Open Access Article
Open Access ArticleCucurbiturils as supramolecular inhibitors of DNA restriction by type II endonucleases†
Cátia
Parente Carvalho
a,
Amir
Norouzy
b,
Vera
Ribeiro
c,
Werner M.
Nau
b and
Uwe
Pischel
*a
aCIQSO-Center for Research in Sustainable Chemistry and Department of Chemical Engineering, Physical Chemistry and Organic Chemistry, University of Huelva, Campus El Carmen s/n, E-21071 Huelva, Spain. E-mail: uwe.pischel@diq.uhu.es; Fax: +34 959 21 99 83; Tel: +34 959 21 99 82
bDepartment of Life Sciences and Chemistry, Jacobs University Bremen, Campus Ring 1, D-28759 Bremen, Germany
cCenter for Molecular and Structural Biomedicine, CBME/IBB, LA, University of Algarve, Faculty of Science and Technology, Campus de Gambelas, 8005-139 Faro, Portugal
First published on 12th January 2015
Abstract
Cucurbiturils (CB6 and CB7) were shown to inhibit the enzymatically catalyzed restriction of plasmids and linear DNA. This effect can be inverted by supramolecular masking of the macrocycles through competitive complexation with polyamines. These experiments provide supramolecular control of biocatalytic processes.
Introduction
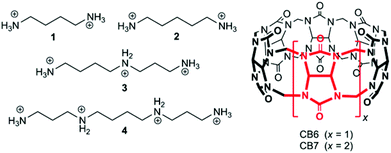
Cucurbit[n]urils (CBn) are water-soluble macrocyclic hosts featuring n glycoluril units linked by methylene bridges (see CB6 and CB7 in Scheme 1).1–5 Thus far the homologues with n = 6, 7, and 8 have received most attention in supramolecular applications, many of which are biologically related.5–21 The potential of CBn macrocycles to control functions in biological chemistry via reversible supramolecular interactions benefits from their good water solubility, high binding constants,22 and low cellular toxicity.23,24According to strategies (a) and (b), shown in Scheme 2, several protocols for biological control by cucurbiturils have been developed. For instance, CB7 is known to protect peptides from protease-catalyzed hydrolytic degradation, which is an example of substrate-related supramolecular control of biological functions by CBn macrocycles (strategy (a)).10,25 As an example for path (b) CB7 can complex inhibitors of bovine carbonic anhydrase or acetylcholinesterase and, thereby, activate catalytic reactions through competitive displacement of the inhibitor from the enzyme.26 In the context of enzymatic transformations of DNA substrates, it was shown that the spermidine·CB6 complex (3·CB6) catalyzes the topoisomerization of supercoiled plasmid DNA and that CB6 can invert the promoting effect of spermine (4) in plasmid DNA restriction (strategy (b)).27 It is noteworthy that while there are case studies of organic hosts that interact with enzymes or other proteins,28–31 the direct modulation of the biocatalytic activity of the enzyme itself by CBn hosts has remained elusive. In the course of our research on cucurbituril–biomolecule interactions, we came across an unprecedented inhibition of DNA restriction, a process that plays a fundamental role in the defense of organisms against foreign DNA.
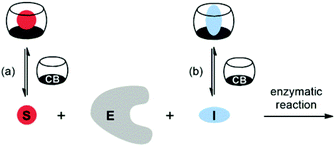 | ||
| Scheme 2 Supramolecular control of enzymatic catalysis by host macrocycles (e.g., CBn) via (a) substrate binding or (b) inhibitor binding. E: enzyme, S: substrate, I: inhibitor. | ||
Results and discussion
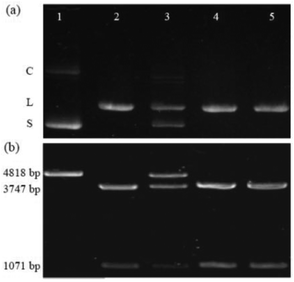
The first example is the hydrolysis of DNA at GGTACC sites catalyzed by the endonuclease KpnI (see Fig. 1a). Fig. 1a shows the corresponding experiments with plasmid pGL3-Basic DNA as a substrate, which were conducted under identical conditions concerning pH, buffer composition, temperature, and digestion time.‡ The electrophoresis of the plasmid DNA alone evidenced its preferential presence in the supercoiled form (lane 1). Upon enzymatic hydrolysis of the plasmid DNA, the linear form appeared (lane 2). However, in the presence of 6 μM CB6 ([CB6]/[KpnI] = 4542, lane 3) the enzymatic reaction was significantly hindered leading to 47% inhibition with respect to the absence of the macrocycle. The incomplete enzymatic reaction was confirmed by the observation of a substantial amount of non-restricted plasmid DNA. The restriction reaction can be re-activated by the addition of the strongly competitive polycationic binder 4 (K = 3.3 × 109 M−1);32 lane 5 with [4]/[CB6] = 1. As a control experiment, the enzymatic hydrolysis was also performed in the presence of 4 but in the absence of CB6 (lane 4 vs. 5). The result confirmed that 4 alone had no effect on the DNA restriction under the selected experimental conditions. The re-activation was also seen for the addition of a stoichiometric amount of the strongly CB6-binding amine 3 (K = 4.1 × 108 M−1); see ESI.†32In another series of experiments,‡ CB7 was used as a supramolecular inhibitor of the restriction of plasmid or linearized pGL3-Basic DNA by various endonucleases (KpnI, SacI, and XapI).§ The results are shown in Table 1. First, the macrocycle concentration was adjusted to affect ca. 50% inhibition, which required 90–500 μM CB7 (Table 1). Second, polyamines 1–3 were screened for their potential to fully re-activate the DNA restriction, which required 1–5 equivalents of a competitor, depending on their relative affinity to the macrocycle (see stoichiometries in Table 1). As a representative example, the electrophoretic analysis of the KpnI-induced hydrolysis of linear pGL3-Basic DNA, its inhibition by CB7, and the re-activation by addition of 1 is shown in Fig. 1b. The DNA (lane 1) showed a band corresponding to the expected 4818 base pairs (bp). The restriction reaction (lane 2) led to fragments with 3747 and 1071 bp. The presence of 200 μM CB7 (lane 3) hindered the enzymatic hydrolysis (48% inhibition), which was reflected by a substantial residual band of unrestricted DNA (4818 bp). The addition of amine 1 ([1]/[CB7] = 5) blocked CB7 and thereby re-activated the process again (lane 5). Lane 4 shows the control in the presence of only 1 for which no effect was observed. While it is known that polyamines also bind to DNA,33 their apparent binding constants are ca. 1–2 orders of magnitude lower than the ones for the respective CB7 complexes (compared to the CB6 complexes they are even ca. 3–4 orders of magnitude lower). This excludes competition between CBn and DNA for the added polyamines under the experimental conditions.‡
| Plasmid DNA | Linear DNAa | |||||
|---|---|---|---|---|---|---|
| KpnIb | SacIc | XapId | KpnIb | SacIc | XapId | |
| a Obtained by total restriction of plasmid pGL3-Basic DNA with PdmI endonuclease. b Restriction at GGTACC sites. c Restriction at GAGCTC sites. d Restriction at RAATTY sites. e Optimized CB7 concentration to achieve ca. 50% inhibition. f Optimized polyamine/macrocycle ratio for complete re-activation (<5% inhibition); binding constants (K/106 M−1) with CB7 in water at pH 7 (this work): 0.11 for 1, 20 for 2, and 1.2 for 3. | ||||||
| [CB7]/μMe | 90 | 250 | 500 | 200 | 350 | 500 |
| Inhibition (%) | 32 | 42 | 54 | 48 | 52 | 58 |
| [1]/[CB7]f | 5 | 5 | 5 | 5 | 5 | 5 |
| [2]/[CB7]f | 1 | 2 | 2 | 1 | 2 | 2 |
| [3]/[CB7]f | 2 | 1 | 1 | 2 | 1 | 1 |
The variety of DNA conformations and endonucleases, for which the inhibition by CBn was observed, points to a general effect. A previous literature report ruled out significant supramolecular interactions between CBn and DNA, based on the absence of competitive displacement of a DNA-intercalating dye.21 In line with this, our own circular dichroism (CD) spectroscopic measurements did not reveal any sign of direct supramolecular interaction between model DNA and CBn (see ESI†). Additional support came from the observation of unchanged gel mobility of circular and linear pGL3-Basic DNA in the absence and presence of excess macrocycles (see ESI†). On this basis, an interaction with the DNA substrate was ruled out as the reason for the inhibitory effect. The possibility of metal-ion sequestration from the buffer solution by CBn is another factor that deserved scrutiny.34 In particular, magnesium ions (Mg2+) are known to serve as an essential cofactor of the type II endonucleases employed herein.35 A control experiment revealed that EDTA (1 mM), a strong Mg2+ chelator, did not lead to endonuclease inhibition as tested for the example of XapI; see ESI.† The EDTA concentration corresponds to double the CB7 concentration used for the experiments with this enzyme. This rules out metal-ion sequestration as a cause of the observed inhibition, at least at the applied CBn concentrations (maximum of 500 μM). It is noteworthy that, bovine serum albumin (BSA) is often present in the digestion solutions, fulfilling the role of a stabilizer and a protective agent against impurities that may be problematic for the restriction reaction. On the one hand, for the BSA concentration used in our assays (ca. 1.5 μM for KpnI and XapI; note that SacI does not require BSA) no protection against the inhibitory action of CBn was noted. Only at much higher BSA concentrations (>30 μM; tested for the example of the XapI assay) the CB-induced inhibition was suppressed. On the other hand, the inhibitory action of CB7 in the absence of BSA was maintained (observed for the example of KpnI in BSA-free reaction buffer; see ESI†). Thereby, the interaction of CBn with BSA can be excluded as well as the underlying reason for the observed inhibitory effect.
There is ample evidence in the literature for host–guest interactions between CBn and peptides or proteins.13,36,37 It is generally accepted that peptides with N-terminal aromatic amino acids (Trp, Phe, Tyr) or long alkylamino chains (Lys) can form strong complexes with CB7 or CB6, respectively.13,14,38–40 However, also amino acids at the C-terminus and even internal ones can form complexes, albeit somewhat weaker.38–41 The binding of proteins to CBn is naturally more complex and depends on the availability of the mentioned amino acids at the surface of the biomolecular structure. One example of an efficient CBn–protein interaction is that with insulin, which features an exposed Phe N-terminal residue.16 Unfortunately, for the investigated restriction enzymes, predictions on accessible binding sites for CBn are not straightforward to make, especially because they are rich in amino acids displaying the required recognition motifs (Lys, Tyr, Trp, and Phe; see amino acid sequences of the enzymes in ESI†). Additionally, their inhibitory action could be caused by both, binding near the DNA binding domain of the enzyme or through a more remote binding associated with an allosteric effect.16
Unfortunately, our efforts to obtain direct experimental evidence (by UV-vis absorption spectroscopy, CD spectroscopy or MALDI-TOF mass spectrometry) for the interaction between the CBn macrocycles and the enzymes were hampered, among others, by the presence of significant concentrations of BSA. However, other possible reasons for the observed inhibitory effect, such as interactions with the substrate itself, metal–ion sequestration, or masking of buffer components were clearly excluded (see above). The combined experimental observations and the literature precedence of protein–CBn interactions provide at least an indirect hint on the origins of the observed effects as proposed in Scheme 3.
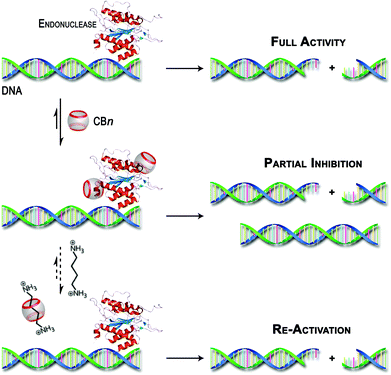 | ||
| Scheme 3 Schematic illustration of the postulated inhibition of DNA restriction enzymes by CBn hosts and their re-activation by competitive binding with polyamines. | ||
Conclusions
In conclusion, we demonstrated that CBn macrocycles can inhibit the enzymatic hydrolysis of DNA substrates. The reversible nature of the observed effects was underpinned by the successful re-activation of the type II endonuclease activity via addition of CBn-binding polyamines. The supramolecular methodology could find potential applications in molecular biology, where42 the use of restriction enzymes plays a crucial role.Acknowledgements
This work was supported by the Spanish MINECO (grant CTQ2011-28390 for U.P.), FEDER, COST (CM1005 “Supramolecular Chemistry in Water”), the DFG (grant NA-686/5 for A.N. and W.M.N.), and the Portuguese FCT (fellowship SFRH/BD/81628/2011 for C.P.C. and grant PEst-OE/EQB/LA0023/2013 for V.R.). We thank Alexandra I. Lazar (Jacobs University Bremen) for the assistance in MALDI-TOF experiments.Notes and references
- J. Lagona, P. Mukhopadhyay, S. Chakrabarti and L. Isaacs, Angew. Chem., Int. Ed., 2005, 44, 4844–4870 CrossRef CAS PubMed.
- W. M. Nau and O. A. Scherman, Isr. J. Chem., 2011, 51, 492 CrossRef CAS (thematic issue about cucurbiturils).
- E. Masson, X. X. Ling, R. Joseph, L. Kyeremeh-Mensah and X. Y. Lu, RSC Adv., 2012, 2, 1213–1247 RSC.
- B. C. Pemberton, R. Raghunathan, S. Volla and J. Sivaguru, Chem. – Eur. J., 2012, 18, 12178–12190 CrossRef CAS PubMed.
- I. Ghosh and W. M. Nau, Adv. Drug Delivery Rev., 2012, 64, 764–783 CrossRef CAS PubMed.
- J. Vázquez, P. Remón, R. N. Dsouza, A. I. Lazar, J. F. Arteaga, W. M. Nau and U. Pischel, Chem. – Eur. J., 2014, 20, 9897–9901 CrossRef PubMed.
- J. P. Da Silva, R. Choudhury, M. Porel, U. Pischel, S. Jockusch, P. C. Hubbard, V. Ramamurthy and A. V. M. Canário, ACS Chem. Biol., 2014, 9, 1432–1436 CrossRef CAS PubMed.
- H. H. Lee, T. S. Choi, S. J. C. Lee, J. W. Lee, J. Park, Y. H. Ko, W. J. Kim, K. Kim and H. I. Kim, Angew. Chem., Int. Ed., 2014, 53, 7461–7465 CrossRef CAS PubMed.
- N. Saleh, I. Ghosh and W. M. Nau, in Supramolecular Systems in Biomedical Fields, ed. H. J. Schneider, RSC Publishing, 2013, pp. 164–212 Search PubMed.
- L. A. Logsdon and A. R. Urbach, J. Am. Chem. Soc., 2013, 135, 11414–11416 CrossRef CAS PubMed.
- D. Ma, B. Zhang, U. Hoffmann, M. Grosse Sundrup, M. Eikermann and L. Isaacs, Angew. Chem., Int. Ed., 2012, 51, 11358–11362 CrossRef CAS PubMed.
- D. Ma, G. Hettiarachchi, D. Nguyen, B. Zhang, J. B. Wittenberg, P. Y. Zavalij, V. Briken and L. Isaacs, Nat. Chem., 2012, 4, 503–510 CrossRef CAS PubMed.
- A. R. Urbach and V. Ramalingam, Isr. J. Chem., 2011, 51, 664–678 CrossRef CAS.
- L. A. Logsdon, C. L. Schardon, V. Ramalingam, S. K. Kwee and A. R. Urbach, J. Am. Chem. Soc., 2011, 133, 17087–17092 CrossRef CAS PubMed.
- D.-W. Lee, K. M. Park, M. Banerjee, S. H. Ha, T. Lee, K. Suh, S. Paul, H. Jung, J. Kim, N. Selvapalam, S. H. Ryu and K. Kim, Nat. Chem., 2011, 3, 154–159 CrossRef CAS PubMed.
- J. M. Chinai, A. B. Taylor, L. M. Ryno, N. D. Hargreaves, C. A. Morris, P. J. Hart and A. R. Urbach, J. Am. Chem. Soc., 2011, 133, 8810–8813 CrossRef CAS PubMed.
- C. Parente Carvalho, V. D. Uzunova, J. P. Da Silva, W. M. Nau and U. Pischel, Chem. Commun., 2011, 47, 8793–8795 RSC.
- R. B. Wang, B. C. MacGillivray and D. H. Macartney, Dalton Trans., 2009, 3584–3589 RSC.
- N. Saleh, A. L. Koner and W. M. Nau, Angew. Chem., Int. Ed., 2008, 47, 5398–5401 CrossRef CAS PubMed.
- A. C. Bhasikuttan, J. Mohanty, W. M. Nau and H. Pal, Angew. Chem., Int. Ed., 2007, 46, 4120–4122 CrossRef CAS PubMed.
- H. Isobe, N. Tomita, J. W. Lee, H.-J. Kim, K. Kim and E. Nakamura, Angew. Chem., Int. Ed., 2000, 39, 4257–4260 CrossRef CAS.
- L. P. Cao, M. Šekutor, P. Y. Zavalij, K. Mlinarić-Majerski, R. Glaser and L. Isaacs, Angew. Chem., Int. Ed., 2014, 53, 988–993 CrossRef CAS PubMed.
- V. D. Uzunova, C. Cullinane, K. Brix, W. M. Nau and A. I. Day, Org. Biomol. Chem., 2010, 8, 2037–2042 CAS.
- G. Hettiarachchi, D. Nguyen, J. Wu, D. Lucas, D. Ma, L. Isaacs and V. Briken, PLoS One, 2010, 5, e10514 Search PubMed.
- A. Hennig, G. Ghale and W. M. Nau, Chem. Commun., 2007, 1614–1616 RSC.
- S. Ghosh and L. Isaacs, J. Am. Chem. Soc., 2010, 132, 4445–4454 CrossRef CAS PubMed.
- H. Isobe, S. Sato, J. W. Lee, H.-J. Kim, K. Kim and E. Nakamura, Chem. Commun., 2005, 1549–1551 RSC.
- D. Bier, R. Rose, K. Bravo-Rodriguez, M. Bartel, J. M. Ramirez-Anguita, S. Dutt, C. Wilch, F.-G. Klärner, E. Sanchez-Garcia, T. Schrader and C. Ottmann, Nat. Chem., 2013, 5, 234–239 CrossRef CAS PubMed.
- R. E. McGovern, H. Fernandes, A. R. Khan, N. P. Power and P. B. Crowley, Nat. Chem., 2012, 4, 527–533 CrossRef CAS PubMed.
- A. Koralewska, W. Augustyniak, A. Temeriusz and M. Kańska, J. Inclusion Phenom. Macrocyclic Chem., 2004, 49, 193–197 CrossRef CAS.
- K. Matsubara, Y. Ando, T. Irie and K. Uekama, Pharm. Res., 1997, 14, 1401–1405 CrossRef CAS.
- M. V. Rekharsky, Y. H. Ko, N. Selvapalam, K. Kim and Y. Inoue, Supramol. Chem., 2007, 19, 39–46 CrossRef CAS.
- A. Kabir, M. Hossain and G. S. Kumar, J. Chem. Thermodyn., 2013, 57, 445–453 CrossRef CAS PubMed.
- V. F. Pais, E. F. A. Carvalho, J. P. C. Tomé and U. Pischel, Supramol. Chem., 2014, 26, 642–647 CrossRef CAS.
- A. Pingoud and A. Jeltsch, Nucleic Acids Res., 2001, 29, 3705–3727 CrossRef CAS PubMed.
- J. W. Lee, S. W. Heo, S. J. C. Lee, J. Y. Ko, H. Kim and H. I. Kim, J. Am. Soc. Mass Spectrom., 2013, 24, 21–29 CrossRef CAS PubMed.
- F. Biedermann and W. M. Nau, Angew. Chem., Int. Ed., 2014, 53, 5694–5699 CrossRef CAS PubMed.
- G. Ghale, N. Kuhnert and W. M. Nau, Nat. Prod. Commun., 2012, 7, 343–348 CAS.
- G. Ghale, V. Ramalingam, A. R. Urbach and W. M. Nau, J. Am. Chem. Soc., 2011, 133, 7528–7535 CrossRef CAS PubMed.
- M. V. Rekharsky, H. Yamamura, Y. H. Ko, N. Selvapalam, K. Kim and Y. Inoue, Chem. Commun., 2008, 2236–2238 RSC.
- S. Sonzini, S. T. J. Ryan and O. A. Scherman, Chem. Commun., 2013, 49, 8779–8781 RSC.
- S.-H. Chan, B. L. Stoddard and S.-Y. Xu, Nucleic Acids Res., 2011, 39, 1–18 CrossRef CAS PubMed.
- C. A. Schneider, W. S. Rasband and K. W. Eliceiri, Nat. Methods, 2012, 9, 671–675 CrossRef CAS.
- R. J. Roberts, T. Vincze, J. Posfai and D. Macelis, Nucleic Acids Res., 2007, 35, D269–D270 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: DNA extraction, details of restriction assays, control experiments (CBn–DNA interaction, EDTA addition), and amino acid sequences of the investigated enzymes. See DOI: 10.1039/c4ob02122c |
| ‡ Reaction conditions: [DNA] = 9.6 nM corresponding to [bp] = 46.3 μM; [KpnI] = 0.5 U μL−1 in a 10 mM Tris-HCl buffer (pH 7.5) containing 10 mM MgCl2, 0.02% Triton X-100, 0.1 mg mL−1 bovine serum albumin (BSA); [SacI] = 0.5 U μL−1 in 10 mM Tris-HCl buffer (pH 7.5) containing 7 mM MgCl2, 50 mM KCl, 1 mM DTT; [XapI] = 0.025 U μL−1 in 33 mM Tris-acetate buffer (pH 7.9) containing 10 mM Mg(OAc)2, 66 mM KOAc, 0.1 mg mL−1 BSA; all at 37 °C for 3 h. The degree of inhibition was evaluated by integration of the optical density of electrophoresis DNA bands by using the ImageJ program.43 Each experiment was carried out at least three times. |
| § The plasmid pGL3-Basic DNA was amplified in DH5α E. coli cells (see ESI†) and purified using the NucleoBond® Xtra Midi EF kit from Macherey-Nagel. The plasmid DNA was linearized by total restriction with PdmI endonuclease ([DNA] = 383.8 nM; [PdmI] = 2 U μL−1, 33 mM Tris-acetate buffer (pH 7.9), 10 mM Mg(OAc)2, 66 mM KOAc, 0.1 mg mL−1 BSA; 37 °C for 12 h) and purification by ethanol precipitation. The enzymes KpnI, XapI, and PdmI were purchased from Thermo Scientific. SacI was available from Promega. KpnI without added BSA was supplied by Sigma-Aldrich. Additional information about the enzymes is available at http://rebase.neb.com.44 |
| This journal is © The Royal Society of Chemistry 2015 |