Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
DNA nanopore translocation in glutamate solutions
C.
Plesa
 ,
N.
van Loo
and
C.
Dekker
*
,
N.
van Loo
and
C.
Dekker
*
Department of Bionanoscience, Kavli Institute of Nanoscience, Delft University of Technology, Lorentzweg 1, 2628 CJ Delft, The Netherlands. E-mail: c.dekker@tudelft.nl
First published on 10th July 2015
Abstract
Nanopore experiments have traditionally been carried out with chloride-based solutions. Here we introduce silver/silver-glutamate-based electrochemistry as an alternative, and study the viscosity, conductivity, and nanopore translocation characteristics of potassium-, sodium-, and lithium-glutamate solutions. We show that it has a linear response at typical voltages and can be used to detect DNA translocations through a nanopore. The glutamate anion also acts as a redox-capable thickening agent, with high-viscosity solutions capable of slowing down the DNA translocation process by up to 11 times, with a corresponding 7 time reduction in signal. These results demonstrate that glutamate can replace chloride as the primary anion in nanopore resistive pulse sensing.
Introduction
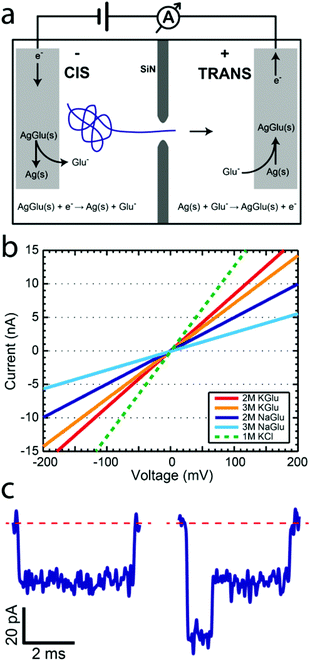
Solid-state nanopores constitute an emerging biophysical technique which has been used to study DNA,1 protein,2 and DNA–protein complexes.3–5 Although this area has been primarily pushed by the promise of its use for low-cost sequencing of long nucleic acid polymers, many other applications have opened up. Numerous studies have been published on methods to improve various aspects of this technique with different pore types,6,7 membrane materials,8–11 new salts,12,13 and passivation techniques.14,15 Although central to their operation, alternative electrochemical reactions for detecting the ionic current have not been pursued. In this study we introduce a novel electrochemical reaction based on glutamate (Glu) anions and demonstrate its potential for studying DNA translocations through solid-state nanopores. This approach provides a way to avoid the use of chloride and slow down the translocation velocity using high-viscosity solutions. Some of the previous attempts to slow down the DNA translocation process16 have utilized thickening agents such as glycerol.17,18 In this study we introduce the concept of redox capable thickening agents, which simultaneously increase the solution viscosity and participate in the electrochemical ionic current readout.We first provide a brief overview of the solid-state nanopore method. Nanopores are typically fabricated in a 20 nm thick free-standing SiN membrane by focusing an electron beam on the membrane using a transmission electron microscope (TEM).19 This allows simultaneous in situ imaging of the pore as it is being created. Membranes with pores are subsequently placed in a flow cell such that it separates two aqueous reservoirs filled with salt solution, as shown in Fig. 1a. Chlorinated silver electrodes are placed into the solution on each side and an electric potential is applied. The resulting ionic current is measured with a low-noise amplifier. Since DNA is negatively charged, it is electrophoretically driven through the nanopore. When a DNA molecule passes through the pore it causes a transient drop in the ionic current, since the molecule reduces the pore volume normally used by ions for transport. The duration of the blockade can be related to the length of the polymer, while the amplitude of the blockade is proportional to the volume of the molecule currently in the pore constriction.
We now address the electrochemistry used for the ionic current detection. In the typical silver/silver-chloride based electrochemistry, a chlorinated silver wire is placed in a chloride-salt-based solution and an electric potential is applied. At the anode, chloride ions are oxidized into solid-phase silver chloride through the reaction Ag(s) + Cl−(aq) → AgCl(s) + e− while at the cathode, silver chloride is reduced and chloride ions are released into the solution via the reaction AgCl(s) + e− → Ag(s) + Cl−(aq). The measured ionic current is linear with respect to the applied voltage at potentials below 1 V. The ease of electrode preparation, made by placing a silver wire in a bleach solution, makes this a simple and effective experimental approach. With only one exception, all nanopore publications that we are aware of use silver-chloride based electrochemistry together with either alkali metal halide solutions (LiCl,12 NaCl,3 KCl,14 RbCl,20 and CsCl21) or organic-salts (BMIM-Cl13), with chlorine always as the halogen constituent. The only non-chloride electrochemistry nanopore study looked at ionic liquids using platinum electrodes.22
L-Glutamic acid (Glu) plays a fundamental role in biology as one of the amino acids used in protein synthesis. In many organisms glutamate, not chloride, is the major intracellular anion.23 Although many of its properties have been studied in detail, we are not aware of any literature on non-enzymatic glutamate-based electrochemistry. In 1974, an article by Tabei et al.24 described a method for making printed circuits through the photolysis of silver glutamate. From this work, we know that silver glutamate is a stable, albeit photosensitive, solid-phase compound, much like silver chloride.
We hypothesized that glutamate could be used to replace the chloride in the electrochemical reactions used in the nanopore studies. Analogous to the silver chloride case, we expect similar electrochemical reactions to take place at the anode
| Ag(s) + Glu−(aq) → AgGlu(s) + e− | (1) |
| AgGlu(s) + e− → Ag(s) + Glu−(aq). | (2) |
In this way, glutamate could be used as a redox-capable thickening agent to increase the viscosity of a solution while still maintaining the required electrochemistry.
Results
We now show that glutamate solutions produce a linear current–voltage response and can be used for detecting DNA nanopore translocations. We prepared silver glutamate electrodes by electrolysis of a 99.99% silver wire (anode) and platinum wire (cathode) in a concentrated 3 M potassium glutamate solution. These electrodes were then used, following standard procedures, in solid-state nanopore experiments. Fig. 1b shows the typical current–voltage curves observed for potassium glutamate (KGlu) and sodium glutamate (NaGlu) solutions at 2 M and 3 M concentrations in 20 nm pores. We have also included a typical response observed for 1 M KCl (dashed green line) for comparison. The IV responses were observed to be linear in the tested range of −800 mV to 800 mV. Additionally, the open pore conductance was observed to be higher at 2 M compared to 3 M, in both KGlu and NaGlu, as explained below while discussing the conductivity measurements. The open pore conductance can be related to several physical characteristics of the pore and the solution using25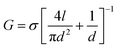 | (3) |
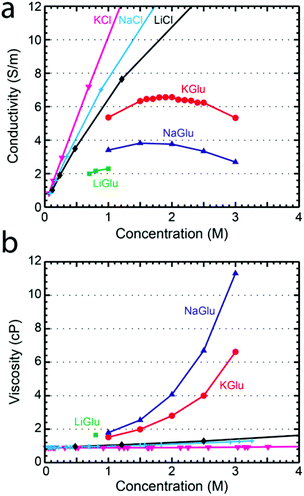
To understand the pore conductance, we measured the conductivity of concentrated solutions of glutamate salts. Fig. 2a shows the conductivity at 23 °C as a function of concentration for KGlu, NaGlu, and LiGlu, alongside literature values26 for KCl, NaCl, and LiCl at 25 °C. The maximum concentrations tested were limited by the solubility of the salts which were found to be around 1 M for LiGlu, 3.5 M for KGlu, and 3.3 M for NaGlu. The conductivity of KGlu was observed to be significantly higher than NaGlu, with values for KGlu of 6.57 S m−1 and 5.33 S m−1 at 2 M and 3 M compared to 3.76 S m−1 and 2.69 S m−1, respectively, for NaGlu. The conductivities of both solutions were observed to have maxima, which was around 2.0 for KGlu and 1.7 for NaGlu. This is a common phenomenon at very high electrolyte concentrations and is attributed to the small inter-ionic separations which lead to strong ion–ion interactions that begin to reduce the conductivity of solutions above a certain concentration. Since we would like to maximize the ionic current signal, larger conductivities are desired and these measurements reveal that KGlu is preferable over NaGlu. As expected, the conductivities observed for the glutamate solutions were smaller than the corresponding chloride solutions at the same concentrations.
We also investigated the viscosity of glutamate salt solutions, relevant for the translocation speed of traversing molecules. A Brookfield DV2TLVCP Wells-Brookfield cone/plate viscometer was used to measure the viscosities of KGlu, NaGlu, and LiGlu at 23 °C. Viscosity is an important parameter in nanopore measurements since increasing viscosity results in a higher drag force and a longer translocation time. In Fig. 2b the viscosity of LiGlu, KGlu, and NaGlu is shown as a function of solution concentration alongside literature values for KCl,27 NaCl,27 and LiCl28 at 25 °C. As expected, the glutamate solutions have a significantly higher viscosity than the chloride solutions at the same concentration. We measured a viscosity of 2.8 cP and 6.6 cP for KGlu at 2 M and 3 M, and even higher viscosities of 4.1 cP and 11.3 cP, respectively, for NaGlu. The larger viscosities of the glutamate solutions compared to the chloride solutions can be explained by the much larger size of the glutamate (MW 147.13 g mol−1) anion compared to chloride (MW 35.45 g mol−1) and highlight its potential as a thickening agent.
Finally, we studied the characteristics of DNA translocation through nanopores in high concentration glutamate solutions to see how much the translocation velocity and conductance blockade were reduced. Linear 20.7 kbp DNA molecules were translocated through nanopores in 1 M, 2 M, and 3 M solutions of KGlu and NaGlu at 100 mV. Fig. 3a shows a typical heat plot showing the event's conductance blockade and translocation time in 2 M KGlu and a 20 nm pore. As expected from the viscosity measurements, events were slower in NaGlu compared to KGlu, as shown in Fig. 3b. In 2 M and 3 M KGlu, events had a 3.3× and 5.8× longer translocation time relative to 1 M KCl, while the translocation in 3 M NaGlu took 11.3× longer. The price paid for this slower translocation is a reduction in the blockade signal, as already observed in the conductivity measurements. In Fig. 3c we show the DNA conductance blockade in 1 M, 2 M, and 3 M glutamate salt solutions. We observe blockade amplitudes of 0.1, 0.35, and 0.31 nS in 1 M, 2 M, and 3 M KGlu, which are around 12.5×, 3.6×, and 4× smaller than the DNA blockade in 1 M KCl (1.25 nS). This becomes worse for the NaGlu, with blockades of 0.22 nS and 0.18 nS, respectively. One common approach to increase the conductance blockade is to decrease the diameter of the nanopores used. Fig. 3d shows the DNA blockade for 3 M KGlu in 10, 16, and 20 nm pores. When the pore diameter is reduced from 20 nm to 10 nm we observe a 1.5× increase in the DNA blockade, to 0.47 nS. The observed trend can be fitted with a simple geometric model25 (solid line in Fig. 3d) based on eqn (3). These DNA translocation experiments demonstrate the viability of using glutamate-based solutions for nanopore measurements.
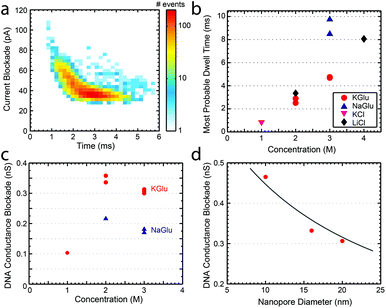 | ||
| Fig. 3 (a) Heat plot showing the conductance blockade and translocation time of 20.7 kbp DNA translocating through a 20 nm pore in 2 M KGlu. (b) The most probable dwell times observed for 20.7 kbp DNA at 100 mV in a variety of buffers. (c) DNA conductance blockade of 1 M, 2 M, and 3 M solutions of KGlu and NaGlu at 100 mV in 20 nm pores. (d) DNA conductance blockade in 3 M KGlu as a function of the pore diameter. The solid line is a fit to the geometric model of Kowalczyk et al.25 | ||
Conclusions
In this study we have demonstrated that glutamate-based electrochemistry can replace traditional chloride-based reactions for use in nanopore measurements. Potassium glutamate provides a higher conductivity, compared to sodium glutamate, with a maximum of 6.57 S m−1 at around 2 M concentration. The use of lithium glutamate is limited by its maximum solubility of around 1 M. DNA translocation through glutamate solutions can be slowed down by 3.3× to 11.3× relative to 1 M KCl, while the conductance blockade signal is reduced by 3.6× and 7× respectively. These numbers are similar to those of a study carried out by adding glycerol into 1.5 M KCl solutions.17 There, a 5.3× increase in the viscosity resulted in a 4.4× increase in the translocation time and a 3.1× decrease in the blockade amplitude, relative to the values for 1.5 M KCl. In addition to slowing down DNA translocation, the use of glutamate instead of chloride may provide a better environment for gold electrodes in three terminal devices.Methods
Free standing SiN membranes were fabricated by using trans-chip illumination lithography.29 Pores were drilled with a Philips CM200-FEG TEM. Ionic currents were measured with an Axopatch 200B and digitized at 500 kHz. Signals were subsequently analysed with the Transalyzer Matlab package.30 All solutions contained 10 mM Tris at pH 8 and 1 mM EDTA. Potassium glutamate, sodium glutamate, and L-glutamic acid were purchased from Sigma-Aldrich (St. Louis, MO, USA). Lithium glutamate was made by Tocris Bioscience (Bristol, UK) following a procedure similar to the one described by Wiesbrock et al.31 Briefly high purity L-α-glutamic acid was dissolved in HPLC grade water, after which an equivalent amount of lithium hydroxide was added. The resulting clear solution was then freeze dried to yield the solid salt. The product was characterized with micro analysis, infrared spectroscopy, proton NMR, and mass spectrometry. High purity silver and platinum wires were purchased from Advent Research Materials (Oxford, UK).Acknowledgements
The authors would like to thank Tim Albrecht for discussions and Meng-Yue Wu for TEM drilling of nanopores. This work was supported by the Netherlands Organisation for Scientific Research (NWO/OCW), as part of the Frontiers of Nanoscience program, and by the European Research Council under the research grant, NanoforBio (no. 247072).References
- M. Wanunu, Phys. Life Rev., 2012, 9, 125–158 CrossRef PubMed.
- C. Plesa, S. W. Kowalczyk, R. Zinsmeester, A. Y. Grosberg, Y. Rabin and C. Dekker, Nano Lett., 2013, 13, 658–663 CrossRef CAS PubMed.
- A. T. Carlsen, O. K. Zahid, J. A. Ruzicka, E. W. Taylor and A. R. Hall, Nano Lett., 2014, 14, 5488–5492 CrossRef CAS PubMed.
- M. Langecker, A. Ivankin, S. Carson, S. R. M. Kinney, F. C. Simmel and M. Wanunu, Nano Lett., 2015, 15, 783–790 CrossRef CAS PubMed.
- C. Plesa, J. W. Ruitenberg, M. J. Witteveen and C. Dekker, Nano Lett., 2015, 15, 3153–3158 CrossRef CAS PubMed.
- L. J. Steinbock, O. Otto, C. Chimerel, J. Gornall and U. F. Keyser, Nano Lett., 2010, 10, 2493–2497 CrossRef CAS PubMed.
- E. A. Heins, Z. S. Siwy, L. A. Baker and C. R. Martin, Nano Lett., 2005, 5, 1824–1829 CrossRef CAS PubMed.
- J. Larkin, R. Henley, D. C. Bell, T. Cohen-Karni, J. K. Rosenstein and M. Wanunu, ACS Nano, 2013, 7, 10121–10128 CrossRef CAS PubMed.
- B. M. Venkatesan, A. B. Shah, J.-M. Zuo and R. Bashir, Adv. Funct. Mater., 2010, 20, 1266–1275 CrossRef CAS PubMed.
- G. F. Schneider, S. W. Kowalczyk, V. E. Calado, G. g. Pandraud, H. W. Zandbergen, L. M. K. Vandersypen and C. Dekker, Nano Lett., 2010, 10, 3163–3167 CrossRef CAS PubMed.
- K. Liu, J. Feng, A. Kis and A. Radenovic, ACS Nano, 2014, 8, 2504–2511 CrossRef CAS PubMed.
- S. W. Kowalczyk, D. B. Wells, A. Aksimentiev and C. Dekker, Nano Lett., 2012, 12, 1038–1044 CrossRef CAS PubMed.
- R. S. S. de Zoysa, D. A. Jayawardhana, Q. Zhao, D. Wang, D. W. Armstrong and X. Guan, J. Phys. Chem. B, 2009, 113, 13332–13336 CrossRef CAS PubMed.
- E. C. Yusko, J. M. Johnson, S. Majd, P. Prangkio, R. C. Rollings, J. Li, J. Yang and M. Mayer, Nat. Nanotechnol., 2011, 6, 253–260 CrossRef CAS PubMed.
- G. F. Schneider, Q. Xu, S. Hage, S. Luik, J. N. H. Spoor, S. Malladi, H. Zandbergen and C. Dekker, Nat. Commun., 2013, 4 Search PubMed.
- B. Luan, G. Stolovitzky and G. Martyna, Nanoscale, 2012, 4, 1068–1077 RSC.
- D. Fologea, J. Uplinger, B. Thomas, D. S. McNabb and J. Li, Nano Lett., 2005, 5, 1734–1737 CrossRef CAS PubMed.
- B. Luan, D. Wang, R. Zhou, S. Harrer, H. Peng and G. Stolovitzky, Nanotechnology, 2012, 23, 455102 CrossRef PubMed.
- A. J. Storm, J. H. Chen, X. S. Ling, H. W. Zandbergen and C. Dekker, Nat. Mater., 2003, 2, 537–540 CrossRef CAS PubMed.
- M. F. Breton, F. Discala, L. Bacri, D. Foster, J. Pelta and A. Oukhaled, J. Phys. Chem. Lett., 2013, 4, 2202–2208 CrossRef CAS.
- G. V. Soni, A. Singer, Z. Yu, Y. Sun, B. McNally and A. Meller, Rev. Sci. Instrum., 2010, 81 Search PubMed.
- M. Davenport, A. Rodriguez, K. J. Shea and Z. S. Siwy, Nano Lett., 2009, 9, 2125–2128 CrossRef CAS PubMed.
- S. Cayley, B. A. Lewis, H. J. Guttman and M. T. Record Jr., J. Mol. Biol., 1991, 222, 281–300 CrossRef CAS.
- H. Tabei, S. Nara and K. Matsuyama, J. Electrochem. Soc., 1974, 121, 67–69 CrossRef CAS PubMed.
- S. W. Kowalczyk, A. Y. Grosberg, Y. Rabin and C. Dekker, Nanotechnology, 2011, 22, 315101 CrossRef PubMed.
- G. D. Fasman, in Practical handbook of biochemistry and molecular biology, ed. G. D. Fasman, CRC Press, Boca Raton, FL, 1989 Search PubMed.
- Z. Hai-Lang and H. Shi-Jun, J. Chem. Eng. Data, 1996, 41, 516–520 CrossRef.
- I. M. Abdulagatov, A. B. Zeinalova and N. D. Azizov, J. Mol. Liq., 2006, 126, 75–88 CrossRef CAS PubMed.
- X. J. A. Janssen, M. P. Jonsson, C. Plesa, G. V. Soni, C. Dekker and N. H. Dekker, Nanotechnology, 2012, 23, 475302 CrossRef PubMed.
- C. Plesa and C. Dekker, Nanotechnology, 2015, 26, 084003 CrossRef CAS PubMed.
- F. Wiesbrock and H. Schmidbaur, CrystEngComm, 2003, 5, 262–264 RSC.
| This journal is © The Royal Society of Chemistry 2015 |