J. L. Ebersole, S. Mohanty, P. Dharshan, M. Griffin, A. Lennart, K. L. Ballard, C. P. King Jr, M. C. Langub, R. J. Kryscio, M. V. Thomas and J. T. McDevitt, Ann. N. Y. Acad. Sci., 2007, 1098, 411–428 CrossRef CAS PubMed.
- B. H. Shadfan, A. R. Simmons, G. W. Simmons, A. Ho, J. Wong, K. H. Lu, R. C. Bast Jr and J. T. McDevitt, Cancer Prev. Res., 2015, 8, 37–48 CrossRef CAS PubMed.
- J. V. Green, T. Kniazeva, M. Abedi, D. S. Sokhey, M. E. Taslim and S. K. Murthy, Lab Chip, 2009, 9, 677–685 RSC.
- S. Kobel, A. Valero, J. Latt, P. Renaud and M. Lutolf, Lab Chip, 2010, 10, 857–863 RSC.
- D. Wlodkowic, J. Skommer, D. McGuinness, S. Faley, W. Kolch, Z. Darzynkiewicz and J. M. Cooper, Anal. Chem., 2009, 81, 6952–6959 CrossRef CAS PubMed.
- S. Faley, K. Seale, J. Hughey, D. K. Schaffer, S. VanCompernolle, B. McKinney, F. Baudenbacher, D. Unutmaz and J. P. Wikswo, Lab Chip, 2008, 8, 1700–1712 RSC.
- G. Hu, Y. Gao and D. Li, Biosens. Bioelectron., 2007, 22, 1403–1409 CrossRef CAS PubMed.
- D. Yan, C. Yang, J. Miao, Y. Lam and X. Huang, Electrophoresis, 2009, 30, 3144–3152 CrossRef CAS PubMed.
- S. Nagrath, L. V. Sequist, S. Maheswaran, D. W. Bell, D. Irimia, L. Ulkus, M. R. Smith, E. L. Kwak, S. Digumarthy, A. Muzikansky, P. Ryan, U. J. Balis, R. G. Tompkins, D. A. Haber and M. Toner, Nature, 2007, 450, 1235–1239 CrossRef CAS PubMed.
- D. E. Hertzog, B. Ivorra, B. Mohammadi, O. Bakajin and J. G. Santiago, Anal. Chem., 2006, 78, 4299–4306 CrossRef CAS PubMed.
- A. L. Page, N. de Rekeneire, S. Sayadi, S. Aberrane, A. C. Janssens, M. Dehoux and E. Baron, Pediatrics, 2014, 133, e363–e370 CrossRef PubMed.
- J. G. Kiang and G. D. Ledney, Oxid. Med. Cell. Longevity, 2013, 2013, 1–10 CrossRef PubMed.
- J. W. L. Cals, C. C. Butler, R. M. Hopstaken, K. Hood and G.-J. Dinant, Br. Med. J., 2009, 338, b1374–b1384 CrossRef PubMed.
- S. K. Hong, BioMed Res. Int., 2014, 2014, 1–10 Search PubMed.
- A. I. Barbosa, A. P. Castanheira, A. D. Edwards and N. M. Reis, Lab Chip, 2014, 14, 2918–2928 RSC.
- A. A. Luderer, Y.-T. Chen, T. F. Soriano, W. J. Kramp, G. Carlson, C. Cuny, T. Sharp, W. Smith, J. Petteway and M. K. Brawer, Urology, 1995, 46, 187–194 CrossRef CAS PubMed.
- D. Bartholomeusz, R. W. Boutte and J. D. Andrade, J. Microelectromech. Syst., 2005, 14, 1364–1374 CrossRef.
- J. V. Jokerst, J. Chou, J. P. Camp, J. Wong, A. Lennart, A. A. Pollard, P. N. Floriano, N. Christodoulides, G. W. Simmons, Y. Zhou, M. F. Ali and J. T. McDevitt, Small, 2011, 7, 613–624 CrossRef CAS PubMed.
- I. Nischang, F. Svec and J. M. Frechet, J. Chromatogr. A, 2009, 1216, 2355–2361 CrossRef CAS PubMed.
- J. Chou, J. Wong, N. Christodoulides, P. N. Floriano, X. Sanchez and J. T. McDevitt, Sensors, 2012, 12, 15467–15499 CrossRef PubMed.
- K. B. Kosto and W. M. Deen, Biophys. J., 2005, 88, 277–286 CrossRef CAS PubMed.
- H. Parsa, C. D. Chin, P. Mongkolwisetwara, B. W. Lee, J. J. Wang and S. K. Sia, Lab Chip, 2008, 8, 2062–2070 RSC.
- P. M. Ridker, Circulation, 2003, 107, 363–369 CrossRef PubMed.
- A. Shrivastava and V. B. Gupta, Chron. Young Sci., 2011, 2, 21–25 CrossRef.
- S. Park, H. J. Lee and W.-G. Koh, Sensors, 2012, 12, 8426–8436 CrossRef CAS PubMed.
- N. Christodoulides, M. Tran, P. N. Floriano, M. Rodriguez, A. Goodey, M. Ali, D. Neikirk and J. T. McDevitt, Anal. Chem., 2002, 74, 3030–3036 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5ra07910a |
| ‡ Authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2015 |