Open Access Article
Open Access ArticleGraphene as a template and structural scaffold for the synthesis of a 3D porous bio-adsorbent to remove antibiotics from water†
Yuan
Zhuang
a,
Fei
Yu
ac,
Jie
Ma
*a and
Junhong
Chen
ab
aState Key Laboratory of Pollution Control and Resource Reuse, School of Environmental Science and Engineering, Tongji University, 1239 Siping Road, Shanghai 200092, P. R. China. E-mail: jma@tongji.edu.cn; Tel: +86-21-6598-1831
bDepartment of Mechanical Engineering, University of Wisconsin–Milwaukee, Milwaukee, WI 53211, USA
cCollege of Chemistry and Environmental Engineering, Shanghai Institute of Technology, Shanghai 2001418, P. R. China
First published on 12th March 2015
Abstract
A graphene–soy protein (GS) aerogel was prepared by a simple thermal reduction method and then used as an adsorbent for the removal of antibiotics. The GS aerogel was characterized by an optical contact angle meter, scanning electron microscopy (SEM), transmission electron microscopy (TEM), X-ray diffraction (XRD), Raman, Branauer–Emmett–Teller (BET) and Fourier transform infrared spectroscopy (FTIR). In GS, graphene acts as a template that loads onto the surface of the protein through hydrogen bonds to form a layered bulk unit and interacts with each other to form self-assembled hydrogels. Moreover, graphene interacts well with protein without obvious structural damage and does not agglomerate. The resulting GS has a high specific area of 30.07 m2 g−1 with abundant microspores and excellent hydrophilic properties, which lead to excellent adsorption properties for tetracycline (500.0 mg g−1) and ciprofloxacin (500.0 mg g−1). This result suggests that the small quantity of graphene assisted the protein to form an excellent bio-adsorbent.
Introduction
Natural polymers such as vegetable proteins have attracted considerable research interest because of their availability, biodegradability, renewability, and interesting functional properties. Proteins extracted from vegetable seeds (e.g., soybean, pea, barley, wheat, rice, oat, sunflower) have been reported to have good emulsifying and foaming capacities, water solubility, and amphiphilic and film-forming properties. Soybeans have been cultivated for more than 3000 years in China and other Asian countries as an important protein source and for use in various forms.1 Despite considerable public and commercial interest in soybean products as food, the proportion of soy protein consumed directly in human nutrition and other industrial uses is quite small, leaving a wide opportunity to seek new uses for soy protein. Utilization of protein will help to overcome environmental problems and create added values to agricultural by-products.2 Due to the presence of abundant functional groups, proteins can interact with various pollutants;3 however, proteins normally have a low specific surface area (SSA) and pore volume. Moreover, they are difficult to separate and recycle after adsorption due to their high solubility.In the past several years, free-standing two-dimensional monolayer graphene with excellent properties has caught global attention and has been adopted for various applications.4–6 Graphene has been considered as an excellent adsorbent in environmental applications due to their high SSA (2630 m2 g−1). With its delocalized π bonds, graphene can potentially adsorb organic contaminants, especially those with molecules containing π-electrons that can interact with the polarized graphene surface via π–π electron coupling or van der Waals interactions.7,8 Compared with polymers, graphene composites may have better hydrophilicity, biological compatibility, and lower cytotoxicity; moreover, gels can be prepared at a large scale by a facile gelation process in a short period of time. However, graphene nanosheets tend to aggregate due to interplanar interactions.9 In addition, most forms of graphene materials are not well dispersible or soluble in most common solvents due to their low hydrophilicity, poor biocompatibility and few functional groups, and high cost. These above factors limit their applications as adsorbents. The use of biocompatible hydrophilic biopolymers may improve water solubility properties of nanomaterials.
Therefore, it is reasonable to predict that the biopolymer-mediated graphene gels may function as porous adsorbents with satisfactory adsorption capacity and limited toxicity for applications in wastewater treatment.10 As an adsorbent, three-dimensional (3D) graphene adsorbents can be easily separated in the aqueous solution. Developing hydrophilic and biocompatible 3D bio-adsorbents with a large SSA and unique mesoporosity will expand their significance in environmental applications.11 Cheng et al.10 prepared three typical graphene oxide (GO)–biopolymer gels (bovine serum albumin, chitosan, and double-stranded DNA) for the first time and investigated the adsorption capabilities of dyes and heavy metals. The GO–biopolymer gels displayed an adsorption capacity as high as 1100 mg g−1 for methylene blue dye and 1350 mg g−1 for methyl violet dye, respectively. Thus, it can be seen that graphene and polymers can form gels to be used as bio-adsorbents; however, the combination ways of graphene with polymers and the characteristics of the resulting materials still need further study.
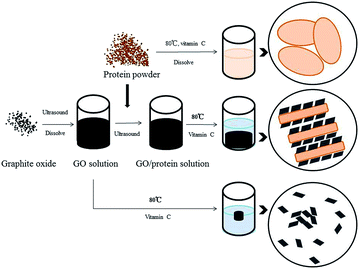
In this paper, a graphene assisted 3D porous soy protein aerogel (GS) is prepared by a simple thermal reduction method, and then the GS is used as an adsorbent for the removal of antibiotics from aqueous solutions. This method is inexpensive, simple, and features a high-yield. In the synthesis process of GS, graphene acts as a template that is loaded onto the surface of the protein to form a layered bulk unit. More importantly, graphene also acts as a structural scaffold to form 3D hydrogels through self-assembly, which can be turned into porous aerogels after freeze drying. The protein combines well with graphene in the GS, and the resulting GS possesses high hydrophilicity and excellent adsorption properties for antibiotics.
Experimental
Materials
All chemicals were purchased from Sinopharm Chemical Reagent Co., Ltd. (Shanghai, China) in analytical purity and were used in the experiments without any further purification. All solutions were prepared using deionized water.Preparation of graphene–soy protein aerogels
Graphite oxide was obtained using the modified Hummers' method,12–14 dispersed in deionized water, and sonicated in an ultrasound bath for 12 h. Soy protein and ascorbic acid were added to the GO dispersion and placed into an ultrasound bath for 5 h to form a uniform solution. The mass ratio of graphene to soy protein was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6; the resulting products were denoted as GS. This mass ratio was selected according to the adsorption performance, as shown in Fig. S1;† hydrogels with six different mass ratios are shown in Fig. S2.† The mixture was heated in a water bath under 90 °C for 12 h to form hydrogels. The aerogels were synthesized after the hydrogels were washed with distilled water for several times and then freeze-dried for 24 h. For comparison, graphene aerogel (GN) without protein was also prepared as described above.
6; the resulting products were denoted as GS. This mass ratio was selected according to the adsorption performance, as shown in Fig. S1;† hydrogels with six different mass ratios are shown in Fig. S2.† The mixture was heated in a water bath under 90 °C for 12 h to form hydrogels. The aerogels were synthesized after the hydrogels were washed with distilled water for several times and then freeze-dried for 24 h. For comparison, graphene aerogel (GN) without protein was also prepared as described above.
Characterization methods
The surface morphologies of GN and GS were visualized using a field-emission SEM (Hitachi, S-4800), AFM (NanoScope III a MultiMode) and TEM (JEOL, JEM-2010). The hydrophilicities were characterized by an optical contact angle meter (Dataphysics, OCA-20). The surface functional groups were observed by FTIR (NEXUS, 670). Measurements of micro-Raman spectra were carried out using a Raman Scope system (LabRam, 1B) with a 532 nm wavelength incident laser light and 20 mW power. XRD were collected on a Bragg–Brentano diffractometer (Rigaku, D/Max-2200) with monochromatic Cu Kα radiation (λ = 1.5418 Å) of a graphite curve monochromator, and the data were collected from 2θ = 2–40° at a continuous scan rate of 2° min−1 for phase identification. The BET isotherms were measured by an Accelerated Surface Area and Porosimetry system (Micromeritics, ASAP 2020). A UV-visible absorption-based approach is used for a direct evaluation of the protein content released from GS.15,16 A calibration plot is then made by monitoring the intensity of the peak as a function of the true concentration of the protein. Earlier reference stated that the adsorption peak at 280 nm is a signature of the soy protein. Thus, we build a calibration curve by monitoring the adsorbance value for this peak at 280 nm as a function of soy protein concentration, and then we are able to estimate the unknown concentration of soy protein in the solution by measuring its optical absorbance.Batch sorption experiments
Batch experiments were conducted to evaluate the adsorption performance of antibiotics on the adsorbents. GN and GS were selected as adsorbents for antibiotics adsorption in an aqueous solution. Then, 100 mg L−1 stock solution was prepared by dissolving 100 mg antibiotics in 1 L deionized water. Working solutions of the required concentrations were obtained by diluting the stock solution with deionized water. All the sorption tests were conducted in well-capped 100 mL flasks containing 20 mL antibiotics solutions with the required concentration. After adding 10 mg GS aerogel, the flasks were shaken in a thermostatic shaker at 150 rpm at 298 K for 24 h. All the adsorption experiments were conducted in duplicate, and only the mean values were reported. The maximum deviation for the duplicates was usually less than 5%. The blank experiments without the addition of adsorbent were conducted to ensure the decrease in the concentration was actually due to the adsorbent rather than by the adsorption on the glass bottle wall. After adsorption, the adsorbent was separated using a 0.45 μm membrane. The residual concentrations in the solution were determined by an ultraviolet spectrophotometer (Tianmei UV-2310(II)) at 360 nm for tetracycline and 270 nm for ciprofloxacin. The adsorption isotherm was studied at pH = 6, and the initial concentration was set from 1 mg L−1 to 50 mg L−1. The adsorption capacity (mg g−1) was calculated using eqn (1).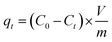 | (1) |
Adsorption isotherms were fitted using Langmuir and Freundlich models, which were used to evaluate the adsorption equilibrium, as shown in eqn (2) and (3). The Langmuir isotherm assumes the adsorbate forms a monolayer around the homogenous surface of the adsorbent and there is no interaction between the adsorbed molecules. The Freundlich model is empirical, which assumes that adsorption takes place on a heterogeneous surface, and proposes multilayer sorption with interactions among the adsorbed molecules. Another important parameter, RL, called the separation factor or equilibrium parameter, can be used to determine the feasibility of adsorption in a given concentration range over adsorbent, as shown by eqn (4).17
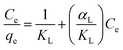 | (2) |
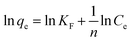 | (3) |
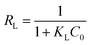 | (4) |
Results and discussion
Morphological and microstructure of GN and GS
Digital images of graphite oxide, protein, GS, and GN hydrogels are shown in Fig. 1a. It can be observed that both GN and GS hydrogels have uniform structures; however, the GN hydrogel is much looser than the GS hydrogel, and the GN hydrogel floats while the GS hydrogel remains at the bottom. There may be two reasons for their different locations in water: first, with a looser porous structure, the GS hydrogel could conserve more water; second, the protein in the GS hydrogel may have helped absorb more water. The GS with a much larger dimension indicates that protein and graphene combine well in this biocomposite and they support each other to form a more porous bulk, under which condition the protein restrains graphene from agglomeration, while graphene combines with protein to form a more ordered and larger structure.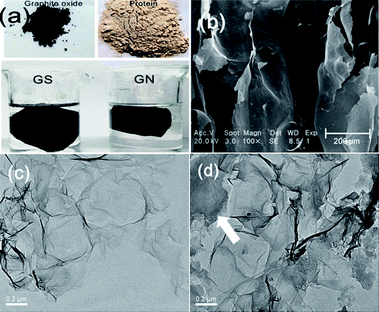 | ||
| Fig. 1 Morphology and microstructure of GN and GS: (a) photographs of GN and GS hydrogels, (b) SEM of GS, (c) TEM of GN, and (d) TEM of GS. | ||
To verify the combination of graphene and protein, we investigated the microstructure of GN and GS aerogels. An SEM of GS is shown in Fig. 1b, and it can be easily seen that GS has a rough structure with layers, indicating pore interactions between nanofillers and the matrix.18 As a result, the proteins are separated into layers of graphene. Because graphene has some functional groups on the edge, it is possible for it to bind macromolecule proteins with its edges. For comparison, we also heated proteins without graphene and found that proteins alone cannot form hydrogels. From Fig. 1d, it can be seen that in GS the protein (as pointed by the arrow in Fig. 1d) exists on the edge of graphene, and in GS the graphene around the protein appears to be much closer together than in GN as shown in Fig. 1c. This phenomenon indicates that there are attraction forces between graphene and protein. Graphene sheets tend to be much closer in GS, indicating there are chemical functions of attraction between graphene and protein. Huang et al.19 has described the combination between graphene and protein as the “bricks and mortar” structure in which the hydrogels can be transformed into a lamellar structure simply by solution casting and drying, and the orientation of protein-coated GO sheets during the drying process led to the formation of the layered structure through water-evaporation-induced self-assembly. Thus, we deduce that in our study graphene acts as a template, as shown in Scheme 1, because graphene loads on the surface of the protein with orientation to form a layered bulk unit, and each graphene sheet interacts with another graphene sheet to form hydrogels through self-assembly. AFM and EDS of GS were shown in Fig. S3.†
Composition and structure analysis
XRD spectra of GN and GS are presented in Fig. 2a, and Raman spectra of GN and GS are presented in Fig. 2b. In Fig. 2a, no peak appears at 2θ of 10.6 degree, proving that graphene oxide is partially reduced during the hydrogel preparation for both GN and GS. In Fig. 3c, 2θ of around 25 degree of GN is the characteristic peak of freeze-dried graphene aerogel,20 which further proves that the nanosheet in this biocomposite is graphene rather than graphene oxide. Moreover, the characteristic peak in GS is to the left of that of GN. Since the lattice parameters of GS are larger than GN, this further proves that the protein prevented the graphene from agglomeration. In Fig. 2b, the presence of disorder sp2-hybridized carbon systems leads to rich and intriguing phenomena in their resonance Raman spectra: point defects are formed and the Raman spectra of the disordered graphene exhibit two new sharp features appearing at 1345 and 1626 cm−1. These two features have, respectively, been called D and G bands, to denote disorder.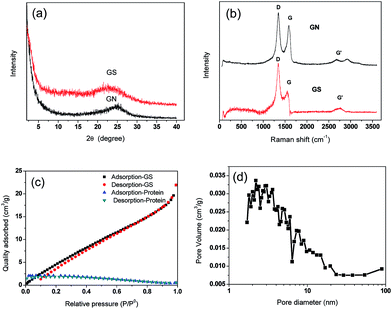 | ||
| Fig. 2 Composition and structure analysis (a) XRD of GN and GS, (b) Raman of GN and GS, (c) N2 adsorption and desorption curves of GS and protein, (d) pore size distribution of GS. | ||
All sp2 carbon materials exhibit a strong Raman feature called G′ band,18 which appears in the range 2500–2800 cm−1. The G′ band, which is assigned as the distinct band of graphene induced by a two-phonon resonant scattering process, becomes broader after composition at 2700 cm−1, and could also explain that GO was partially reduced.21 The slight increase of the intensity ratio of the D peak to G peak (ID/IG) for the GS compared with GN demonstrated the successful incorporation of graphene into protein without obvious structural damage.22Fig. 2c shows the N2 adsorption–desorption isotherms of GS and protein. The N2 adsorption–desorption isotherms of GN are shown in Fig. S3a,† and the pore distributions are shown in Fig. S3b.† The isotherms of GN exhibit a typical type-I curve and a hysteresis loop at a relative pressure of 0.4, indicating the presence of slit-shaped pores between parallel layers of graphene.23 The protein has nearly no adsorption ability for N2, as it does not contain any pores. Although the GS primarily contains protein, the GS has an obvious increase in N2 adsorption. The specific areas of GS and GN are 30.07 m2 g−1 and 119.17 m2 g−1 respectively, while the protein specific area is about 0 m2 g−1. The pore size distribution of GS is also shown in Fig. 2d. The GS and GN have the same tendency in the curves of pore volume and have peaks in the same position of around 2 nm, indicating the graphene maintains its nanopores in GS. Moreover, because the protein did not close the pores of graphene, it can be deduced that the combination force between the graphene and the protein may be a hydrogen bond. The macropore, mesopore, and micropore of GS are 0.009, 0.714, and 0.120 cm3 g−1, respectively. The average pore size of GS is 10.23 nm.
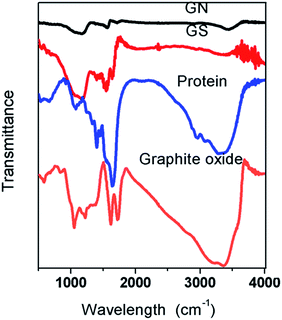
Fig. 3 shows the FTIR spectra of graphite oxide, protein, GS, and GN. It can be seen that both graphite and protein have a broad band between 3200 cm−1 and 3400 cm−1, which is attributed to the O–H stretching vibrations.21 The band nearly disappears in the two aerogels, thereby proving that GO has been reduced. In graphite oxide, the absorption band at 1720 cm−1 is the characteristic band of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O groups in carbonyl and carboxyl moieties and the band at 1089 cm−1 is assigned to the C–O bonds; these peaks are weak in both GN and GS aerogels. Peaks at 1630 cm−1 (asymmetric stretching vibration of the –COO− groups) were observed for graphene and 1400 cm−1 (symmetric stretching vibration of the –COO− groups) was observed for proteins;24 these peaks are in GS. In the aerogels, the disappearance of oxygen-containing functional groups shows that the GO has been reduced.25 In GS, there is a strong band at 1227 cm−1 which is assigned to the C–OH, while in the GN and graphite oxide this band is relatively weaker, which indicates that hydrogen bonds are formed between protein and graphene. Bands at 1562 cm−1 in protein are assigned to the N–H bending of –NH2 groups;26 in GS, they are much weaker than in protein and thus it can be inferred that –NH2 groups in protein and –COO− groups in graphene formed hydrogen bonds in GS.
O groups in carbonyl and carboxyl moieties and the band at 1089 cm−1 is assigned to the C–O bonds; these peaks are weak in both GN and GS aerogels. Peaks at 1630 cm−1 (asymmetric stretching vibration of the –COO− groups) were observed for graphene and 1400 cm−1 (symmetric stretching vibration of the –COO− groups) was observed for proteins;24 these peaks are in GS. In the aerogels, the disappearance of oxygen-containing functional groups shows that the GO has been reduced.25 In GS, there is a strong band at 1227 cm−1 which is assigned to the C–OH, while in the GN and graphite oxide this band is relatively weaker, which indicates that hydrogen bonds are formed between protein and graphene. Bands at 1562 cm−1 in protein are assigned to the N–H bending of –NH2 groups;26 in GS, they are much weaker than in protein and thus it can be inferred that –NH2 groups in protein and –COO− groups in graphene formed hydrogen bonds in GS.
Antibiotics adsorption
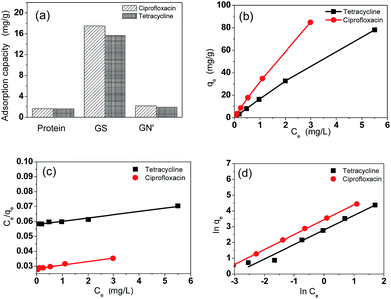
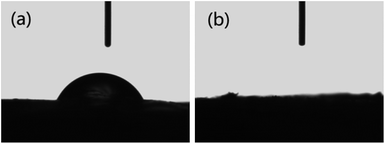
Fig. 4a shows the adsorption capacities of protein, GS and GN′. GN′ is a hypothetical material which has the same composition as GS, but its adsorption capacity was calculated by algebraic summing of each composition. It can be seen that the adsorption capacity of GS is obviously much larger than the algebraic summation of each component in GS. This result suggests that the small quantity of graphene assisted the protein to form an excellent bio-adsorbent. The adsorption isotherm indicates the distribution relationship of the adsorbate molecules between the liquid phase and the solid phase when the adsorption process reaches equilibrium. Fig. 4b shows the relationship between Ce and qe. The adsorption isotherms were calculated by Langmuir and Freundlich models, as shown in Fig. 4c and d, respectively. The relative parameters calculated from the Langmuir and Freundlich models are listed in Table 1. Based on the R2 values, it can be seen from Table 1 that the adsorption isotherms fit well by both the Langmuir and the Freundlich models. It could be calculated from the Langmuir isotherm equation that the maximum theoretical adsorption capacities of the two antibiotics on GS were both 500.0 mg g−1, which were relatively higher than other adsorbents, as shown in Tables 2 and 3. The excellent adsorption ability of the GS is due to the large specific surface area, abundant functional groups, and high hydrophilicity as suggested by BET, FTIR and contact angle measurements. Contact angles of GN aerogels and GS aerogels are shown in Fig. 5a and b. The contact angle of GS aerogel was 0 degrees, which further proved the hydrophilic protein helped the hydrogels to achieve high hydrophilicity. The good regression coefficients of the Langmuir isotherm model, which can be seen in Table 1, suggest a good affinity between tetracycline and aerogel. The values of RL (0.85 for ciprofloxacin and 0.74 for tetracycline) between 0 and 1 indicate that the adsorption is favorable. Moreover, the n values for the Freundlich isotherm (1.1 for both ciprofloxacin and ciprofloxacin) are between 1 and 10, further proving that the adsorption is favourable under the studied conditions. Fig. S4† shows the loss of adsorption capacity of adsorbent which was reused 10 times after regeneration. The loss of adsorption capacity in the second cycle was around 40% and after 10 times it was around 60% which demonstrated an extended useful lifetime for the adsorbent.| Isotherm | Isotherm | Ciprofloxacin | Tetracycline |
|---|---|---|---|
| Langmuir | q m (mg g−1) | 500 | 500 |
| K L | 17.5 | 35.7 | |
| R 2 | 0.978 | 0.967 | |
| Freundlich | K F | 16.3 | 31.3 |
| n | 1.1 | 1.1 | |
| R 2 | 0.972 | 0.999 |
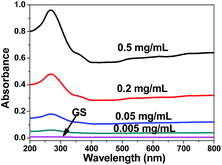
After GS separation from aqueous solution, the concentration of residual soy protein in an aqueous solution was estimated using a UV-visible absorption-based approach, it is noteworthy to observe that the concentration of residual soy protein has very low absorbance, as shown in Fig. 6, which clearly indicated the protein had good combination with graphene, soy protein have not been deserted in the adsorption experiment of antibiotics. Hence, it can be seen that the GS has significant potential to be used as an adsorbent.
Therefore, the GS biocomposite obtained many advantages through composition, such as high hydrophilicity, high specificity, and abundant functional groups, which is beneficial for the adsorption of pollutants. Graphene not only brings nano-pores into protein, but also forms a template and a scaffold for protein to form a self-assembled porous hydrogel. With a low graphene content, this biocomposite also is a cost-effective solution.
Conclusions
A graphene-assisted 3D porous soy protein aerogel (GS) is prepared by a simple method and then used as an adsorbent for the removal of antibiotics. The GS aerogel has good hydrophilicity and abundant functional groups. Moreover, unlike protein which does not contain any nano-pores, the GS has a large specific surface area. The graphene in GS turned out to be separated by protein and was prevented from aggregation. The resulting GS possesses excellent adsorption properties. As protein is inexpensive and nontoxic, the porous biocomposite aerogel has significant potential for use as an adsorbent for biological applications.Acknowledgements
This research was supported by the National Natural Science Foundation of China (no. 21207100).References
- K. Nishinari, Y. Fang, S. Guo and G. O. Phillips, Food Hydrocolloids, 2014, 39, 301–318 CrossRef CAS PubMed.
- R. Kumar, V. Choudhary, M. Saroj, I. K. Varma and B. Mattiason, Ind. Crops Prod., 2002, 16, 155–172 CrossRef CAS.
- A. L. Caroni, C. R. de Lima, M. R. Pereira and J. L. Fonseca, J. Colloid Interface Sci., 2009, 340, 182–191 CrossRef CAS PubMed.
- S. Vadahanambi, S. H. Lee, W. J. Kim and I. K. Oh, Environ. Sci. Technol., 2013, 47, 10510–10517 CAS.
- K. Yu, Z. Bo, G. Lu, S. Mao, S. Cui, Y. Zhu, X. Chen, R. S. Ruoff and J. Chen, Nanoscale Res. Lett., 2011, 6, 202 CrossRef PubMed.
- Z. H. Wen, S. M. Cui, H. J. Kim, S. Mao, K. H. Yu, G. H. Lu, H. H. Pu, O. Mao and J. H. Chen, J. Mater. Chem., 2012, 22, 3300–3306 RSC.
- Y. Lin, S. Xu and J. Li, Chem. Eng. J., 2013, 225, 679–685 CrossRef CAS PubMed.
- H. Chen, B. Gao and H. Li, J. Environ. Chem. Eng., 2014, 2, 310–315 CrossRef CAS PubMed.
- S. Chowdhury and R. Balasubramanian, Adv. Colloid Interface Sci., 2014, 204, 35–56 CrossRef CAS PubMed.
- C. S. Cheng, J. Deng, B. Lei, A. He, X. Zhang, L. Ma, S. Li and C. Zhao, J. Hazard. Mater., 2013, 263, 467–478 CrossRef CAS PubMed.
- Z. Zhao, X. Wang, J. Qiu, J. Lin, D. Xu, C. A. Zhang, M. Lv and X. Yang, Rev. Adv. Mater. Sci., 2014, 36, 137–151 Search PubMed.
- W. S. Hummers and R. E. Offeman, J. Am. Chem. Soc., 1958, 80, 1339–1340 CrossRef CAS.
- M. Hirata, T. Gotou, S. Horiuchi, M. Fujiwara and M. Ohba, Carbon, 2004, 42, 2929–2937 CAS.
- S. Mao, H. H. Pu and J. H. Chen, RSC Adv., 2012, 7, 2643–2662 RSC.
- S. Attal, R. Thiruvengadathan and O. Regev, Anal. Chem., 2006, 78, 8098–8104 CrossRef CAS PubMed.
- S. H. Jeong, K. K. Kim, S. J. Jeong, K. H. An, S. H. Lee and Y. H. Lee, Synth. Met., 2007, 157, 570–574 CrossRef CAS PubMed.
- R. Li, L. Liu and F. Yang, Chem. Eng. J., 2013, 229, 460–468 CrossRef CAS PubMed.
- C. Rodríguez-González, A. L. Martínez-Hernández, V. M. Castaño, O. V. Kharissova, R. S. Ruoff and C. Velasco-Santos, Ind. Eng. Chem. Res., 2012, 51, 3619–3629 CrossRef.
- L. Huang, C. Li, W. Yuan and G. Shi, Nanoscale, 2013, 5, 3780–3786 RSC.
- Y. Xu, K. Sheng, C. Li and G. Shi, ACS Nano, 2010, 7, 4324–4330 CrossRef PubMed.
- J. N. Tiwari, K. Mahesh, N. H. Le, K. C. Kemp, R. Timilsina, R. N. Tiwari and K. S. Kim, Carbon, 2013, 56, 173–182 CrossRef CAS PubMed.
- C. Gao, T. Liu, C. Shuai and S. Peng, Sci. Rep., 2014, 4, 4712 Search PubMed.
- A. C. Ferrari, Solid State Commun., 2007, 143, 47–57 CrossRef CAS PubMed.
- X. Shi, W. Wang, Y. Kang and A. Wang, J. Appl. Polym. Sci., 2012, 125, 1822–1832 CrossRef CAS.
- T. Ma, P. R. Chang, P. Zheng, F. Zhao and X. Ma, Mater. Chem. Phys., 2014, 146, 446–451 CrossRef CAS PubMed.
- S. K. Yadav, Y. C. Jung, C.-M. Yang, Y.-I. Ko, K.-S. Yang, Y. A. Kim and J. W. Cho, RSC Adv., 2014, 4, 5382 RSC.
- Y. Chao, W. Zhu, B. Yan, Y. Lin, S. Xun, H. Ji, X. Wu, H. Li and C. Han, J. Appl. Polym. Sci., 2014, 131, 40561 CrossRef.
- P. Liu, W. J. Liu, H. Jiang, J. J. Chen, W. W. Li and H. Q. Yu, Bioresour. Technol., 2012, 121, 235–240 CrossRef CAS PubMed.
- L. Huang, M. Wang, C. Shi, J. Huang and B. Zhang, Desalin. Water Treat., 2014, 52, 2678–2687 CrossRef CAS.
- J. Wang, J. Dai and M. Meng, J. Appl. Polym. Sci., 2014, 131, 40310 Search PubMed.
- W. T. Jiang, P. H. Chang, Y. S. Wang, Y. Tsai, J. S. Jean, Z. Li and K. Krukowski, J. Hazard. Mater., 2013, 250–251, 362–369 CrossRef CAS PubMed.
- S. Wu, X. Zhao, Y. Li, C. Zhao, Q. Du, J. Sun, Y. Wang, X. Peng, Y. Xia, Z. Wang and L. Xia, Chem. Eng. J., 2013, 230, 389–395 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c4ra12413h |
| This journal is © The Royal Society of Chemistry 2015 |