Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Defensive symbioses of animals with prokaryotic and eukaryotic microorganisms†
Laura V.
Flórez‡
,
Peter H. W.
Biedermann‡
,
Tobias
Engl‡
and
Martin
Kaltenpoth‡§
*
Max Planck Institute for Chemical Ecology, Insect Symbiosis Research Group, Hans-Knöll-Str. 8, 07745 Jena, Germany. E-mail: mkaltenpoth@ice.mpg.de
First published on 20th April 2015
Abstract
Covering: through 2014
Many organisms team up with microbes for defense against predators, parasites, parasitoids, or pathogens. Here we review the described protective symbioses between animals (including marine invertebrates, nematodes, insects, and vertebrates) and bacteria, fungi, and dinoflagellates. We focus on associations where the microbial natural products mediating the protective activity have been elucidated or at least strong evidence for the role of symbiotic microbes in defense is available. In addition to providing an overview of the known defensive animal–microbe symbioses, we aim to derive general patterns on the chemistry, ecology, and evolution of such associations.
1 Introduction
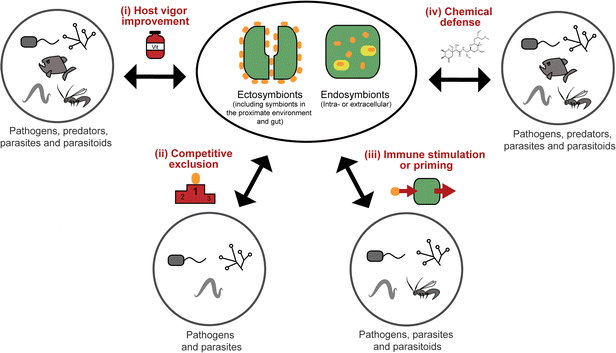
All organisms are threatened by antagonistic encounters with predators, pathogens, parasites, and/or parasitoids, which exert strong selective pressures on evolving efficient defense strategies. Such protective adaptations include behavioral, mechanical, and structural defenses against predators,1 as well as a sophisticated immune system providing protection from microbial intruders and parasitoids.2 In addition, many animals across a broad range of taxa use an arsenal of chemicals to defend themselves against various antagonists.3,4 Many of these defensive compounds are produced by the animals themselves, but it is becoming increasingly evident that microbial symbionts can make important contributions to their host's defense.5,6While symbiosis research has traditionally focused on the nutritional aspects of mutualistic associations between animals and microorganisms, more recent research has revealed the importance of defensive alliances with microorganisms for their hosts' ecology and evolution.5,6 In general, there are four different ways in which microbial symbionts can contribute to their host's protection from antagonists (Fig. 1): (i) microbial partners can improve the overall vigor of their host and thereby enable it to allocate an increased amount of resources into defense. This is likely true for many, if not all, nutritional symbioses, even though it is not often discussed in this context, given the usually more obvious (and more dramatic) direct effects of nutritional symbiosis on host survival and fecundity. (ii) Microbial symbionts can provide protection to their host by competitively excluding pathogenic microbes.7 (iii) The interaction with symbiotic microorganisms can stimulate or prime the host's immune system and thereby enhance resistance against pathogens, parasites, or parasitoids.8 (iv) Microbes can produce bioactive compounds or their precursors and thereby contribute to their host's defensive chemistry.9,10 In the context of natural products chemistry, defensive symbioses of the last category are the most interesting, as they often involve novel compounds of potential interest for application in human medicine, agriculture, or food technology.
In the present review, we aim to provide an overview of the known defensive symbioses between Metazoa and microorganisms, with an emphasis on associations where host protection is mediated by symbiont-produced secondary metabolites. We are building on previous reviews of microbial protective symbioses in particular groups of animals, including marine organisms,11–14 insects,15–19 and nematodes,20–22 as well as on reviews covering the metabolites produced by symbiotic bacteria.9,10 Generally, we focus particularly on symbioses for which the defensive chemistry has been elucidated, and a protective benefit for the host has been demonstrated or is at least very likely. Most of these involve associations with bacteria, but a few defensive alliances with fungi and dinoflagellates have also been described. As might be expected, bioactive compounds derived from polyketide synthases (PKS) and non-ribosomal peptide synthetases (NRPS) are particularly widespread in defensive symbioses, occurring in marine systems like sponges, corals, ascidians and bryozoans, as well as in terrestrial associations involving nematodes and insects. However, a diverse range of other compound classes with interesting activities occur across symbiotic associations and habitats, including organic acids, phenolics, ribosomal peptides and terpenes (Table S1†). Following our review of the literature on defensive microbial symbioses in animals, we conclude with a synthesis section aimed at deriving general patterns on the chemistry, ecology and evolution of defensive animal–microbe symbioses.
2 Defensive animal–microbe symbioses
2.1 Marine invertebrates
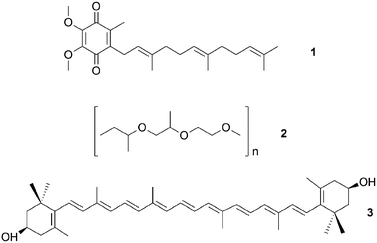
Culture-dependent approaches to isolate defensive symbionts. Some of the first insights in sponge defensive symbioses were gained by culture-dependent approaches. Konya et al.37 followed the reports of surface-associated compounds influencing the settlement of invertebrate larvae causing fouling, and the idea that bacteria might produce these compounds. Concordantly, they succeeded in isolating an Alteromonas strain from the sponge Halichondria okadai that inhibited the settlement of Balanus amphitrite cyprids. The active compound was identified by bioassay-guided fractionation as ubiquinone-8 1. Several structurally related compounds like other ubiquinones but also vitamin K inhibited larval settlement as well.37 Using a similar approach, Dash et al.38 isolated Winogradskyella poriferum from Lissodendoryx isodictyalis, which directly inhibits the settlement of B. amphitite and Hydroides elegans larvae and additionally reduces the growth and biofilm formation of several bacteria that are known to induce larval settlement on sponges. The active compound was identified as a poly-ether 2 of variable chain length.39 However, the specificity and prevalence of both associations and their effect on host fitness remain unknown. A different function was reported by Miki et al.40 for two Flexibacter sp. isolated from the sponge Reniera japonica. The bacteria produce the carotenoid 3R,3′R-zeaxanthine 3, which is a potent quencher of singlet molecular oxygen and a scavenger of free organic radicals, suggesting a protective role against reactive oxygen species (ROS).
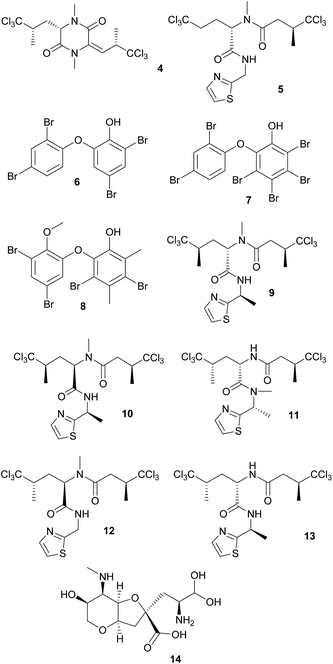
Defensive cyanobacterial symbionts in dictyoceratid sponges. The first culture-independent approaches for the identification of symbiont-produced defensive chemicals in sponges relied on the physical separation of host and symbiont cells. Unson and Faulkner used flow cytometric cell sorting and subsequently located the sesquiterpenes herbadysidolide and spirodysin only in tissue of the sponge Dysidea herbacea itself, whereas the polychlorinated diketopiperazides dihydrodysamide C 4 and demethyl-dihydrodysamide C, as well as 13-demethylisodysidenin 5 were only present in the fraction containing the symbiotic cyanobacterium Oscillatoria spongeliae.41 Flowers et al. repeated the experiment using density gradient centrifugation, verifying the earlier findings and additionally locating didechlorodihydrodysamide C within O. spongeliae.42 They further found a cyanobacterial cell fraction devoid of the chlorinated compounds, indicating either different physiological states or strains of the symbionts. The symbiont-derived compounds were tested for their bioactive potential and shown to strongly deter fish-feeding, suggesting that they are involved in defense against predators in the natural environment.41 Interestingly, D. herbacea can also carry a different strain of O. spongeliae that – instead of the chlorinated compounds – produces polybrominated biphenyl ethers 6–8 that not only deter fish-feeding,43 but also show antimicrobial activity.44 Importantly, these results provided the first description of different sponge chemotypes due to variation in the metabolic profiles of their symbionts, a pattern that was subsequently found repeatedly across several sponge taxa as well as other marine invertebrates.44
Using fluorescence in situ hybridization (FISH) and polymerase chain reaction (PCR) amplification, O. spongeliae was also identified as the producer of polychlorinated peptides like dysidenin 9, iso- 10 and neodysidenin 11, 13-demethylisodysidenin 12 and nor-dysidenin 13 in D. herbacea,45 some of which have been shown to be toxic for fish.46 The primers and probes used for the detection of the dysidenins were based on the biosynthetic gene cluster derived from the cyanobacterium Lyngbya majuscula that produces the homologous compounds barbamide and nordysidenin. The PCR results revealed that not all O. spongeliae strains contain the dysidenin gene cluster, resulting in different chemotypes of the sponge host depending on the symbiont strain.45 In a more extensive screen, Ridley et al. found species-specific secondary metabolite profiles in four dictyoceratid sponge species, comprising either chlorinated peptides, brominated diphenyl ethers or nonhalogenated compounds, mainly sterols.47 Phylogenetic analyses supported a general pattern of co-speciation of the sponges with their respective O. spongeliae symbionts, but also revealed a likely host switch. Additional studies confirmed that the presence of unique symbiont strains in different sponge species of the family Dysideidae48 conferred the characteristic chemical profiles to their hosts and supported the occurrence of host switches and independent infection events.49 Furthermore, D. herbacea individuals can harbor an additional symbiont of the genus Synechocystis, which produces the potent neurotoxin dysiherbaine 14.50 In analogy to the Oscillatoria symbionts, Synechocystis strains vary in their ability to synthesize dysiherbaine, thereby resulting in different host chemotypes. However, the ecological significance of symbiont-mediated dysiherbaine production for the host remains elusive.
Production of bioactive polyketides by sponge symbionts. Polyketide synthases (PKS)51 and non-ribosomal peptide synthetases (NRPS)52 are enzyme complexes that synthesize secondary metabolites based on a stepwise elongation of the product, catalyzed by often repetitive and conserved modules that are encoded in a single operon. The conserved nature of the individual modules provides the opportunity for PCR-based screens with degenerate primers and allows for in silico predictions of possible metabolite structures based on the architecture of the gene cluster.28–30,53 PKS and NRPS gene clusters and/or their products have been reported for several different sponge taxa. From the sponge Pseudoceratina clavata, Kim et al. isolated multiple Salinispora spp. that contained a rifamycin-like PKS gene cluster and showed strong in vitro antibiotic activity.54 Concordantly, rifamycin B and SV could be isolated in vitro, and specific primers detected the biosynthetic genes in most isolated strains. The carribbean sponge Plakortis simplex contains the polyketide plakortin and several derivatives, in addition to the glycosphingolipids plakosides and simplexides, as well as the crasserides and bacteriohopanoids, all of which are mainly or exclusively known from Sphingomonas bacteria.55 Together, these compounds exhibit a wide spectrum of biological activities that might be involved in chemical defense of the sponge against microbes (plakortins: antimicrobial/antimalarial56–58), fish or other predators (crasserides,59 plakortethers60), or in regulating its microbial community by modulating the host's immune system (plakosides,61 simplexides62). An attempt to isolate the plakortin biosynthesis genes failed, but yielded an unusual polyketide–fatty acid synthase hybrid that supposedly synthesizes an acyl chain with various functional groups, probably containing a sulfate group.63 Fisch et al. also exploited the conserved sequence of the ketosynthase (KS) domain to screen metagenomic fosmid libraries from the sponges Psammocinia aff. bulbosa and Mycale hentscheli for candidate bacterial gene clusters involved in the production of psymberin (=ircinastatin A) and mycalamide A, respectively.64 These compounds were long known to exert antiviral65,66 and selective cytotoxic activity against certain tumor cell lines.67 KS sequences were successfully amplified from both sponge metagenomes, and the entire psymberin locus from P. aff. bulbosa was sequenced, but the producing bacteria have not been identified.
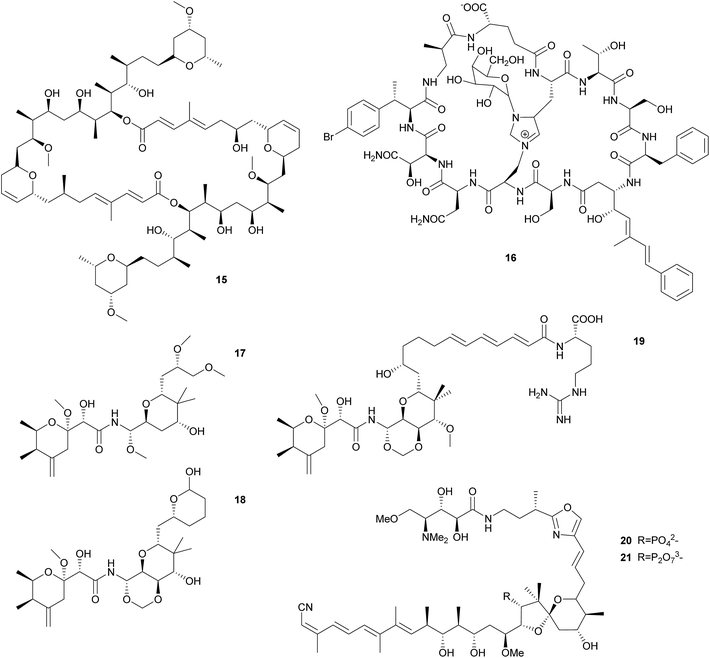
Sponge-associated symbionts in the candidate genus Entotheonella proved to be an especially rich source of polyketides. Using differential centrifugation, Bewley et al. were able to separate the bacteria associated with the lithistid sponge Theonella swinhoei into three fractions, containing unicellular cyanobacteria, unicellular heterotrophic bacteria, and filamentous heterotrophic bacteria, respectively.68 The antifungal and cytotoxic69 macrolide swinholide A 15 was isolated from the unicellular heterotrophic fraction, while a cyclic peptide was isolated from the filamentous heterotrophic bacteria. The latter shows high structural similarity to the antifungal theonegramid70 and was later named theopalauamide 16 and also characterized as antifungal.71 Schmidt et al. characterized the filamentous symbiont from different T. swinhoei chemotypes on the 16S rRNA level and found very closely related species in the chemotypes containing theopalauamide, theonegramide and theonellamide A, respectively.72 The name ‘Candidatus Entotheonella palauensis’ was proposed for the strain from the theopalauamide producing chemotype.
The subsequent exploration of the Entotheonella symbionts in T. swinhoei revealed an extraordinarily large biosynthetic repertoire, including the potential for the production of theopederin A, onnamide A, polytheonamides, as well as keramamides, cyclotheonamides, nazumamide, and proteusins.73 Interestingly, the identification of a bacterium of the genus Pseudomonas as the producer of the polyketide pederin 17 in a beetle and the elucidation of its biosynthesis (see 2.2.2) was a useful starting point to identify the genes responsible for polyketide biosynthesis in T. swinhoei, due to the structural similarity of pederin and the cytotoxic theopederin A 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 74 as well as the cytotoxic and antiviral onnamide A 19.75 PCR-based screening and subsequent sequencing of metagenomic cosmid libraries of different T. swinhoei chemotypes revealed the onnamide gene cluster,53 which was confirmed to be of bacterial origin and closely resembles the pederin cluster. This cluster was only detected within the sponge Y chemotype, which contains solely pederin-like metabolites.76 Later, Freeman et al. reported the ribosome-produced polytheonamides as additional bacterial products from T. swinhoei,77 which form unimolecular ion channels78 and are active against Gram-positive bacteria. Wilson et al. finally attributed the metabolic genes of both onnamide and the polytheonamides to Entotheonella by analyzing single cells via differential centrifugation and fluorescence-assisted cell sorting, followed by multiple displacement amplification and whole genome sequencing of individual bacterial cells.73 Interestingly, the genome sequences of Entotheonella revealed two very similar strains that both carried a plasmid containing the onnamide and polytheonamide genes, but differed remarkably with regard to chromosomally encoded secondary metabolite gene clusters. In addition to the plasmid-localized clusters, the biosynthetically rich TSY1 strain carried 28 secondary metabolite biosynthetic gene clusters, including those for the synthesis of keramamides, cyclotheonamides, nazumamide, and proteusins, as well as a non-functional konbamide cluster. By contrast, the TSY2 strain carried ‘only’ seven additional biosynthetic gene cluster, with nearly no overlap in the secondary metabolite repertoire with TSY1. This diversity in biosynthetic potential was found to extend to the Entotheonella symbionts across several sponge taxa, indicating that Entotheonella strains in the newly described bacterial phylum ‘Tectomicrobia’ will likely serve as a rich source for future discoveries of novel natural products.
74 as well as the cytotoxic and antiviral onnamide A 19.75 PCR-based screening and subsequent sequencing of metagenomic cosmid libraries of different T. swinhoei chemotypes revealed the onnamide gene cluster,53 which was confirmed to be of bacterial origin and closely resembles the pederin cluster. This cluster was only detected within the sponge Y chemotype, which contains solely pederin-like metabolites.76 Later, Freeman et al. reported the ribosome-produced polytheonamides as additional bacterial products from T. swinhoei,77 which form unimolecular ion channels78 and are active against Gram-positive bacteria. Wilson et al. finally attributed the metabolic genes of both onnamide and the polytheonamides to Entotheonella by analyzing single cells via differential centrifugation and fluorescence-assisted cell sorting, followed by multiple displacement amplification and whole genome sequencing of individual bacterial cells.73 Interestingly, the genome sequences of Entotheonella revealed two very similar strains that both carried a plasmid containing the onnamide and polytheonamide genes, but differed remarkably with regard to chromosomally encoded secondary metabolite gene clusters. In addition to the plasmid-localized clusters, the biosynthetically rich TSY1 strain carried 28 secondary metabolite biosynthetic gene clusters, including those for the synthesis of keramamides, cyclotheonamides, nazumamide, and proteusins, as well as a non-functional konbamide cluster. By contrast, the TSY2 strain carried ‘only’ seven additional biosynthetic gene cluster, with nearly no overlap in the secondary metabolite repertoire with TSY1. This diversity in biosynthetic potential was found to extend to the Entotheonella symbionts across several sponge taxa, indicating that Entotheonella strains in the newly described bacterial phylum ‘Tectomicrobia’ will likely serve as a rich source for future discoveries of novel natural products.
Similar to Theonella, the sponge genus Discodermia contains a diversity of bioactive secondary metabolites produced by symbiotic microbes. In fact, Entotheonella symbionts have been reported from different Discodermia species,79–81 which present a large diversity of PKS clusters.79,81 Additionally, the cytotoxic cyclic peptides calyxamide A and B, structurally similar to the above mentioned keramamides, were isolated from Discodermia calyx.81 However, it has only been possible in a single case to unambiguously connect secondary metabolite production to a specific bacterium in a Discodermia host: Wakimoto et al. sequenced the gene cluster responsible for the production of calyculins from the metagenome of D. calyx, localized the PKS cluster using FISH within filamentous bacteria and isolated these by laser microdissection.82 PCR on the isolated bacteria confirmed the PKS localization and identified the symbionts via 16S rRNA analysis as an Entotheonella species. Interestingly, the authors were also able to characterize a means for storage of a defensive compound in a form that is harmless for the host. The usually cytotoxic calyculin A 20 is phosphorylated by the Entotheonella symbionts and stored as the less toxic diphosphate 21. Upon wounding of the sponge, the phosphocalyculin is rapidly converted by a host-derived enzyme to the more than a thousand times more toxic calyculin, thus representing an activated chemical defense mechanism.82
Fungal defensive symbioses in sponges. In contrast to the wealth of knowledge on protective bacterial symbionts in sponges, convincing evidence for defensive fungal symbionts is lacking.25,83 This is insofar surprising as the number of fungal species isolated from sponges84 and their potential for secondary metabolite production is tremendous.33,85 A few studies have addressed the symbiotic aspect of sponge–fungi relationships, and shown maternal transmission of a yeast in the sponge Chondrilla,86 horizontal gene transfer between fungi and sponge mitochondria,87 as well as fungal recognition proteins in sponges.88 Another indication of the potentially symbiotic nature of fungi in sponges is the presence of specific fungal 18S rRNA sequences in sponge databases.24 Furthermore, sponge-associated fungi were found to contain a large diversity of PKS and NRPS genes,89 but their possible roles in the defense of the host remain enigmatic.
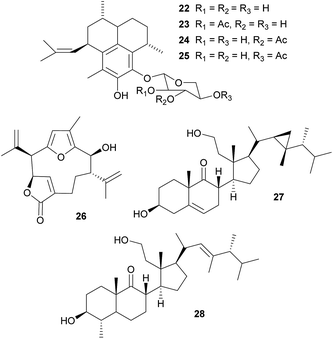
Defensive diterpenes. Like many other sessile marine animals, corals are a rich source of bioactive secondary metabolites that play an important role in the defense against predators.99–101 Among these, the pseudopterosins 22–25 are a group of tricyclic diterpene glycosides with potent antiinflammatory and analgesic activity that were originally isolated from the soft coral Pseudopterogorgia elisabethae.102–104 Enrichment of P. elisabethae's symbiotic dinoflagellates of the genus Symbiodinium by differential centrifugation revealed the predominant localization of the pseudopterosins in the symbiont fraction, suggesting that they are produced by the dinoflagellates.105 Concordantly, incubation of this fraction with either NaH14CO3 or tritiated geranylgeranyl diphosphate (3H-GGDP) resulted in labeled pseudopterosins.105 A similar strategy of symbiont cell enrichment and subsequent radioactive labeling with 3H-GGDP revealed the Symbiodinium-mediated production of kallolide A 26 in Pseudopterogorgia bipinnata.106 Interestingly, only one out of four different Symbiodinium strains exhibited kallolide production in vivo, indicating differences in chemical properties and defensive capabilities across different symbionts.106 Even though the adaptive significance of the symbiont-produced pseudopterosins and kallolides for the coral hosts has not yet been demonstrated in vivo, extracts of both coral species (P. elisabethae and P. bipinnata) were unpalatable to the generalist fish predator Thalassoma bifasciatum,101 highlighting the potential importance of the Symbiodinium-produced bioactive compounds for the antipredator defense of the coral host.
Secosterols. Secosterols isolated from corals, sponges, and ascidians can exhibit a diverse range of biological activities, including antiproliferative, antifouling, antiinflammatory, antimicrobial, ichthyotoxic and antiviral.107 In the octocoral Pseudopterogorgia americana, bioassay-guided fractionation revealed the deterrent activity of 9,11-secogorgosterol 27 and 9,11-secodinosterol 28 against predatory fish in laboratory and field assays.108 Even though the source of the secosterols in P. americana has not been unambiguously identified, zooxanthellae isolated from other marine organisms (including a coral) were reported to produce gorgosterol and dinosterol.109 Furthermore, gorgosterol is transformed to 9,11-secogorgosterol by enzyme preparations of P. americana colonies.110 Thus, it seems likely that dinoflagellate symbiont-produced precursors are modified by host enzymes to synthesize the defensive secosterols.
Protective symbionts in Hydra. The epithelial surfaces of freshwater polyps in the genus Hydra harbor stable and species-specific bacterial assemblages111,112 that are shaped by the host via antimicrobial peptides.113 By generating germ-free animals and reinfecting them with individual bacterial taxa or combinations thereof, a recent study revealed that the symbiotic community of Hydra vulgaris plays an important role in protecting the host against fungal infestation.114 Although the mechanistic basis of the protective effect remains to be elucidated, both in vitro and in vivo studies point to a combined activity of the host and its microbiota in pathogen defense.114
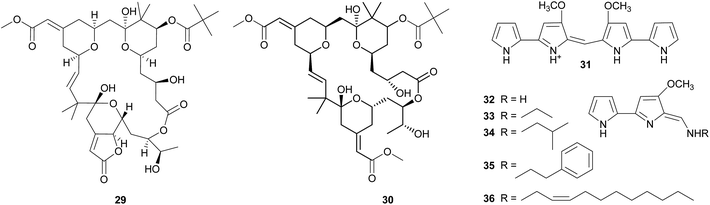
Bryostatins. The cosmopolitan bryozoan Bugula neritina is chemically defended against predators by a cocktail of cyclic polyketides, the bryostatins.123,124 While these compounds are present in low concentrations in adult B. neritina colonies, the abundance of bryostatin 10 29 and bryostatin 20 30 is strongly increased in young larvae.125 By binding to the diacylglycerol binding site of protein kinase C's regulatory domain,126 the bryostatins exert toxicity and deterrence to fish, corals, and sea anemones and thereby protect B. neritina larvae from predation.125,127–130 Importantly, attacked and rejected larvae show high rates of settlement, demonstrating a direct fitness benefit from chemical protection.129,130 After settlement and metamorphosis, bryostatin levels rapidly decrease, indicating a switch from chemical to structural defense as the colony matures.125
Soon after the structure elucidation of bryostatin 1, this compound was suspected to be of bacterial origin rather than produced by B. neritina itself.131 Concordantly, earlier studies had already reported on rod-shaped bacteria that are consistently associated with adult and larval B. neritina.132,133 Based on the 16S rRNA sequence, these bacteria were later described as a new taxon within the γ-Proteobacteria and named ‘Candidatus Endobugula sertula’.134 A series of subsequent studies provided convincing evidence that the bryostatins are indeed produced by ‘Ca. E. sertula’, thereby constituting one of the best documented cases of defensive symbiosis between animals and microorganisms in the marine environment. Davidson et al.135 used in situ hybridization to co-localize the symbiotic bacteria and a polyketide synthase (PKS) gene fragment putatively involved in bryostatin synthesis. Simultaneous fluorescent detection of ‘Ca. E. sertula’ and the bryostatins later revealed the dynamics of bryostatin production during the life cycle of B. neritina.115 As expected under the hypothesis of symbiont-mediated bryostatin synthesis, reduction of symbiont titers in adult B. neritina by antibiotic treatment resulted in a strong decrease in bryostatin concentrations.135 The offspring of antibiotic-treated colonies likewise showed strong reductions in symbiont abundance and bryostatin concentrations, and symbiont-free larvae failed to deter predatory fish.128 Interestingly, however, settlement and growth of juvenile B. neritina was not affected by symbiont elimination, indicating that the defensive capacities of the symbionts are the only or at least the most important benefit for the host.128
Efforts to elucidate the genomic basis of bryostatin production resulted in the discovery of a single large PKS gene cluster (bry) in a B. neritina genomic library enriched for bacterial DNA.136,137 This gene cluster is expressed in ‘Ca. E. sertula’ cells in the pallial sinus of B. neritina larvae, and expression is not detectable after symbiont elimination through antibiotic treatment, providing further evidence that it is indeed encoded by the ‘Ca. E. sertula’ genome.135 Bioinformatic predictions supported the biosynthesis of the bryostatin core structure by the bry gene cluster,124,138 and heterologous expression of bryP and bryA confirmed the functionality of these genes.139,140 The symbionts of two sibling species of B. neritina exhibited high similarity in structure and sequence (98%) of the bry gene cluster, indicating a common ancestry.137
The occurrence of bryostatin-producing symbionts was confirmed for two sibling species of B. neritina as well as for Bugula simplex.141,142 Surprisingly, a third sibling species of B. neritina was devoid of bryostatin-producing symbionts,143 but still exhibited deterrence to a fish predator, providing evidence for additional defensive compounds produced by the bryozoan itself or an as yet unknown symbiont.128 In Bugula pacifica and B. turbinata, symbionts closely related to ‘Ca. E. sertula’ and ‘Ca. E. glebosa’ (the symbiont of B. simplex) were discovered, but no bryostatin activity could be detected.144 Interestingly, extracts from B. pacifica showed broad-spectrum antibacterial activity, suggesting that defensive compounds other than bryostatins are present and may be produced by the symbionts.145 Three additional Bugula species – B. dentata, B. stolonifera, and B. turrita – appeared to be devoid of the symbionts.144 The patchy occurrence of Endobugula symbionts across host species indicates a dynamic symbiotic association with frequent host switches or symbiont acquisitions/losses. Given the deficiency in recombination of the symbionts,124 changes in defensive chemistry by symbiont switches or replacements might be advantageous in the arms race against co-adapting predators. Alternatively, the symbiotic partnership may respond by changing the absolute or relative composition of the bryostatin cocktail, which can influence its activity against predators.128
Tambjamines. The tambjamines 31–36 are a group of 4-methoxypyrolic natural products that occur across several taxonomically distinct groups of marine organisms, including bryozoans,146,147 nudibranchs,148 and ascidians.149,150 Based on this disparate distribution and the occurrence of identical or closely related compounds in bacteria,151,152 the tambjamines were suspected to be of microbial origin. The discovery of the tambjamine-producing marine bacterium Pseudoalteromonas tunicata152 and its association with a range of marine animals153 – including bryozoans, mussels, ascidians, fish, corals, and sponges153–155 – support this hypothesis. Recently, the molecular basis of tambjamine production in P. tunicata was elucidated by heterologous expression of the tam gene cluster in E. coli.156 The tambjamines show toxicity and/or deterrence against predatory fish as well as antimicrobial activity, indicating that they might confer protection from both pathogens and predators.152,157–161 Some predatory nudibranchs, however, are resistant to the adverse effects of tambjamines; in fact, they sequester the bioactive compounds from their bryozoan or ascidian diet and use them for their own defense.148,160
Gastropods. An interesting case of symbiont-mediated structural protection has been described in the scaly snail Crysomallon squamiferum, a gastropod occurring at hydrothermal vents.171 The snail's foot is covered in hardened scales of multiple layers that likely confer protection against predation.171,172 The outer layer is composed of pyrite (FeS2) and greigite (Fe3S4), whose biosynthesis has not been described in metazoans. Interestingly, a community of δ- and ε-Proteobacteria, which are known for their ability to recycle sulfur and mineralize iron sulfides, were found to live in association with the snail.171 Thus, it was suggested that the bacterial partners are responsible for depositing the outer scale layer and thereby confer protection to the snail host.171 However, another study based on the structural and chemical composition of the scales suggests that the snail itself controls the biomineralization via sulfur compounds derived from the hydrothermal vents.173 To our knowledge, no study to date has taken an experimental approach that aims to manipulate the bacterial community associated with the snail, so the case remains unresolved.
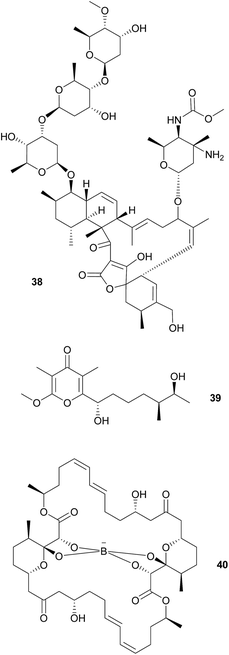
As cone snails are well-known for their arsenal of protective peptide toxins, further microbe-derived defensive compounds were not expected. Surprisingly, however, Peraud et al. found a diverse actinomycete community associated with different cone snails of the genus Conus that displayed bioactive properties.174Streptomyces sp. CP32 isolated from C. pulicarius produces several benzyl thiazole and thiazoline compounds (aerugine, pulicatins A–G and watasemycins A & B) that exhibit antimicrobial, anti-inflammatory and antihypotensive activity.175 Another Streptomyces isolate from C. tribblei that also produces pulicatin A was hypothesized to protect the snail surface against microbial colonization.175 Eight nobilamides and two related compounds were identified in further isolates from C. tribblei and Chicoreus nobilis, some of which inhibit the TRPV1 cation channel that is a major mediator of pain and inflammation in vertebrates.176 A Gordonia sp. isolate from a different Conus species produces a number of circumcin derivatives that show neuroactivity or broad antimicrobial activity.177 Also, another Streptomyces sp. isolated from the recently discovered turrid gastropod Lienardia totopotens produces the antibacterial and cytotoxic lobophorins 38.178 However, for the majority of these compounds, evidence for a beneficial effect on the host's fitness is lacking, so the possible mutualistic nature of the associations remains to be established. Unlike the previous cases, nocapyrones 39 are already long known from mollusk secreted mucus. Some are either toxic for various predators or induce escape reactions in conspecifics.179 Interestingly, the ncp PKS gene cluster for three derivatives of this class of compounds, which are secreted in the mucus of C. tribblei and C. rolani, were identified in the bacterium Nocardiopsis alba.180
Wood boring bivalve mollusks in the family Teredinidae (“shipworms”) harbor various symbionts in their gills181 and gastric caeca,182 that are known to contribute to the host's carbon metabolism by providing cellulose degrading enzymes.183 Furthermore, Teredinibacter turnerae, found in the gills of the shipworms, seems to be involved in structuring the community of shipworm-associated bacteria. The sequenced genome contains three PKS and six NRPS gene clusters,184 one of which encodes for the biosynthesis of tartrolons 40 that occur across all shipworm tissues.185 While the two isolated tartrolons (one as the free form and the other chelating a boron atom) show no activity against eukaryotic cells or the shipworm's native microbial community, they inhibit the growth of B. subtilis and marine pathogenic bacteria.185
Cephalopods. The association of the Hawaiian bobtail squid, Euprymna scolopes, with the luminescent bacterium Vibrio fischeri is undoubtedly one of the best-studied symbiotic model systems, particularly with regard to the molecular basis of host–symbiont interactions mediating the specific establishment and maintenance of the association.186,187 The squid carries V. fischeri bacteria in a specialized light organ that helps to disguise the squid from predators and prey through ‘counterillumination’.188 While not a chemical defense per se, the symbionts' light emission is a by-product of a biochemical reaction in which luciferase catalyzes the reaction between an aliphatic aldehyde substrate (reduced flavin mononucleotide) and molecular oxygen.189 The association with bioluminescent V. fischeri is not confined to E. scolopes, but also occurs in several other squid as well as fish species.189
Presumably, symbiotic bacteria also reside within the accessory nidamental gland (ANG) of several squid genera, including Loligo, Sepia and Euprymna. The ANG houses a highly specific bacterial community of α- and γ-Proteobacteria as well as Bacteroidetes, with Roseobacter dominating in Loligo and Sepia species, and Phaeobacter in Euprymna scolopes.190–192 Sexual maturity in these squids is accompanied by the enrichment of symbiont-synthesized carotenoids, although the exact function of those carotenoids remains unknown. It is also uncertain whether a specific carotenoid-producing physiological stage in the bacteria is required for maturity of the females, or whether maturing females induce the bacteria to produce the carotenoids.193–195 During oviposition, the bacteria are transferred from the ANG to the eggs and likely serve as an inoculum resulting in dense bacterial populations within the egg capsules.192 However, no symbiotic bacteria were found on hatched embryos, indicating that the squids acquire their symbiotic microbiota de novo from the environment in every generation.196,197 Extracts from the ANG contained high amounts of unsaturated fatty acids and exhibited antimicrobial activity, as did egg extracts and bacterial isolates.198–200 In addition to active inhibition, the secreted bacteria might provide colonization resistance of the egg capsules by depleting nutritional resources.201
A symbiosis with both nutritional and defensive benefits occurs in marine isopods of the genus Santia.206 These crustaceans harbor a photosynthetically active episymbiotic community comprising Cyanobacteria of the genus Synechocystis. In order to provide their symbionts with suitable conditions for photosynthesis, the isopods occupy exposed areas with sufficient sunlight. Two investigated populations or species (the actual status has not been determined) showed remarkable differences regarding their symbionts and the defense against predators. One population, whose large epibiotic Synechocystis symbionts confer a characteristic red coloration to their hosts, is usually ignored or rejected by predatory fish, while the other population carrying an inconspicuous brown Synechocystis strain is readily consumed.206 The symbiont seems to be vertically transmitted from mothers to newly emerged juveniles and – in addition to the difference in color – shows morphological strain variation across the two host populations.206 When experimentally removed from their surface, the isopods were equally consumed by fish. Methanol extracts of isopods with their red symbionts partially restored protection, indicating that symbiont-produced bioactive metabolites are involved in their host's defense against predators.206 However, the chemical basis of the protective effect remains to be elucidated.
Didemnid ascidians. Colonial ascidians of the family Didemnidae have been studied extensively as producers of a rich repertoire of bioactive secondary metabolites, many of which are produced by microbial symbionts.12,207–210 We will focus here on five groups of compounds, for which a symbiotic origin has been demonstrated or is at least very likely: the cyanobactins (including patellamides, trunkamide, lissoclinamides, patellins, and many others), didemnins, patellazoles, bistramides, and palmerolides.
Many didemnid ascidians live in an obligate symbiosis with vertically transmitted Cyanobacteria of the genera Prochloron or Synechocystis.211–213Prochloron symbionts have been found on the surface and/or in the common cloacal cavity of colonial didemnids such as Lissoclinum patella, L. bistratum, L. voeltzkowi, L. punctatum, Trididemnum cyclops, T. clinides, Didemnum molle, and Diplosoma virens,212,213 while Synechocystis is associated with ascidians of the genus Trididemnum.211 Through photosynthesis, the cyanobacterial symbionts make a major contribution to the hosts' energy demands, and they play an important role in the recycling of nitrogenous compounds.214 In addition to these nutritional contributions, the symbionts have been implicated in the production of bioactive secondary metabolites that play a role in the defense of the host.207,209
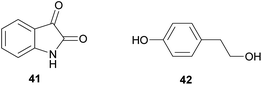
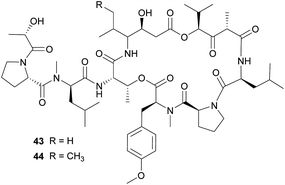
The didemnins, potent antiviral and antitumor cyclic peptides, were first isolated from the Caribbean ascidian Trididemnum solidum,215,216 which hosts the cyanobacterial symbiont Synechocystis trididemni.211 Behavioral assays demonstrated that T. solidum larvae are distasteful to predatory fish species, and two isolated didemnins (didemnin B 43 and nordidemnin B 44) significantly deterred predators when applied at naturally occurring concentrations.158,217,218 Since didemnin B was also found in a phylogenetically distant ascidian and shows structural similarity to metabolites from free-living cyanobacteria, it was suspected to be of symbiotic origin in T. solidum.209 While there is to our knowledge no direct evidence supporting a cyanobacterial source of the didemnins in T. solidum, the recent discovery of a plasmid-localized didemnin biosynthetic gene cluster in the free-living α-Proteobacteria Tistrella mobilis and T. bauzanensis219,220 raises the possibility that S. trididemni has acquired the potential for didemnin biosynthesis via horizontal gene transfer.
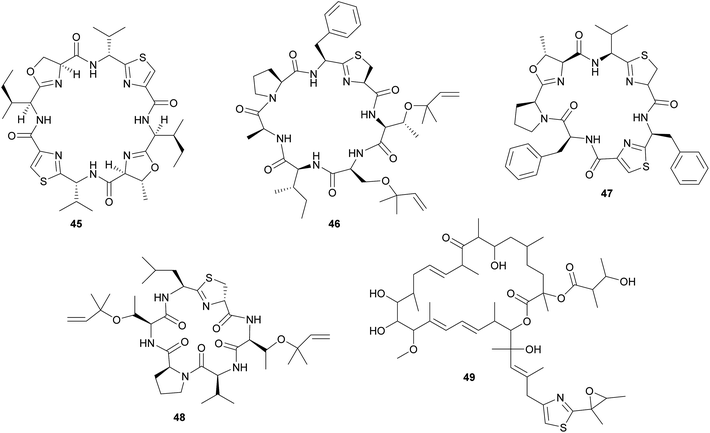
In analogy to the didemnins, it was long suspected that another group of cyclic peptides in ascidians, the cyanobactins (including the patellamides 45, trunkamide 46, lissoclinamides 47, patellins 48, and many others), are produced by cyanobacterial symbionts. This hypothesis was based on the co-occurrence of Prochloron symbionts and cyanobactins in several didemnid ascidians, particularly those of the genus Lissoclinum.221 Indeed, more recent studies identified the Prochloron gene cluster responsible for patellamide production (pat) and demonstrated its activity by heterologous expression in Escherichia coli.222–224 Notably, the discovery of the pat gene cluster224 represents one of the first examples to elucidate the biosynthetic pathway for the production of a symbiont-produced defensive metabolite in a marine system by whole genome sequencing. Interestingly, the pat cluster is highly conserved across Prochloron symbionts of diverse hosts, but hypervariable cassettes in the precursor peptide result in the large diversity of cyclic peptides.222 Analogously, the tru cluster is responsible for the synthesis of diverse patellins, including trunkamide, and it shares a high degree of similarity with the pat genes, except for the region that is likely involved in the prenylation of the patellins.225 Thus, the variability of the cyanobactin gene clusters confers the metabolic versatility to the ascidian symbiosis as well as to free-living cyanobacterial relatives.225 Even though the fitness benefits of symbiont-mediated cyanobactin production for the host have not been demonstrated, their abundance in ascidian tissues and toxicity against eukaryotic cells strongly imply a protective function.11,209
In addition to the cyanobactins, individuals of the ascidian Lissoclinum patella are occasionally found to contain the toxic patellazoles 49, a group of thiazole-containing polyketides.226,227 Metagenomic approaches towards the identification of the patellazole-producing organisms excluded the Prochloron symbionts as possible candidates and rather pointed to a proteobacterial origin of these secondary metabolites.228 Subsequent studies verified this by identifying the patellazole gene cluster (ptz) in the intracellular α-proteobacterial symbiont ‘Candidatus Endolissoclinum faulkneri’.229 Interestingly, apart from the trans-AT PKS gene cluster responsible for patellazole synthesis, the genome of ‘Ca. E. faulkneri’ shows clear signs of erosion, with a strongly reduced size and coding density, an AT-biased nucleotide composition, and the loss of regulatory genes involved in DNA replication and cell division.229 Thus, ‘Ca. E. faulkneri’ appears to be an obligate defensive mutualist of L. patella, similar to the recently discovered ‘Candidatus Profftella armatura’ in the asian citrus psyllid, which retained the complete pathway for the putatively defensive compound diaphorin in an otherwise eroded genome230 (see 2.2.2). As for the cyanobactins, the role of the patellazoles in the defense of the symbiosis against antagonists still needs to be established.
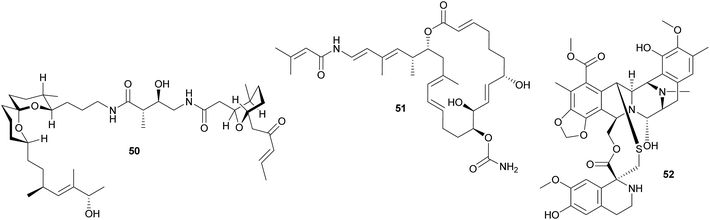
Other polyketides in didemnid ascidians include the bistramides 50 of Lissoclinum bistratum,231–234 and the palmerolides 51 of Synoicum adareanum.235 While the former were localized to the Prochloron symbionts by cell fractionation,231 the evidence for a microbial origin of the latter is limited to the sequencing of bacterial trans-AT PKS ketosynthase domain fragments putatively involved in palmerolide synthesis.235 Finally, it should be noted that metagenomic analyses of Prochloron symbionts in L. patella revealed further secondary metabolite gene clusters, which may be involved in the synthesis of as yet unknown bioactive compounds for protection against antagonists.236
Other ascidians. The intracellular γ-proteobacterial symbiont ‘Candidatus Endoecteinascidia frumentensis’ was identified in the mangrove ascidian Ecteinascidia turbinata (Perophoridae).237,238 The bacteria are probably vertically transmitted, and recent studies identified the core of an NRPS biosynthetic gene cluster that could be tied to the intracellular symbiont through analyses of the codon usage.239 This cluster is likely responsible for the synthesis of the secondary metabolite ecteinascidin 743 52 (ET-743),239 a promising anti-cancer agent that is highly toxic to eukaryotic cells and may therefore serve as an anti-predator defense in the ascidian symbiosis.
2.2 Terrestrial invertebrates
The two entomopathogenic nematode families Steinernematidae and Heterorhabditidae are characterized by their obligate association with bacteria in the γ-proteobacterial genera Xenorhabdus and Photorhabdus, respectively. Although some of these symbionts can occur in multiple hosts, most strains are species-specific and essential for growth and reproduction of their nematode hosts.20 Specifically, they assist the nematode in overcoming the immune system of the insect prey, killing it, and protecting the cadaver against microbial and animal competitors.246 To this end, an arsenal of diverse bacterial metabolites do not only repel insect scavengers like ants, but are also active against viruses, con- and hetero-specific bacteria, saprobic fungi, protozoa and nematode competitors. Their defensive chemistry enables the bacteria to essentially monopolize the insect for 1–2 weeks after colonization, which ensures optimal resource use by the nematode-symbiont consortium as well as successful acquisition of the symbiont by the host offspring.20 Here we review the protection of the insect cadaver through defensive chemical compounds synthesized by the bacteria, but do not discuss the chemistry involved in killing the insect host, which is an offensive rather than defensive symbiont-provided benefit and has been reviewed extensively elsewhere.20,241,243
All Steinernema and Heterorhabditis nematodes go through an infective free-living juvenile phase, during which they carry the bacterial symbionts in their intestinal tract. After location of a suitable prey by active search or ambushing, the nematode enters the insect through the respiratory or digestive system, penetrates the hemocoel and releases the bacterial symbionts.247 The host insect is typically killed 24–48 hours after infection, which is when the bacteria reach high abundances. Most defensive compounds are produced by the bacteria during the following post-exponential phase of growth. For both nematodes and their symbionts, successful colonization of the insect host is crucial, as nematodes cannot re-emerge from an insect after infection and thus have only a single chance to colonize a host.247 This may explain why both Xenorhabdus and Photorhabdus independently evolved extraordinarily effective insect-killing and carcass-defending abilities.248 However, although functionally similar by conferring protection against the same enemies, the defensive metabolites of both groups are structurally very different.
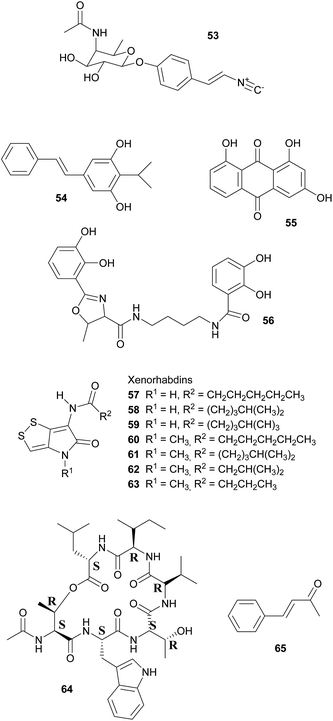
Broad-spectrum antibiotic activity of metabolites from the bacterial symbionts of nematodes against Gram-positive and Gram-negative bacteria, yeasts and filamentous fungi was already demonstrated in the 1980's, long before the chemical nature of most of the antibiotic substances was known.249–251 Subsequently, researchers attributed these antimicrobial effects to a series of (i) highly specific proteinaceous bacteriocins, i.e. lumicins252 and photorhabdicins253 from Photorhabdus spp., and xenorhabdicins254–256 from Xenorhabdus spp.; (ii) broad-spectrum antibacterial and antifungal compounds, including isopropylstilbenes,245,257,258 antraquinones,242,259 and a carbapenem260 from Photorhabdus spp., as well as fabclavines,261 xenorhabdins,262,263 xenorxides,242 and nematophins,245,264 from Xenorhabdus spp.; and (iii) narrow-spectrum anti-Gram-positive xenobactin,265 xenematide266 and xenocoumacins,267 and anti-Gram-negative benzylideneacetone268 from Xenorhabdus spp. Additionally, several other metabolites including xenoamicin,265 taxlllaids,269 cyclohexandione,270 chaiyaphumine271 and szentiamide272,273 with activity against human-disease causing protozoa (Plasmodium falciparum, Trypanosoma brucei) have been identified. While it is possible that these compounds defend the nematodes against competitors within the insect cadaver, direct evidence for their ecological role is thus far lacking. Finally, both Photorhabdus and Xenorhabdus spp. produce rhabduscin 53 and chitinases, both with a dual function – the promotion of prey killing/digestion and defense against fungal competitors274–276 – and small molecules that deter animal scavengers.20 In the following paragraphs, we will expand on the nature and function of the defensive compounds, with a focus on the bacteriocins and the anti-bacterial small molecules.
Bacteriocins are killer proteins used by bacteria to defend themselves against closely related competitors.20,253 In Photorhabdus and Xenorhabdus, three kinds of bacteriocins, the lumicins, photorhabdicins and xenorhabdicins, have been described. Normally detectable in low quantities, their production is strongly induced when bacterial cells are lysed.255 Xenorhabdicins were first described from X. nematophila and shown to be active against strains of Xenorhabdus, Photorhabdus, and related sister taxa.256 Likewise, Photorhabdus spp. synthesize photorhabdicins and lumicins.252 The biosynthetic genes for lumicins have been shown to be co-localized with the respective resistance genes, which together are highly diverse between symbiont strains.253 This likely ensures specificity of the bacteria–nematode partnership, if multiple founder nematodes colonize the same insect. Indeed, assays with different Xenorhabdus strains showed that their bacteriocins are primarily active against conspecific rather than heterospecific strains.254
Defense against unrelated bacterial competitors (e.g. the insect's gut community), fungi and animals is mediated by extracellular, non-proteinaceous small molecules with variable narrow- to broad-spectrum activity.20,242,250 Together, these compounds assure that the insect carcass does not putrefy for several weeks until the nematodes disperse.277 In Photorhabdus, carbapenem-like molecules, as well as isopropylstilbenes and anthraquinone pigments are mainly responsible for this effect.245,258,259,277,278 Carbapenems are β-lactam antibiotics that are best known from Enterobacteria. In P. luminescens, a gene cluster responsible for the production of a carbapenem-like molecule with specific activity against Gram-negative bacteria has been identified.277 This strain also synthesizes isopropylstilbene antibiotics that generally suppress bacterial growth by inhibiting RNA synthesis, of which one, 3,5-dihydroxy-4-isopropylstilbene 54, is also strongly fungicidal, nematicidal and insecticidal.20,251,258,279 This compound is probably of crucial importance for defense, as large amounts are synthesized by the symbionts from days 2–5 after colonization of the insect prey throughout the following weeks until the cadaver is abandoned.257,280 Anthraquinone pigments 55 produced by a type II PKS281 are responsible for the red color of insects killed by Photorhabdus.278 Several of these pigments have been isolated from the bacterial symbionts, which is remarkable as these compounds normally occur only in higher plants, lichens and fungi.242,258,259,282 Anthraquinone derivatives have antibiotic and nematicidal properties, thus indicating a defensive function.241,259 This is also assumed for photobactin 56, a catechol siderophore from P. luminescens, although its exact function remains to be determined.260
Xenorhabdus spp. synthesize a different array of bioactive small molecules, including xenorhabdins, xenorxides, fabclavines, indole derivatives, xenocoumacins, xenematide, xenobactin, and benzylideneacetone.20,241,251,261 Xenorhabdins 57–63, the largest group among these, are dithiolopyrrolone derivatives (compounds also known from Streptomyces) with suppression of Gram-positive bacteria and fungi by inhibition of RNA and protein synthesis.251,262,283 In many cases, several xenorhabdins are produced by the same bacterial strain, and as some are also insecticidal, they fulfill a double function by killing the insect and preserving/protecting the carcass against competitors.242 Oxidized xenorhabdins, the so-called xenorxides, are broad-spectrum defensive metabolites against both Gram-positive and Gram-negative bacteria as well as fungi.242 Four types of fabclavines have been identified from X. budapestensis and X. szentirmaii and are active against a broad spectrum of bacteria, fungi and protozoa.261 Indole derivatives, like nematophin from X. nematophilus,264 likewise have a broad activity spectrum and are comparable to isopropylstilbenes in terms of their mode of action.245,251 By contrast, xenocoumacins, xenematide and xenobactin inhibit Gram-positive bacteria,241,266,267 with xenobactin 64, a hexadepsipeptide, also being active against protozoa.265 Complementary to xenobactin, benzylideneacetone (trans-4-phenyl-3-buten-2-one) 65 specifically suppresses growth of Gram-negative bacteria.268
Ant-deterrent factors (ADFs) are small extracellular molecules that protect insect cadavers infected by both the Heterorhabditis–Photorhabdus and the Steinernema–Xenorhabdus symbiotic complexes against scavenging arthropods, particularly ants.284 ADF repellency depends on the strain and age of the bacteria and the ant species tested,285 with the Heterorhabditis–Photorhabdus association being the better protected complex.284 To date, however, the chemicals responsible for ant-deterrent effects have not been identified.
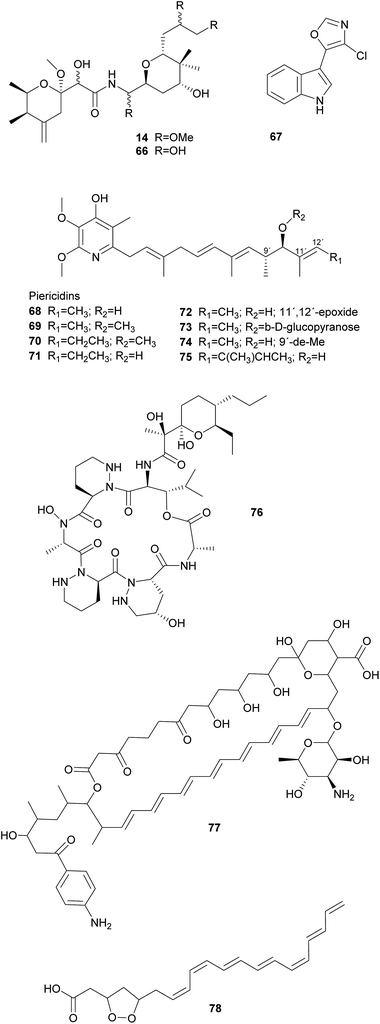
Symbiotic antipredator defense. Natural enemies of insects include predators, parasitoids, and microbial pathogens, as well as nematodes and viruses. Examples of symbiont-conferred protection have been discovered against all of these antagonists. However, an anti-predator function has so far only been demonstrated for the association between rove beetles (Paederus spp.) and a close relative of Pseudomonas aeruginosa.287 These γ-Proteobacteria are capable of producing pederin 14, a potent toxin that is synthesized using enzymes of the trans-AT PKS family and resembles onnamide-type natural products found in sponges (see 2.1.1).10 The ecological relevance of this defensive compound is supported by the observation that beetle larvae hatching from pederin-containing eggs experience reduced predation from wolf spiders as compared to pederin-free larvae.288 Interestingly, there is evidence that the symbionts have horizontally acquired the genes required for the production of pederin, suggesting that mobile genetic elements may explain the widespread capability of producing highly similar bioactive metabolites in a range of phylogenetically distant symbiotic partners.289 In fact, a recently described case of a probable defensive symbiosis between the asian citrus psyllid and the β-Proteobacterium ‘Candidatus Profftella armatura’ further supports this hypothesis.230 The highly reduced genome of the bacterial symbiont encodes the complete gene cluster for the synthesis of diaphorin 66, a toxin that is structurally very similar to onnamides and pederin. Thus, the gene cluster might have been transferred to or from the rove beetle symbiont. Notably, ‘Ca. P. armatura’ and the production of diaphorin are observed without exception among individuals within and across geographically distant psyllid populations. This high prevalence suggests an obligate mutualistic association and diverges from the usually intermediate infection frequencies described for the majority of defensive symbioses.230
Symbiont-mediated protection against parasitoids, fungi, and nematodes in aphids and fruit flies. One of the earliest known cases of symbiont-mediated defense in insects involves the protection against parasitoid wasps in aphids. In the aphid Acyrthosiphon pisum, Hamiltonella defensa bacteria confer protection against the wasp Aphidius ervi.290 However, this defensive action depends on the presence of the bacteriophage APSE (A. pisum secondary endosymbiont) in the symbiont, which encodes toxins that are likely candidates for the defensive activity. Concordantly, three APSE variants that confer different degrees of protection carry distinct toxin genes, encoding for the production of shiga toxin, cytolethal distending toxin, and YD-repeat toxin, respectively.19H. defensa also protects other aphid species against parasitoids, i.e. Aphis fabae and likely also Aphis craccivora,19 although the same defense mechanism might not operate in other host species like the grain aphid (Sitobion avenae). Interestingly, however, an alternative strategy for protection by this secondary symbiont in S. avenae is still likely, as parasitoid wasps preferentially oviposit in H. defensa-free eggs.291 In addition to Hamiltonella, the secondary symbionts Regiella insecticola and Serratia symbiotica can provide resistance against parasitic wasps in aphids. These cases, however, are not bacteriophage-mediated, suggesting alternative strategies for protection.19 Symbiont-conferred protection against parasitoids has also been reported in other insects, e.g. Drosophila hydei, in which Spiroplasma can defend the larvae against the wasp Leptopilina heteroma.292 Additionally, some studies suggested that Arsenophonus in psyllids293 and Wolbachia in the weevil Hypera postica294 can similarly enhance the resistance of the host against parasitoids. In both cases, however, further experimental evidence is required to confirm the existence of a defensive symbiosis and to elucidate the mechanistic basis of protection.
The role of facultative symbionts in the defense against pathogenic fungi has also been studied in aphids. While Hamiltonella appears to have no effect on aphid susceptibility to fungal pathogens, at least four other secondary symbionts of the pea aphid (Rickettsia, Rickettsiella, Regiella and Spiroplasma) are capable of increasing survival chances of aphids exposed to the entomopathogen Pandora neoaphidis.295,296 In addition, the presence of these symbionts also reduces sporulation efficiency of the fungus in those cases where the pathogen kills the aphid. This may be adaptive for the aphids by reducing the spread of infection among groups of clonal aphids, thereby enhancing the inclusive fitness of the clone.295,296 However, the mechanistic basis of the symbiont-mediated protection against pathogenic fungi in aphids remains to be elucidated.
Little is known about symbiont-mediated defense against nematodes, with only one reported case in Drosophila neotestacea, in which Spiroplasma symbionts significantly enhance the reproductive output of flies that are parasitized by the nematode Howardula aoronymphium both in laboratory and wild populations.297,298 The presence of Spiroplasma results in reduced growth of the adult female nematodes within the host and ultimately in impaired fertility of the parasite as well as a reduced virulence against the host.298 Although the mechanistic basis of Spiroplasma's protective activity is not yet fully known, transcriptional profiling suggests the production of toxins that may inactivate the ribosomes of parasitic nematodes.297
Protection against pathogens: Actinobacteria as defensive symbionts. Actinobacteria are of great importance for humans – most of our antibiotics today originate from these bacteria, specifically from members in the genus Streptomyces. But also other organisms make use of Actinobacteria and their defensive capabilities through protective symbioses.16 Interestingly, however, it remains a matter of debate whether antibiotics primarily evolved to defend their producers in nature. Instead, their immense diversity and occurrence in often sub-inhibitory concentrations in nature suggest that they may be used as signaling molecules, which modulate gene expression in the recipient organisms at low dosage.299 Thus, an increase in antibiotic production may have evolved secondarily in interactions with other organisms.300 Independent of their original function, antibiotics of Actinobacteria play a crucial role for the protection of several animals against pathogens. In insects, their roles are best understood in beewolf digger wasps and fungus-growing ants.
Solitary wasps of the tribe Philanthini within the Crabronidae (“beewolves”) dig underground nests in soil, mass provision individual progeny in brood cells with insect prey and engage in a defensive symbiosis with ‘Candidatus Streptomyces philanthi’ bacteria to protect their larvae against mold fungi from the surrounding soil.301–305 Uniquely, bacterial symbionts are applied to the brood chambers from antennal reservoirs of the females.306S. philanthi strains display their protective abilities after incorporation into the cocoon by the larvae.302 For the following two weeks, the symbionts produce a cocktail of streptochlorin 67 and eight piericidin 68–75 derivatives, which are distributed all over the surface of the cocoon and protect the immature wasp against opportunistic pathogens until its emergence several months later.302,307–309
Like beewolves, fungus-farming ants nest in the soil and are confronted with environmental pathogens that threaten their brood and the fungal cultivars. Moreover, leaf-cutter ant gardens are challenged by specialized Escovopsis fungal pathogens and endophytic fungi, brought in by the ants with the plant substrate supplying the cultivars with nutrition.310 To counteract these threats, ant workers combine continuous fungus-weeding and -tending behavior with the application of antimicrobial secretions from their metapleural glands311 as well as antimicrobials produced by symbiotic Actinobacteria.312,313 These Actinobacteria comprise vertically and occasionally horizontally transmitted Pseudonocardia symbionts310,314–316 as well as environmentally acquired members of the genera Streptomyces and Amycolatopsis.317–319 The Pseudonocardia symbionts defend the fungus garden against the specialized Escovopsis cultivar pathogens, by producing dentigerumycin 76 and five angucyclines (in a Pseudonocardia isolate from Apterostigma dentigerum),320,321 or a nystatin-like compound (in a Pseudonocardia isolate from Acromyrmex octospinosus), respectively.317Streptomyces and Amycolatopsis, on the other hand, produce candicidin 77 and antimycin with broad-spectrum activities against fungal competitors of the cultivars (e.g. endophytic fungi in the leaf substrate).322,323 Furthermore, Streptomyces in small crypts on the body surface of adult ants may also protect the ants themselves against pathogens by producing actinomycins and valinomycin.317,323,324
Apart from leaf-cutter ants, other Myrmicinae ants in the genus Allomerus possibly make use of Streptomyces and Amycolatopsis as defensive symbionts. These ants farm Chaetothyriales mould fungi within their ant-plant nests, but instead of food, these fungi give structure to the ant galleries.325 The galleries are used to trap and catch insect prey for nutrition.326 Several Actinobacteria showing antifungal activities were isolated from the cuticle of Allomerus ants, and these bacteria were hypothesized to play a role in the defense of the galleries against fungal pathogens and competitors.327 The examples of attine and Allomerus ants indicate that defensive secondary metabolites of Actinobacteria can play an important role in ant fungiculture. Given that only a handful of ant symbionts has been studied, it is likely that many more antibiotics may be isolated from such symbioses.328
Compared to fungus-growing ants, much less is known about the role of defensive bacterial symbionts in the gardens of the other fungus-farming insect groups: termites and bark/ambrosia beetles.329 Fungus-farming termites occupy the same ecological niche in the Paleotropics as leaf-cutter ants in the New World. As in leaf-cutter ants, Actinobacteria have been isolated from termite nests, but in vitro assays showed antifungal activity against Pseudoxylaria and Trichoderma fungal competitors as well as the termites' Termitomyces cultivar.330 This indicates that antifungals are either applied in a targeted fashion by the termites, or unspecific Actinobacteria were isolated that do not act as defensive symbionts in fungus-farming termites. The activity of the two microtermolides A and B that were identified from termite-associated Streptomyces spp. was not tested.331 Instead, it is possible that fungus-farming termites are associated with a Bacillus sp. as a defensive symbiont. This strain produces bacillaene A, which specifically inhibits several cultivar competitors in vitro.332
As in termites, comparatively little is known about the possible role of Actinobacteria in the defense of bark and ambrosia beetle nests. These beetles bore tunnels in the phloem (bark beetles) or xylem (ambrosia beetles), on the walls of which they cultivate food fungi in the orders Microascales and Ophiostomatales.333 Females transmit spores of their cultivars to new nests in highly specialized organs called mycetangia.334 As beetles typically nest in recently dead trees, cultivars are usually confronted with competition from other wood-colonizing fungi. Actinobacterial symbionts are typically isolated in very low abundance from beetles and their nests. In a study on Dendroctonus frontalis bark beetles, however, Scott et al.335 found Streptomyces thermosacchari to be present in the beetle's mycetangia as well as on the cultivars. These bacteria specifically inhibited the growth of Ophiostoma minus, a prevalent antagonist of the beetles, by producing the antifungal metabolite mycangimycin as well as other compounds that were not identified. In vitro, mycangimycin 78 turned out to be 20 times more effective against O. minus than against the beetle's cultivar Entomocorticium sp. A.335,336 Another Streptomyces strain displayed no activity in competition assays with associates of D. frontalis, but produces frontalamides A and B under certain culture conditions.337 However, Streptomyces are not consistently present in D. frontalis nests and are generally isolated at very low frequencies from other North American bark and ambrosia beetles.338 This underlines the importance of further in vivo studies to investigate the relevance of Actinobacteria for bark beetle defense in nature.
Protection against pathogens: gut and nutritional resources. Gut bacteria can play an important role in defense against invading microbial pathogens. In the locust Schistocerca gregaria, members of the intestinal microbiota can produce phenolic compounds with antimicrobial properties that have been suggested to derive from the conversion of plant secondary metabolites by microbes. Hydroquinone 79, as well as 3,4-hydroxybenzoic and 3,5-hydroxybenzoic acids 80 and 81, are usually present in the guts and feces of locusts, while absent in insects lacking their normal gut microbiota.339 Interestingly, the entomopathogenic fungus Metarhizium anisopliae is inhibited by these compounds and fails to invade locust guts when the symbiotic microbiota is present.340 While Pantoea agglomerans appears to be responsible for producing at least one of the three antimicrobial phenols found in the locust gut,339 there is evidence from in vitro experiments that Klebsiella pneumoniae and Enterococcus cloacae may also contribute to the production of defensive compounds.340 Furthermore, a greater diversity of the bacterial community in the locust gut is associated with improved resistance against pathogens, suggesting that multiple players contribute to the efficient defense.341
Besides protecting against direct pathogen colonization in or on the insect body, gut-associated microbes can also contribute to the preservation of nutritional resources, as is the case for bacteria in honeybees and stingless bees, and for yeasts in drosophilids (see below, Defensive symbioses with fungi). In bees, a number of lactic acid bacteria including Lactobacillus and Bifidobacterium frequently occur in propolis and in the honey crop, both of which exhibit antimicrobial properties. These lactic acid bacteria participate in the fermentation and preservation of an essential food source, the beebread. In addition, the bees line their hive with a layer of propolis, which serves as a sterilization mechanism protecting the brood against pathogens.17In vitro, a set of compounds with antimicrobial properties were produced by lactic acid bacteria isolated from the honey crop of the honeybee Apis mellifera, including organic acids (lactic, formic, and acetic acid), hydrogen peroxide, different volatiles (benzene, toluene, octane, ethylbenzene and nonane), 3-OH fatty acids, 2-heptanone and various peptides.342 These substances inhibit a number of bacteria and fungi that are commonly found on bee-visited flowers. However, the strongest inhibitory effects were observed when several different lactic acid bacteria were co-cultivated with the potential pathogens, suggesting a synergistic activity of the microbial consortium.342 Along with the sterilizing effects on resources and the hive, lactic acid bacteria can also enhance survival of honeybee larvae by conferring protection against the American and European foulbrood diseases, caused by Paenibacillus larvae343 and Melisococcus plutonius,344 respectively.
In addition to lactic acid bacteria, there are other gut-associated microbes that can play important protective roles in bees, particularly bumblebees. By experimentally manipulating the gut microbiota of the bumblebee Bombus terrestris, Koch and colleagues provided evidence that the bacterial community plays a role in reducing infection rates by the trypanosomatid parasite Crithidia bombi.345 Furthermore, the abundance of this parasite was shown to correlate negatively with the presence of the gut symbiont Gilliamella apicola (γ-Proteobacteria) in natural bumblebee populations.346 These and other studies on the bacterial community in different bees indicate that a balanced and stable microbiota plays a substantial role in bee health by reducing pathogen susceptibility.17
Similar to the aforementioned gut microbes, bacteria present on the egg surface of house flies are also involved in the preservation of nutrient provisions. As house flies lay their eggs on manure that their offspring will use for nutrition, the larvae will most likely encounter fungal competitors that have been shown to reduce their chances of reaching adulthood. However, the bacterial community on the surface of the fly eggs can suppress the growth of these fungi on the manure and thereby play an important protective role for the developing larvae.347
Antiviral protection. Viruses can also pose a significant threat to many different insect species. The Drosophila C virus (DCV) is common in natural populations of Drosophila melanogaster and causes high mortality under laboratory conditions. However, D. melanogaster frequently carries the α-Proteobacterium Wolbachia pipientis, which can reduce host susceptibility to DCV and other RNA viruses.348,349 These findings have stimulated investigation of other insect–symbiont–virus systems, particularly those involving vectors of human pathogenic viruses.350 In Culex quinquefasciatus, the natural occurrence of Wolbachia resulted in reduced titers and impaired transmission capacity of West Nile virus.351 Although the mechanistic basis underlying this effect is not yet completely understood, significant progress in this area has been made in non-naturally infected vectors of arboviruses and other human parasites. In Aedes aegypti mosquitoes, infections with dengue and chikungunya viruses, as well as the malaria-causing protozoan parasite Plasmodium, are restrained when the insect is artificially infected with a Wolbachia strain from D. melanogaster.352 This protective effect is achieved through activation of the host's immune system, which involves stimulating the expression of several Toll-pathway genes as well as defensins and cecropins.8 In addition, the presence of symbiont genes potentially involved in the production of antimicrobial compounds might also play a role in inhibiting mosquito pathogens.352,353
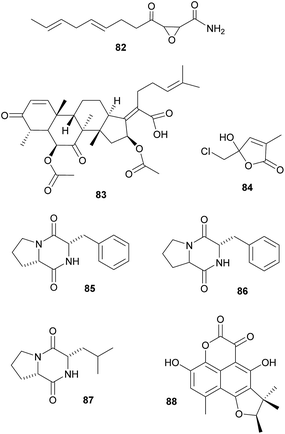
Defensive symbioses with fungi: protection of food or the nesting environment. Most of the defensive fungal symbionts of animals have been described from fungus-farming insects, specifically from leaf-cutter ants, fungus-growing termites and bark and ambrosia beetles. All three groups farm their fungi in social societies and show behavioral adaptations to protect their fungi against fungal competitors and pathogens.329 Furthermore, in addition to defensive actinobacterial symbionts (see above), several studies implicated fungi in the protection of the host or its fungal cultivar against pathogens. Specifically, ‘killer yeasts’ were shown to inhibit the growth of Escovopsis cultivar pathogens within the gardens of Atta ants,354,355 and Ogataea pini, a yeast associated with fungus-growing Dendroctonus bark beetles, produces volatiles (ethanol, carbon disulfide and delta-3-carene) that inhibit the growth of Beauvaria bassiana entomopathogens.356 Additionally, in some cases the cultivar fungi themselves produce defensive secondary metabolites. Among ambrosia beetles, Euwalecea validus is associated with a cultivar (likely an unidentified Fusarium sp.) that produces cerulenin 82 and helvolic acid 83 – antibiotics that inhibit the growth of mould fungi in vitro and likely also suppress bacterial contaminations.357 Similarly, the Lepiota and Tyridiomyces cultivars of Cyphomyrmex fungus-growing ants produce lepiochlorin 84
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 358,359 and several diketopiperazines 85–87,360 respectively, which may be active against bacterial and fungal pathogens. Likewise, Leucocoprinus cultivars of Atta ants show in vitro suppression of fungi endophytic to the leaves that the ants provision as substrate for the cultivar.361 The active secondary metabolites, however, have not been identified yet. The importance of host protection by the cultivars of leaf-cutter ants is supported by the observation that almost all species cover their broods with the cultivar fungus.362,363 In fungus-growing termites, unknown myocins produced by the Termitomyces cultivars suppress the growth of related strains in vitro364 – thereby reducing competition and ensuring the specificity of the symbiosis, analogous to the bacteriocins inhibiting close relatives in the bacterial symbionts of nematodes (see 2.2.1).
358,359 and several diketopiperazines 85–87,360 respectively, which may be active against bacterial and fungal pathogens. Likewise, Leucocoprinus cultivars of Atta ants show in vitro suppression of fungi endophytic to the leaves that the ants provision as substrate for the cultivar.361 The active secondary metabolites, however, have not been identified yet. The importance of host protection by the cultivars of leaf-cutter ants is supported by the observation that almost all species cover their broods with the cultivar fungus.362,363 In fungus-growing termites, unknown myocins produced by the Termitomyces cultivars suppress the growth of related strains in vitro364 – thereby reducing competition and ensuring the specificity of the symbiosis, analogous to the bacteriocins inhibiting close relatives in the bacterial symbionts of nematodes (see 2.2.1).Beyond fungus-farming insects, leaf-rolling weevils in the genus Euops (Attelabidae) are associated with polysaccharide-degrading Penicillium symbionts that are planted on leaves in which eggs and larvae are rolled. Penicillium herquei, the associate of Euops chinensis, has been shown to produce (+)-scleroderolide 88in vivo.365 This antibiotic inhibits the growth of several bacterial and fungal pathogens in competition assays on plates and keeps larval cradles free of other microbes365,367 In honeybees, Penicillium spp., Aspergillus spp. and several Mucorales have been shown to decrease colony failure due to chalkbrood disease caused by the fungus Ascosphaera apis, by competitive exclusion due to the production of antimycotic substances.368 Several mold fungi that are typically regarded as insect pathogens are also potent producers of antimycotic substances369 and are potentially more common defensive symbionts than currently apparent. Finally, Drosophila melanogaster fruit flies strongly benefit from their association with yeasts that – in addition to their nutritional role – also inhibit the growth of fungal food competitors, like the noxious mold Aspergillus nidulans, by producing as yet unknown secondary metabolites.370
2.3 Vertebrates
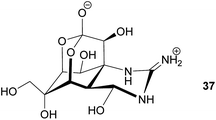
Antipredator defense. In addition to the presence of tetrodotoxin (TTX) 37 in marine invertebrates like nemerteans and mollusks, this highly potent neurotoxin also occurs in fish as well as in amphibians. While the case of bacteria-mediated TTX production seems to be strongly supported in some marine organisms, there is little evidence for a bacterial origin of TTX in frogs, newts and salamanders. Several of these species possess unique analogs of TTX, which are absent or only present in very low quantities in marine animals or in bacteria.165 Despite the lack of conclusive evidence for the source organism, there are clear indications of its antipredator functions. For example, some amphibians actively secrete the toxin upon predator encounter.165 Also, coevolutionary signatures are observed between newts of the genus Taricha and garter snakes (Thamnophis spp.), where the spatial dynamics of TTX levels in the newt and the corresponding resistance in the predator are suggestive of an evolutionary arms race.164,165
Protection against microbial pathogens on the skin and in the gut. Besides predators, microbial pathogens are a major threat to vertebrates and exert a strong selective pressure on evolving efficient defense mechanisms. Amphibians, which are particularly vulnerable to infectious diseases,371 have recently began to suffer devastating effects from chytridiomycosis caused by the fungus Batrachochytrium dendrobatidis. Populations from Australia as well as North, Central and South America have been affected,372 but resistance to the chytrid fungus infection varies both within and across species.373,374 One of the hypotheses that have been proposed to explain this variability in resistance is the presence of symbiotic bacteria, mainly on the skin, that can produce compounds capable of inhibiting the pathogen. Concordantly, the β-Proteobacterium Janthinobacterium lividum, isolated from the skin of the red-back salamander Plethodon cinereus, inhibits the growth of the chytrid fungus in vitro by producing indole 3-carboxaldehyde 89 and violacein 90.373 From the same salamander species, an isolate of Lysobacter gummosus was shown to produce 2,4-diacetylphloroglucinol 91, which also exhibits in vitro activity against B. dendrobatidis.375 Further work on protective bacteria in P. cinereus revealed that 63% of field collected individuals harbor J. lividum or other bacteria capable of violacein production.376 Such frequencies are in line with the often facultative nature of defensive symbioses. In addition to its in vitro activity, the presence of violacein was shown to be associated with increased survival in P. cinereus376 as well as in the frog Rana muscosa.377,378 These results open the possibility for applying bacterial violacein-producers directly on infected amphibians or their natural environment in order to mitigate the effects of the pathogen.373,374 An interesting aspect of this defensive symbiosis is the potential synergism between compounds produced by the bacteria and AMPs from the host, as demonstrated in vitro for the inhibitory effect of 2,4-diacetylphloroglucinol and a mixture of AMPs from R. muscosa against B. dendrobatidis.372 Furthermore, co-cultures of four different bacterial isolates from the red-back salamander including Janthinobacterium sp. resulted in synergistic inhibition of B. dendrobatidis.379 Thus, the pathogen defense of amphibians likely relies on a combined protective effect of the bacterial community and the host's immune response.
As in amphibians, the skin is also a potential entry gate for pathogens in other vertebrates including humans. In fact, human skin is one of the main habitats accommodating microbial partners. The variety of physicochemical conditions on the skin in terms of temperature, humidity, oiliness, and oxygen availability contribute to the high bacterial diversity on its surface. One of the main constituents on the healthy human skin is Staphylococcus epidermidis, a bacterium capable of producing a number of AMPs, including epidermin 92, Pep5 and epilancin K7, which are classified as lantibiotics owing to the presence of lanthionine and/or methyllanthionine in their structures.380,381 Both are unusual thioether amino acids,382 which account for the multiple rings in the structure of lantibiotics and are considered essential for their antibacterial activity.381 However, evidence for the efficacy of lantibiotics against pathogens is limited to in vitro studies. Phenol-soluble modulins (PSMs) are a second group of AMPs produced by S. epidermidis. In contrast to the lantibiotics, there is in vivo evidence for a role of PSMs in the protection of the skin surface, as their inoculation on mouse skin resulted in a significant reduction of the commonly pathogenic group A Streptococcus, while it does not affect the presence of S. epidermidis.381,383 In addition to the direct antimicrobial action, PSMs can also support the host's immune system. Specifically, the application of PSMs to isolated neutrophils resulted in increased eradication of pathogenic bacteria, co-localization with host AMPs and enhancement of extracellular trap formation by the neutrophils.381,384 Other mechanisms for pathogen inhibition by S. epidermis include blocking of quorum sensing via a thiolactone-containing peptide and its derivatives,381,385 as well as inhibition of biofilm formation in the nasal cavity by the production of serine proteases.381,386 Despite the numerous examples for a mutualistic potential of S. epidermidis on human skin, this symbiosis provides a good example for a context-dependent host–microbe association, as there are also clear indications of the potential for pathogenicity by S. epidermidis: an unbalanced microbiota composition possibly associated with an impairment of the host's innate immune response can allow for S. epidermidis' access to internal tissues and result in pathogenesis, which is often reflected in severe nosocomial infections.387
Similarly, while Bacterioides fragilis is frequently isolated from clinical samples and can be involved in human disease,388 it has also been recognized as a native member of the human gut microbiome and can be beneficial for the host. Its potential for a mutualistic role has been extensively investigated in mice, where there is strong evidence that the production of polysaccharide A by B. fragilis prevents intestinal inflammatory disease caused by the opportunistic pathogen Helicobacter hepaticus. Although the mechanistic details of this protective effect are not yet fully understood, the abundances of both symbiont and pathogen do not differ between healthy and diseased mice, so an immunomodulatory effect suppressing disease development (i.e. absence of detrimental consequences for the host), rather than pathogen clearance, appears to be the key to this bacteria-mediated protection.389 Concordantly, there is a growing body of evidence in humans suggesting that the microbial community, including both bacteria and fungi, plays a crucial role in immunoregulatory processes.390–392 In fact, the presence of a healthy microbiota cannot only regulate inflammatory responses but also train the host's immune system and thus confer an indirect defense to the action of pathogens.391
Gut pathogens are also a frequent threat for fish populations, which is particularly problematic in the case of intensive fish culture that facilitates the emergence of pathogens as well as the spread of antibiotic resistant bacterial strains.393 However, several lactic acid bacteria (LAB) such as Carnobacterium and Lactobacillus strains isolated from different fish species are capable of inhibiting pathogenic bacteria in vitro, in particular Aeromonas salmononicida and Vibrio anguillarum.394,395 The production of specific inhibitory substances among fish-associated LAB has only been identified in Carnobacterium strains, which synthesize carnocin UI49, piscicocin V1, and divercin V41. However, the microbes' inhibitory activity has also been attributed to the production of additional compounds such as organic acids, hydrogen peroxide and siderophores.395 In addition to the synthesis of bioactive compounds, there is evidence for the stimulation of the host's innate immune response caused by the LAB, as demonstrated in vivo in gilthead seabream individuals infected with Lactobacillus delbrueckii.396
Protection against microbial pathogens on bird eggs. Perhaps the most remarkable defensive symbiosis between microbes and vertebrates is that of Enterococcus bacteria inhabiting the uropygial gland secretions of hoopoe birds (Upupa epops). This system is particularly interesting in terms of the elaborate set of behavioral and morphological adaptations underlying the evolution and maintenance of the partnership. While male and non-breeding female hoopoes produce a white and odorless uropygial gland secretion with only the occasional presence of few bacteria, secretions from breeding females and nestlings are brown, emit a strong smell and contain high numbers of Firmicutes in the genus Enterococcus, mainly E. faecalis.397 Interestingly, female birds actively collect the secretions from the uropygial gland and deposit them on both feathers and eggs, the latter of which contain specialized structures to harbor the symbiont-containing secretions.398 Recent studies provide evidence that the bacteria in the secretions play a protective role by preventing the growth of detrimental microbes on the eggs. Notably, there is a positive correlation between hatching success and Enterococcus loads in the uropygial secretions and on the egg shells.398 Additionally, E. faecalis can inhibit the keratin-degrading action of pathogenic Bacillus licheniformis, thus playing a protective role on the feathers of adult hoopoes.399 When first isolated from the gland secretions, E. faecalis was shown to produce enterocins, later specified as enterocins MR10 and AS-48, which present in vitro inhibitory activity against a range of Gram-positive and Gram-negative bacteria.400,401 Later on, it was discovered that also the volatile fraction of the brown secretions contained bacteria-produced compounds with antimicrobial activity, primarily butanoic acid, 2-methyl butanoic acid, 4-methyl pentanoic acid, indole, 3-phenyl propanoic acid and 4-chloroindole.402 While individual volatile compounds showed differential efficacy against various bacteria, a mixture resembling the composition of the brown secretions consistently inhibited a broad spectrum of microbes.402
3 Ecological and evolutionary implications
The previous sections illustrate the expanding body of literature describing symbiont-mediated chemical defense across diverse animal taxa inhabiting a broad range of habitats. In this section, we aim to draw some general conclusions on the key ecological and evolutionary factors shaping this diversity and point to novel directions for future research.3.1 Implications of host ecology and lifestyle
Protective associations between microorganisms and animals are present across a range of phylogenetically distant metazoan taxa (Fig. 2 and Table S1†). This ubiquity not only reflects the general potential to evolve defensive symbioses but also its occurrence in organisms with markedly different natural histories. However, the identity and diversity of an organism's antagonists strongly depends on its environment, lifestyle and life history, so these factors are likely to have a major impact on the evolution of defensive symbioses. In marine environments, many organisms have a sessile life-stage that must be especially well defended due to its often conspicuous nature, predictability in space and time, and incapability to escape from predators. Thus, chemical protection is widespread among marine invertebrates, especially in taxa with soft bodies that lack structural protection. Concordantly, sponges, bryozoans, tunicates, some corals and mollusks exhibit an arsenal of defensive chemicals, more and more of which are being discovered to be of symbiotic origin (Table S1†).9–11,208 As the sessile lifestyle often goes along with filter feeding, these animals have a high probability to get in contact with microbes, thereby increasing the chances to acquire a beneficial symbiont, while at the same time risking exposure to pathogens.12 In terrestrial environments, by contrast, sessile animals are exceptionally rare. However, many terrestrial organisms have either a valuable, but immobile resource (nest, food) or immobile developmental stages (eggs, pupae) that are vulnerable to threats from predators, pathogens and parasites due to their predictability in time and space. Food resources that are available en masse, like fungus gardens of insects, insect cadavers killed by nematodes or mass-provisioned insect nests, for example, and eggs or brood that are exposed to a hostile environment for a long time (e.g. in nests within soil) run a high risk of pathogen infection.16,17 While there are a few known examples of a symbiotic protection in these immobile life stages of terrestrial animals,302,397 the abundance of such associations in the marine environment suggests that there are likely many more to be discovered.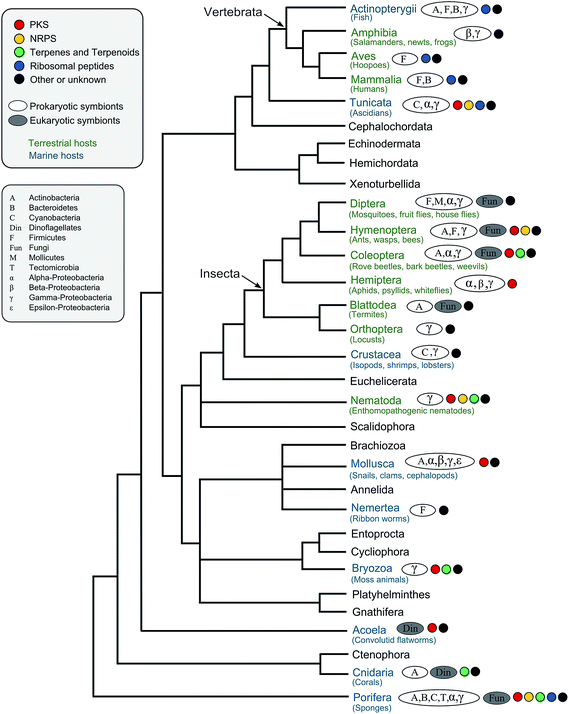 | ||
| Fig. 2 Cladogram of selected animal groups highlighting those with described defensive microbial symbionts and the corresponding symbiont taxa. Colored circles represent the major biosynthetic pathways reported for symbiont-produced compounds. Branch lengths are not to scale and branch order is adapted from previous phylogenetic analyses for the deep branches,403 hexapods,404 and all other bilaterians.405,406. Common names in brackets denote selected groups within the respective taxon that harbor known defensive symbionts. | ||
The transition between mobile and immobile stages during the life cycle of an animal can incur a trade-off between chemical and mechanical defenses, resulting in life-stage specific benefits of defensive symbioses. In bryozoans, for example, the larvae and early post-settlement stages are associated with higher mortality given the increased vulnerability to predation, competition and disease, as well as desiccation, temperature stress, and radiation.12 Concordantly, the concentration of symbiont-produced defensive bryostatins is particularly high in bryozoan larvae, while adults switch to a predominantly structural defense, with only reproductive tissues being chemically defended by the bryostatins.125 Analogously, in a terrestrial system, high concentrations of symbiont-provided antibiotics are only produced during the cocoon stage of the European beewolf and provide protection during the immobile phase of hibernation.302 Thus, the symbionts' investment in chemical defense can vary during the host's life cycle, being complemented by alternative defenses of the host.12
Another important aspect of an organism's exposure to pathogens and thus its benefit to engage in defensive microbial symbiosis is its degree of sociality. Social or gregarious behavior in combination with an often high relatedness and low genetic variability among group members makes colonies and aggregations of members of an individual species particularly exploitable by pathogenic microbes.407,408 On the other hand, group-living also confers the benefit of social immunity407 and can facilitate the transmission and maintenance of beneficial microorganisms, e.g. such that provide protection against the colony's pathogens.409 Hence, it is not surprising that defensive symbionts are commonly found in social insect colonies and clonal groups of organisms (e.g. aphid colonies, fungal monocultures of farming insects), in which social behaviors commonly ensure the spreading of defensive symbionts.344,345,410,411
3.2 Diversity of defensive symbionts and protective chemicals
Across animals, a wide diversity of microbial partners have been identified as defensive symbionts (Fig. 2 and Table S1†). However, particular groups of microbial taxa stand out among these partnerships. Non-surprisingly, the high biosynthetic potential of Actinobacteria renders them effective defenders for a number of animals including sponges,54 corals,412 mollusks,176,177,180 and insects.16 However, other bacterial groups like Proteobacteria and Firmicutes, as well as eukaryotic organisms like dinoflagellates, are also important and widespread players in defensive partnerships (Fig. 2 and Table S1†). Likewise, despite the predominance of PKS- and NRPS-derived compounds in protective symbioses across many taxa, an array of other chemical classes also appear repeatedly, particularly ribosomal peptides and terpenes, but also β-lactam and oligosaccharide antibiotics (Fig. 2 and Table S1†).3.3 Implications of symbiont localization
For cases in which symbiont-mediated defense relies on the production of a bioactive compound, it has been suggested that an external localization of the microbial partner, i.e. on the body surface or in the lining of body cavities of the host, is more effective than an endosymbiotic localization, since the protective substances are readily exposed to potential enemies.12 In fact, a majority of the microbial symbionts described to play an anti-pathogenic role are located directly on the body surface of the host,373,375,381 on specific superficial structures,202,302,310,398 within the gut,341,343,344,396 or externally on food provisions or the nesting environment.367 In such localizations, defensive symbionts can exert their protective activity before antagonists breach the host's surfaces, thereby reducing detrimental effects to a minimum. In addition, the localization outside of the host's body may reduce potentially harmful side effects that the noxious defensive chemicals may have on the host itself. There are, however, a number of symbionts providing chemical defense that are located within the host's body,47,287,298 sometimes even within the host's cells.105,229,230 Interestingly, for many of these endosymbionts, the relevant antagonists identified so far are predators, so various types of antagonists may exert different selective pressures on symbiont localization. Yet, a correspondence between symbiont localization and type of enemy remains speculative, given the often limited information on the complete range of relevant antagonists.3.4 Evolutionary dynamics
As opposed to many intracellular symbionts conferring nutritional benefits, those playing a defensive role are often found at intermediate infection frequencies in host populations.47,76,229,287,288,290,296,298,348,349,413 The underlying reasons are probably multiple, a primary one being the context-dependent nature of protective functions. In the absence of relevant antagonists, the host still pays a cost for harboring the symbiotic partners,414 which can outweigh the benefits and shift the selective balance in favor of symbiont loss. A similar situation can occur when only certain life stages are protected and thus the relative benefit of carrying the symbionts changes during the life cycle of the host. In the long run, however, balancing selection is most likely responsible for maintaining the partnerships.19,290Another cause for intermediate infection rates may be the comparatively low degree of intimacy and stability observed in many – but not all230 – defensive symbioses, which stands in stark contrast to the known intracellular nutritional mutualists that have been associated with some groups of invertebrates for hundreds of millions of years.415 The often exposed localization of ecto- and extracellular symbionts increases the chances of environmental acquisition of other microbial strains and thus symbiont replacement, resulting in the general lack of strict co-cladogenesis in many systems.416 Although a vertical transmission route does exist in several examples of defensive animal–bacteria symbioses, occasional horizontal transmission often occurs, e.g. in antibiotic-producing actinomycetes of fungus-growing ants314,315,417 and beewolves,303 in the secondary symbionts of aphids,418 and in the defensive symbionts of bryozoans.144 Also, in many other marine symbioses, intraspecific variation in both the associated microbial communities and the symbiont-provided defensive chemistry indicate a high probability of symbiont exchange by occasional horizontal transfer or environmental determination.47,76,413 The flexible acquisition of defensive symbionts might represent a fast and versatile adaptive process for defense against coevolving antagonists, but also requires sophisticated partner choice mechanisms to ensure the evolutionary stability of the symbiotic partnership.419
The acquisition of genetic material from unrelated microbes through horizontal gene transfer (HGT) can also mediate rapid chemical changes and thereby facilitate adaptations in defense against coevolving antagonists. While rare in nutritional symbioses, there are several examples of defensive traits that were likely acquired via HGT. The Pseudomonas symbiont of rove beetles shows strong indications of HGT of the genes for the defense toxin pederin. The striking similarity of the biosynthetic genes of pederin and diaphorin, the toxin produced by the intracellular symbiont of the asian citrus psyllid, ‘Ca. Profftella armatura’, suggests that a horizontal transmission event, led to the convergent characteristics in distant lineages.230 The horizontal acquisition in the Paederus symbiont is also supported by the localization of the gene on a genomic island,289 and the occurrence of a similar gene cluster for onnamide biosynthesis in the marine sponge Theonella swinhoei.73,76,420 Although still lacking direct evidence, cases of potential HGT have also been suspected in several marine symbioses, particularly those of ascidians and bryozoans. The recent discovery of a plasmid-localized didemnin biosynthetic gene cluster in the free-living α-proteobacteria Tistrella mobilis and T. bauzanensis219,220 raises the possibility that the Synechocystis trididemni cyanobacterial symbiont of didemnid ascidians has acquired genes for didemnin biosynthesis via HGT.12 Furthermore, the synthesis of bioactive tambjamines by microbial partners of the distant marine groups of ascidians and bryozoans might be explained by horizontal gene transfer.12 Nevertheless, our understanding of the prevalence of HGT in defensive symbioses and its impact on the evolutionary dynamics of the host's interaction with antagonists remains rudimentary.
An interesting feature found repeatedly across different animal–microbe protective associations is the simultaneous employment of multiple defensive chemicals, produced by either a single or several symbiotic partners. In the beewolf-Streptomyces symbiosis, for example, a “cocktail” of compounds produced by a single symbiotic strain per host species is capable of providing an efficient protection against an array of opportunistic bacterial and fungal pathogens.309 In a similar fashion, animal hosts with strikingly different life history strategies including hoopoe birds,400–402 locusts,339 entomopathogenic nematodes,20 didemnid ascidians,222,225 salamanders,373 and bryozoans,124 are associated with a mixture of symbiont-derived compounds likely involved in defense. While in some cases, individual symbionts produce a range of different chemicals, in others – like the didemnid ascidians228 – multiple bacterial partners are responsible for the production of the defensive compounds. In both cases, effects are often not only complementary, but also synergistic.309,342 These combined strategies are in line with the aforementioned versatility, as they are more likely effective against a range of antagonists. Additionally, co-application of several antibiotic substances at the same time is known to strongly hamper the evolution of resistance in the targeted antagonists.421
3.5 Outlook: current status, challenges and opportunities of defensive symbiosis research
Compared to nutritional symbioses, defensive ones are generally more difficult to detect,17 because they are often of facultative nature and their effects are only perceived in the presence of the relevant antagonists, which are in many cases not known, not available under laboratory conditions, and/or not reliably detectable in short-term or site-restricted observations.19 Furthermore, defensive symbiont localization can be varied and unexpected, including occurrence of the symbionts on the surface of the host, within the food resource or the nesting environment, which makes distinction of symbionts from environmental contaminants challenging.17Additional challenges of characterizing defensive symbioses are habitat-specific. In fact, defensive symbioses in marine and terrestrial animals have been explored from evidently different perspectives. A majority of the studies on marine associations has been motivated by the prospect to discover novel bioactive compounds, while the recognition of their bacterial origin has come much later. Hence, with few exceptions, research on marine defensive symbioses is characterized by a strong background on the chemical basis of defense, whereas the link to the producing microorganisms and the fitness consequences of the symbiosis for the host often remain enigmatic. Obviously, this is also due to the limitations for experimental manipulation in marine habitats, specifically the assessment of fitness benefits by artificially generating aposymbiotic hosts. On the other hand, terrestrial systems – represented to a great extent by insects and nematodes – have been most often approached from an ecological perspective and usually first described based on the identification of the key partners. However, there is often less information about the mode of action of protective symbionts in terrestrial animals and – with a few notable exceptions – on the chemistry involved in defense (Table S1†). Thus, while gaps in the ecological knowledge of many of the marine symbioses remain to be filled, terrestrial studies could take advantage of the advances in natural product discovery accomplished in the marine world. Certainly, an interdisciplinary approach integrating mechanistic and ecological studies, molecular characterization and natural product research is and will remain to be of utmost importance for the field.
In this context, current technological developments will continue to play an important role for the progress in defensive symbiosis research. Molecular biology tools, particularly next-generation-sequencing of microbial communities, RNAseq, and single-cell genomics can rapidly provide strong links between natural products and their producers, especially in systems not amenable to manipulative experimentation. Additionally, increasing sensitivity and resolution in mass spectrometry (MS) as well as improvements in MS-imaging (like nanoSIMS, MALDI imaging, and DESI imaging422,423) allow for the detection and quantification of bioactive compounds in situ, which is currently missing for most (but not all309,323) terrestrial defensive symbioses.
In the search for novel defensive symbioses, special attention should be directed towards systems that exhibit ecological and evolutionary conditions predisposing them towards defensive alliances with microbes. For example, sessile or ground-nesting animals, those that have developed food domestication habits, or have gregarious or social lifestyles in combination with high relatedness, stand out as promising candidates. Beyond associations with bacteria, defensive symbiotic partnerships between animals and fungi remain heavily understudied.25,83,424 Fungal partners are common nutritional symbionts of various animal groups, but have been rarely screened for their bioactive potential. This is surprising as fungi have a vast biosynthetic potential and are a rich source of antibiotics.25,83,424–426 Likewise, only few cases of defensive symbiotic viruses have been described. Polydnaviruses in Microplitis demolitor and other parasitoid wasps (families Braconidae and Ichneumonidae) aid in suppressing the immune response of the parasitized host and thereby confer protection to the wasp.427,428 In another intriguing example, an RNA virus of the parasitoid wasp Dinocampus coccinellae manipulates the behavior of the wasp's coccinellid beetle host, inducing it to protect the wasp pupa from predation until it emerges from the cocoon.429 Finally, bacteriophages have been shown to adhere to metazoan mucosal surfaces and limit bacterial infections to their own and the host's benefit.430 The potentially high degree of specificity of the interaction between viruses/phages and bacteria, along with the simplicity of acquiring viruses and the low cost of their maintenance make them appear as ideal defensive symbionts. In summary, it is quite possible that other cases of symbiotic relationships with fungi and viruses await discovery for those who venture to look beyond bacterial symbionts.
In conclusion, animal–microbe defensive symbioses are widespread, ecologically diverse and evolutionarily dynamic. They are a promising research target for the field of natural products discovery, due to their immense chemical potential and the advantages of studying the microbial producers directly embedded in an ecological context (i.e. fulfilling a role for their eukaryotic host), as opposed to free-living microorganisms.29,431 Furthermore, discovered natural products are more likely to be applicable in medical contexts, since they have been naturally tested for side effects on, at least some, eukaryotes. Although significant gaps in our understanding of symbiont-mediated defenses remain, the fast pace of technological advances and the momentum currently experienced by symbiosis research promise to quickly deepen our insights into these fascinating and promising associations.
4 Acknowledgements
We thank all colleagues in symbiosis research for their contributions to the knowledge on defensive associations, and we sincerely apologize to those whose work we may have overlooked during our literature searches or could not cite due to space constraints. We are grateful to Julia Kubanek, Jörn Piel, and Helge Bode for insightful comments on the manuscript, and we acknowledge financial support from the Max Planck Society (to LVF, PHWB, TE, MK), the German Science Foundation (to MK, DFG KA2846/2-1), and the Swiss National Science Foundation (to PHWB, P300P3_151134).5 References
- D. L. Evans and J. O. Schmidt, Insect defenses, State University of New York Press, Albany, NY, USA, 1990 Search PubMed.
- S. Yuan, X. Tao, S. Huang, S. Chen and A. Xu, Annu. Rev. Anim. Biosci., 2014, 2, 235–258 CrossRef CAS PubMed.
- T. Eisner, M. Eisner and M. V. S. Siegler, Secret weapons: defenses of insects, spiders, scorpions, and other many-legged creatures, Belknap Press, Cambridge, MA, USA, 2005 Search PubMed.
- M. E. Hay, J. Exp. Mar. Biol. Ecol., 1996, 200, 103–134 CrossRef CAS.
- K. Clay, Funct. Ecol., 2014, 28, 293–298 CrossRef.
- J. F. White and M. S. Torres, Defensive Mutualism in Microbial Symbiosis, CRC Press, Boca Raton, FL, USA, 2009 Search PubMed.
- R. J. Dillon and V. M. Dillon, Annu. Rev. Entomol., 2004, 49, 71–92 CrossRef CAS PubMed.
- X. Pan, G. Zhou, J. Wu, G. Bian, P. Lu, A. S. Raikhel and Z. Xi, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, E23–E31 CrossRef PubMed.
- J. Piel, Nat. Prod. Rep., 2004, 21, 519–538 RSC.
- J. Piel, Nat. Prod. Rep., 2009, 26, 338–362 RSC.
- E. W. Schmidt, in Defensive Mutualism in Microbial Symbiosis, ed. J. F. White and M. S. Torres, Boca Raton, FL, USA, 2009, pp. 65–83 Search PubMed.
- N. B. Lopanik, Funct. Ecol., 2014, 28, 328–340 CrossRef.
- G. M. Konig, S. Kehraus, S. F. Seibert, A. Abdel-Lateff and D. Muller, ChemBioChem, 2006, 7, 229–238 CrossRef PubMed.
- T. L. Simmons, R. C. Coates, B. R. Clark, N. Engene, D. Gonzalez, E. Esquenazi, P. C. Dorrestein and W. H. Gerwick, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 4587–4594 CrossRef CAS PubMed.
- J. C. Brownlie and K. N. Johnson, Trends Microbiol., 2009, 17, 348–354 CrossRef CAS PubMed.
- M. Kaltenpoth, Trends Microbiol., 2009, 17, 529–535 CrossRef CAS PubMed.
- M. Kaltenpoth and T. Engl, Funct. Ecol., 2014, 28, 315–327 CrossRef.
- K. M. Oliver and N. A. Moran, in Defensive mutualism in microbial symbiosis, ed. J. F. White and M. S. Torres, CRC Press, Boca Raton, FL, USA, 2009, pp. 129–148 Search PubMed.
- K. M. Oliver, A. H. Smith and J. A. Russell, Funct. Ecol., 2014, 28, 341–355 CrossRef.
- H. S. Koppenhoefer and R. Gaugler, in Defensive Mutualism in Microbial Symbiosis, ed. J. F. White and M. S. Torres, CRC Press, Boca Raton, FL, USA, 2009, vol. 27, pp. 99–116 Search PubMed.
- N. Morales-Soto, H. Snyder and S. Forst, in Defensive Mutualism in Microbial Symbiosis, ed. J. F. White and M. S. Torres, CRC Press, Boca Raton, FL, USA, 2009, pp. 117–127 Search PubMed.
- H. B. Bode, Curr. Opin. Chem. Biol., 2009, 13, 224–230 CrossRef CAS PubMed.
- S. S. Ebada and P. Proksch, in Handbook of Marine Natural Products, ed. E. Fattorusso, W. H. Gerwick and O. Taglialatela-Scafati, Springer, London, UK, 2012, pp. 191–293 Search PubMed.
- R. L. Simister, P. Deines, E. S. Botte, N. S. Webster and M. W. Taylor, Environ. Microbiol., 2012, 14, 517–524 CrossRef CAS PubMed.
- N. S. Webster and M. W. Taylor, Environ. Microbiol., 2012, 14, 335–346 CrossRef CAS PubMed.
- N. S. Webster and L. L. Blackall, ISME J., 2008, 3, 1–3 CrossRef PubMed.
- U. R. Abdelmohsen, K. Bayer and U. Hentschel, Nat. Prod. Rep., 2014, 31, 381–399 RSC.
- A. Uria and J. Piel, Phytochem. Rev., 2009, 8, 401–414 CrossRef CAS.
- J. Piel, Annu. Rev. Microbiol., 2011, 65, 431–453 CrossRef CAS PubMed.
- M. C. Wilson and J. Piel, Chem. Biol., 2013, 20, 636–647 CrossRef CAS PubMed.
- U. Hentschel, J. Piel, S. M. Degnan and M. W. Taylor, Nat. Rev. Microbiol., 2012, 10, 641–654 CrossRef CAS PubMed.
- R. Thacker and C. Freeman, in Adv. Mar. Biol., ed. M. Becerro, M. Uriz, M. Maldonado and T. Xavier, Elsevier, London, UK, 2012, vol. 62, pp. 57–111 Search PubMed.
- J. W. Blunt, B. R. Copp, R. A. Keyzers, M. H. G. Munro and M. R. Prinsep, Nat. Prod. Rep., 2014, 31, 160–258 RSC.
- T. R. A. Thomas, D. P. Kavlekar and P. A. LokaBharathi, Mar. Drugs, 2010, 8, 1417–1468 CrossRef CAS PubMed.
- O. K. Radjasa, Y. M. Vaske, G. Navarro, H. C. Vervoort, K. Tenney, R. G. Linington and P. Crews, Bioorg. Med. Chem., 2011, 19, 6658–6674 CrossRef CAS PubMed.
- J. A. Fuerst, Appl. Microbiol. Biotechnol., 2014, 98, 1–17 CrossRef PubMed.
- K. Konya, N. Shimidzu, N. Otaki, A. Yokoyama, K. Adachi and W. Miki, Experientia, 1995, 51, 153–155 CrossRef CAS.
- S. Dash, C. L. Jin, O. O. Lee, Y. Xu and P. Y. Qian, J. Ind. Microbiol. Biotechnol., 2009, 36, 1047–1056 CrossRef CAS PubMed.
- S. Dash, Y. Nogata, X. J. Zhou, Y. F. Zhang, Y. Xu, X. R. Guo, X. X. Zhang and P. Y. Qian, Bioresour. Technol., 2011, 102, 7532–7537 CrossRef CAS PubMed.
- W. Miki, N. Otaki, A. Yokoyama and T. Kusumi, Experientia, 1996, 52, 93–96 CrossRef CAS.
- M. D. Unson and D. J. Faulkner, Experientia, 1993, 49, 349–353 CrossRef CAS.
- A. E. Flowers, M. J. Garson, R. I. Webb, E. J. Dumdei and R. D. Charan, Cell Tissue Res., 1998, 292, 597–607 CrossRef CAS.
- J. Faulkner, M. D. Unson and C. A. Bewley, Pure Appl. Chem., 1994, 66, 1983–1990 CrossRef.
- M. D. Unson, N. D. Holland and D. J. Faulkner, Mar. Biol., 1994, 119, 1–11 CrossRef CAS.
- P. Flatt, J. Gautschi, R. Thacker, M. Musafija-Girt, P. Crews and W. Gerwick, Mar. Biol., 2005, 147, 761–774 CrossRef CAS PubMed.
- J. Vansande, F. Deneubourg, R. Beauwens, J. C. Braekman, D. Daloze and J. E. Dumont, Mol. Pharmacol., 1990, 37, 583–589 CAS.
- C. P. Ridley, P. R. Bergquist, M. K. Harper, D. J. Faulkner, J. N. A. Hooper and M. G. Haygood, Chem. Biol., 2005, 12, 397–406 CrossRef CAS PubMed.
- R. Thacker, M. Diaz, K. Ruetzler, P. Erwin, S. Kimble, M. Pierce and S. Dillard, in Porifera Research: Biodiversity, Innovation and Sustainability, ed. M. Custódio, G. Lôbo-Hajdu, E. Hajdu and G. Muricy, Série Licros, Rio de Janeiro, Brazil, 2007, pp. 621–626 Search PubMed.
- D. Erpenbeck, J. N. A. Hooper, I. Bonnard, P. Sutcliffe, M. Chandra, P. Perio, C. Wolff, B. Banaigs, G. Worheide, C. Debitus and S. Petek, Mar. Biol., 2012, 159, 1119–1127 CrossRef CAS.
- R. Sakai, H. Kamiya, M. Murata and K. Shimamoto, J. Am. Chem. Soc., 1997, 119, 4112–4116 CrossRef CAS.
- B. J. Rawlings, Nat. Prod. Rep., 2001, 18, 231–281 RSC.
- M. A. Marahiel, T. Stachelhaus and H. D. Mootz, Chem. Rev., 1997, 97, 2651–2674 CrossRef CAS PubMed.
- J. Piel, D. Q. Hui, N. Fusetani and S. Matsunaga, Environ. Microbiol., 2004, 6, 921–927 CrossRef CAS PubMed.
- T. K. Kim, A. K. Hewavitharana, P. N. Shaw and J. A. Fuerst, Appl. Environ. Microbiol., 2006, 72, 2118–2125 CrossRef CAS PubMed.
- M. Laroche, C. Imperatore, L. Grozdanov, V. Costantino, A. Mangoni, U. Hentschel and E. Fattorusso, Mar. Biol., 2007, 151, 1365–1373 CrossRef CAS.
- M. D. Higgs and D. J. Faulkner, J. Org. Chem., 1978, 43, 3454–3457 CrossRef CAS.
- E. Fattorusso, S. Parapini, C. Campagnuolo, N. Basilico, O. Taglialatela-Scafati and D. Taramelli, J. Antimicrob. Chemother., 2002, 50, 883–888 CrossRef CAS PubMed.
- C. Campagnuolo, E. Fattorusso, A. Romano, O. Taglialatela-Scafati, N. Basilico, S. Parapini and D. Taramelli, Eur. J. Org. Chem., 2005, 2005, 5077–5083 CrossRef PubMed.
- V. Costantino, E. Fattorusso and A. Mangoni, J. Org. Chem., 1993, 58, 186–191 CrossRef CAS.
- C. Campagnuolo, E. Fattorusso, O. Taglialatela-Scafati, A. Ianaro and B. Pisano, Eur. J. Org. Chem., 2002, 2002, 61–69 CrossRef.
- V. Costantino, E. Fattorusso, A. Mangoni, M. Di Rosa and A. Ianaro, J. Am. Chem. Soc., 1997, 119, 12465–12470 CrossRef CAS.
- V. Costantino, E. Fattorusso, A. Mangoni, M. Di Rosa and A. Ianaro, Bioorg. Med. Chem. Lett., 1999, 9, 271–276 CrossRef CAS.
- G. Della Sala, T. Hochmuth, V. Costantino, R. Teta, W. Gerwick, L. Gerwick, J. Piel and A. Mangoni, Environ. Microbiol. Rep., 2013, 5, 809–818 CrossRef CAS PubMed.
- K. M. Fisch, C. Gurgui, N. Heycke, S. A. van der Sar, S. A. Anderson, V. L. Webb, S. Taudien, M. Platzer, B. K. Rubio, S. J. Robinson, P. Crews and J. Piel, Nat. Chem. Biol., 2009, 5, 494–501 CrossRef CAS PubMed.
- N. B. Perry, J. W. Blunt, M. H. G. Munro and L. K. Pannell, J. Am. Chem. Soc., 1988, 110, 4850–4851 CrossRef CAS.
- N. B. Perry, J. W. Blunt, M. H. G. Munro and A. M. Thompson, J. Org. Chem., 1990, 55, 223–227 CrossRef CAS.
- R. H. Cichewicz, F. A. Valeriote and P. Crews, Org. Lett., 2004, 6, 1951–1954 CrossRef CAS PubMed.
- C. A. Bewley, N. D. Holland and D. J. Faulkner, Experientia, 1996, 52, 716–722 CrossRef CAS.
- M. R. Bubb, I. Spector, A. D. Bershadsky and E. D. Korn, J. Biol. Chem., 1995, 270, 3463–3466 CrossRef CAS PubMed.
- C. A. Bewley and D. J. Faulkner, J. Org. Chem., 1994, 59, 4849–4852 CrossRef CAS.
- E. W. Schmidt, C. A. Bewley and D. J. Faulkner, J. Org. Chem., 1998, 63, 1254–1258 CrossRef CAS.
- E. W. Schmidt, A. Y. Obraztsova, S. K. Davidson, D. J. Faulkner and M. G. Haygood, Mar. Biol., 2000, 136, 969–977 CrossRef CAS.
- M. C. Wilson, T. Mori, C. Ruckert, A. R. Uria, M. J. Helf, K. Takada, C. Gernert, U. A. E. Steffens, N. Heycke, S. Schmitt, C. Rinke, E. J. N. Helfrich, A. O. Brachmann, C. Gurgui, T. Wakimoto, M. Kracht, M. Crusemann, U. Hentschel, I. Abe, S. Matsunaga, J. Kalinowski, H. Takeyama and J. Piel, Nature, 2014, 506, 58–62 CrossRef CAS PubMed.
- N. Fusetani, T. Sugawara and S. Matsunaga, J. Org. Chem., 1992, 57, 3828–3832 CrossRef CAS.
- S. Sakemi, T. Ichiba, S. Kohmoto, G. Saucy and T. Higa, J. Am. Chem. Soc., 1988, 110, 4851–4853 CrossRef CAS.
- J. Piel, D. Q. Hui, G. P. Wen, D. Butzke, M. Platzer, N. Fusetani and S. Matsunaga, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 16222–16227 CrossRef CAS PubMed.
- M. F. Freeman, C. Gurgui, M. J. Helf, B. I. Morinaka, A. R. Uria, N. J. Oldham, H. G. Sahl, S. Matsunaga and J. Piel, Science, 2012, 338, 387–390 CrossRef CAS PubMed.
- M. Iwamoto, H. Shimizu, I. Muramatsu, S. Matsunaga and S. Oiki, J. Physiol. Sci., 2010, 60, S121 Search PubMed.
- A. Schirmer, R. Gadkari, C. D. Reeves, F. Ibrahim, E. F. DeLong and C. R. Hutchinson, Appl. Environ. Microbiol., 2005, 71, 4840–4849 CrossRef CAS PubMed.
- W. M. Bruck, S. H. Sennett, S. A. Pomponi, P. Willenz and P. J. McCarthy, ISME J., 2008, 2, 335–339 CrossRef CAS PubMed.
- M. Kimura, T. Wakimoto, Y. Egami, K. C. Tan, Y. Ise and I. Abe, J. Nat. Prod., 2012, 75, 290–294 CrossRef CAS PubMed.
- T. Wakimoto, Y. Egami, Y. Nakashima, Y. Wakimoto, T. Mori, T. Awakawa, T. Ito, H. Kenmoku, Y. Asakawa, J. Piel and I. Abe, Nat. Chem. Biol., 2014, 10, 648–655 CrossRef CAS PubMed.
- T. S. Suryanarayanan, Bot. Mar., 2012, 55, 553–564 Search PubMed.
- M. Henríquez, K. Vergara, J. Norambuena, A. Beiza, F. Maza, P. Ubilla, I. Araya, R. Chávez, A. San-Martín, J. Darias, M. Darias and I. Vaca, World J. Microbiol. Biotechnol., 2014, 30, 65–76 CrossRef PubMed.
- M. E. Rateb and R. Ebel, Nat. Prod. Rep., 2011, 28, 290–344 RSC.
- M. Maldonado, N. Cortadellas, M. I. Trillas and K. Rutzler, Biol. Bull., 2005, 209, 94–106 CrossRef CAS.
- C. Rot, I. Goldfarb, M. Ilan and D. Huchon, BMC Evol. Biol., 2006, 6, 71 CrossRef PubMed.
- S. Perovic-Ottstadt, T. Adell, P. Proksch, M. Wiens, M. Korzhev, V. Gamulin, I. M. Muller and W. E. G. Muller, Eur. J. Biochem., 2004, 271, 1924–1937 CrossRef CAS PubMed.
- K. Zhou, X. Zhang, F. L. Zhang and Z. Y. Li, Microb. Ecol., 2011, 62, 644–654 CrossRef PubMed.
- R. D. Gates and T. D. Ainsworth, J. Exp. Mar. Biol. Ecol., 2011, 408, 94–101 CrossRef PubMed.
- R. H. Burris, Mar. Biol., 1983, 75, 151–155 CrossRef CAS.
- M. P. Lesser, L. I. Falcon, A. Rodriguez-Roman, S. Enriquez, O. Hoegh-Guldberg and R. Iglesias-Prieto, Mar. Ecol.: Prog. Ser., 2007, 346, 143–152 CrossRef CAS.
- M. P. Lesser, C. H. Mazel, M. Y. Gorbunov and P. G. Falkowski, Science, 2004, 305, 997–1000 CrossRef CAS PubMed.
- K. B. Ritchie, Mar. Ecol.: Prog. Ser., 2006, 322, 1–14 CrossRef CAS.
- K. L. Rypien, J. R. Ward and F. Azam, Environ. Microbiol., 2010, 12, 28–39 CrossRef CAS PubMed.
- J. Mao-Jones, K. B. Ritchie, L. E. Jones and S. P. Ellner, PLoS Biol., 2010, 8, e1000345 Search PubMed.
- M. Shnit-Orland and A. Kushmaro, FEMS Microbiol. Ecol., 2009, 67, 371–380 CrossRef CAS PubMed.
- M. Shnit-Orland, A. Sivan and A. Kushmaro, Microb. Ecol., 2012, 64, 851–859 CrossRef CAS PubMed.
- W. Fenical and J. R. Pawlik, Mar. Ecol.: Prog. Ser., 1991, 75, 1–8 CrossRef CAS.
- A. D. Rodriguez, Tetrahedron, 1995, 51, 4571–4618 CrossRef CAS.
- W. O'Neal and J. R. Pawlik, Mar. Ecol.: Prog. Ser., 2002, 240, 117–126 CrossRef.
- S. A. Look, W. Fenical, R. S. Jacobs and J. Clardy, Proc. Natl. Acad. Sci. U. S. A., 1986, 83, 6238–6240 CrossRef CAS.
- S. A. Look, W. Fenical, G. K. Matsumoto and J. Clardy, J. Org. Chem., 1986, 51, 5140–5145 CrossRef CAS.
- W. Fenical, J. Nat. Prod., 1987, 50, 1001–1008 CrossRef CAS.
- L. D. Mydlarz, R. S. Jacobs, J. Boehnlein and R. G. Kerr, Chem. Biol., 2003, 10, 1051–1056 CrossRef CAS PubMed.
- J. M. Boehnlein, L. Z. Santiago-Vazquez and R. G. Kerr, Mar. Ecol.: Prog. Ser., 2005, 303, 105–111 CrossRef CAS.
- D. Sica and D. Musumeci, Steroids, 2004, 69, 743–756 CrossRef CAS PubMed.
- R. D. A. Epifanio, L. F. Maia, J. R. Pawlik and W. Fenical, Mar. Ecol.: Prog. Ser., 2007, 329, 307–310 CrossRef CAS.
- N. W. Withers, W. Kokke, W. Fenical and C. Djerassi, Proc. Natl. Acad. Sci. U. S. A., 1982, 79, 3764–3768 CrossRef CAS.
- R. G. Kerr, L. C. Rodriguez and J. Kellman, Tetrahedron Lett., 1996, 37, 8301–8304 CrossRef CAS.
- S. Franzenburg, S. Fraune, P. M. Altrock, S. Kuenzel, J. F. Baines, A. Traulsen and T. C. G. Bosch, ISME J., 2013, 7, 781–790 CrossRef CAS PubMed.
- S. Fraune and T. C. G. Bosch, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 13146–13151 CrossRef CAS PubMed.
- S. Franzenburg, J. Walter, S. Kuenzel, J. Wang, J. F. Baines, T. C. G. Bosch and S. Fraune, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, E3730–E3738 CrossRef CAS PubMed.
- S. Fraune, F. Anton-Erxleben, R. Augustin, S. Franzenburg, M. Knop, K. Schröder, D. Wolloweit-Ohl and T. C. G. Bosch, ISME J., 2014 DOI:10.1038/ismej.2014.239.
- K. H. Sharp, S. K. Davidson and M. G. Haygood, ISME J., 2007, 1, 693–702 CrossRef PubMed.
- M. Tischler, S. W. Ayer and R. J. Andersen, Comp. Biochem. Physiol., Part B: Biochem. Mol. Biol., 1986, 84, 43–45 CrossRef.
- C. K. Narkowicz, A. J. Blackman, E. Lacey, J. H. Gill and K. Heiland, J. Nat. Prod., 2002, 65, 938–941 CrossRef CAS PubMed.
- J. T. Walls, A. J. Blackman and D. A. Ritz, J. Chem. Ecol., 1991, 17, 1871–1881 CrossRef CAS PubMed.
- J. T. Walls, A. J. Blackman and D. A. Ritz, Hydrobiologia, 1995, 297, 163–172 CrossRef CAS.
- Y. H. Choi, A. Park, F. J. Schmitz and I. Vanaltena, J. Nat. Prod., 1993, 56, 1431–1433 CrossRef CAS.
- F. J. Schmitz, F. S. Deguzman, Y. H. Choi, M. B. Hossain, S. K. Rizvi and D. Vanderhelm, Pure Appl. Chem., 1990, 62, 1393–1396 CrossRef CAS.
- L. Peters, A. D. Wright, A. Krick and G. M. Konig, J. Chem. Ecol., 2004, 30, 1165–1181 CrossRef CAS.
- G. R. Pettit, C. L. Herald, D. L. Doubek, D. L. Herald, E. Arnold and J. Clardy, J. Am. Chem. Soc., 1982, 104, 6846–6848 CrossRef CAS.
- A. E. Trindade-Silva, G. E. Lim-Fong, K. H. Sharp and M. G. Haygood, Curr. Opin. Biotechnol., 2010, 21, 834–842 CrossRef CAS PubMed.
- N. B. Lopanik, N. M. Targett and N. Lindquist, Mar. Ecol.: Prog. Ser., 2006, 327, 183–191 CrossRef CAS.
- T. J. Nelson and D. L. Alkon, Trends Biochem. Sci., 2009, 34, 136–145 CrossRef CAS PubMed.
- N. Lopanik, K. R. Gustafson and N. Lindquist, J. Nat. Prod., 2004, 67, 1412–1414 CrossRef CAS PubMed.
- N. Lopanik, N. Lindquist and N. Targett, Oecologia, 2004, 139, 131–139 CrossRef PubMed.
- N. Lindquist and M. E. Hay, Ecol. Monogr., 1996, 66, 431–450 CrossRef.
- N. Lindquist, Mar. Biol., 1996, 126, 745–755 CrossRef.
- U. Anthoni, P. H. Nielsen, M. Pereira and C. Christophersen, Comp. Biochem. Physiol., Part B: Biochem. Mol. Biol., 1990, 96, 431–437 CrossRef.
- R. M. Woollacott, Mar. Biol., 1981, 65, 155–158 CrossRef.
- R. M. Woollacott and R. L. Zimmer, J. Morphol., 1975, 147, 355–377 CrossRef PubMed.
- M. G. Haygood and S. K. Davidson, Appl. Environ. Microbiol., 1997, 63, 4612–4616 CAS.
- S. K. Davidson, S. W. Allen, G. E. Lim, C. M. Anderson and M. G. Haygood, Appl. Environ. Microbiol., 2001, 67, 4531–4537 CrossRef CAS.
- M. Hildebrand, L. E. Waggoner, H. B. Liu, S. Sudek, S. Allen, C. Anderson, D. H. Sherman and M. Haygood, Chem. Biol., 2004, 11, 1543–1552 CrossRef CAS PubMed.
- S. Sudek, N. B. Lopanik, L. E. Waggoner, M. Hildebrand, C. Anderson, H. B. Liu, A. Patel, D. H. Sherman and M. G. Haygood, J. Nat. Prod., 2007, 70, 67–74 CrossRef CAS PubMed.
- T. Nguyen, K. Ishida, H. Jenke-Kodama, E. Dittmann, C. Gurgui, T. Hochmuth, S. Taudien, M. Platzer, C. Hertweck and J. Piel, Nat. Biotechnol., 2008, 26, 225–233 CrossRef CAS PubMed.
- T. J. Buchholz, C. M. Rath, N. B. Lopanik, N. P. Gardner, K. Hakansson and D. H. Sherman, Chem. Biol., 2010, 17, 1092–1100 CrossRef CAS PubMed.
- N. B. Lopanik, J. A. Shields, T. J. Buchholz, C. M. Rath, J. Hothersall, M. G. Haygood, K. Hakansson, C. M. Thomas and D. H. Sherman, Chem. Biol., 2008, 15, 1175–1186 CrossRef CAS PubMed.
- S. K. Davidson and M. G. Haygood, Biol. Bull., 1999, 196, 273–280 CrossRef CAS.
- G. E. Lim and M. G. Haygood, Appl. Environ. Microbiol., 2004, 70, 4921–4929 CrossRef CAS PubMed.
- T. M. McGovern and M. E. Hellberg, Mol. Ecol., 2003, 12, 1207–1215 CrossRef CAS.
- G. E. Lim-Fong, L. A. Regali and M. G. Haygood, Appl. Environ. Microbiol., 2008, 74, 3605–3609 CrossRef CAS PubMed.
- J. S. Shellenberger and J. R. P. Ross, Northwest Sci., 1998, 72, 23–33 Search PubMed.
- S. Matsunaga, N. Fusetani and K. Hashimoto, Experientia, 1986, 42, 84 CrossRef CAS.
- A. J. Blackman and C. P. Li, Aust. J. Chem., 1994, 47, 1625–1629 CrossRef CAS.
- B. Carte and D. J. Faulkner, J. Org. Chem., 1983, 48, 2314–2318 CrossRef CAS.
- R. Kazlauskas, J. F. Marwood, P. T. Murphy and R. J. Wells, Aust. J. Chem., 1982, 35, 215–217 CrossRef CAS.
- N. Lindquist and W. Fenical, Experientia, 1991, 47, 504–506 CrossRef CAS.
- H. H. Wasserman, D. J. Frieland and D. A. Morrison, Tetrahedron Lett., 1968, 6, 641–644 CrossRef CAS.
- A. Franks, P. Haywood, C. Holmstrom, S. Egan, S. Kjelleberg and N. Kumar, Molecules, 2005, 10, 1286–1291 CrossRef CAS.
- C. Holmstrom and S. Kjelleberg, FEMS Microbiol. Ecol., 1999, 30, 285–293 CrossRef CAS.
- H. Heindl, J. Wiese, V. Thiel and J. F. Imhoff, Syst. Appl. Microbiol., 2010, 33, 94–104 CrossRef CAS PubMed.
- J. P. Galkiewicz, Z. A. Pratte, M. A. Gray and C. A. Kellogg, FEMS Microbiol. Ecol., 2011, 77, 333–346 CrossRef CAS PubMed.
- C. Burke, T. Thomas, S. Egan and S. Kjelleberg, Environ. Microbiol., 2007, 9, 814–818 CrossRef CAS PubMed.
- D. M. Pinkerton, M. G. Banwell, M. J. Garson, N. Kumar, M. O. de Moraes, B. C. Cavalcanti, F. W. A. Barros and C. Pessoa, Chem. Biodiversity, 2010, 7, 1311–1324 CAS.
- N. Lindquist, M. E. Hay and W. Fenical, Ecol. Monogr., 1992, 62, 547–568 CrossRef.
- V. J. Paul, N. Lindquist and W. Fenical, Mar. Ecol.: Prog. Ser., 1990, 59, 109–118 CrossRef CAS.
- B. Carte and D. J. Faulkner, J. Chem. Ecol., 1986, 12, 795–804 CrossRef CAS PubMed.
- A. Franks, S. Egan, C. Holmstrom, S. James, H. Lappin-Scott and S. Kjelleberg, Appl. Environ. Microbiol., 2006, 72, 6079–6087 CrossRef CAS PubMed.
- V. Bane, M. Lehane, M. Dikshit, A. Riordan and A. Furey, Toxins, 2014, 6, 693–755 CrossRef CAS PubMed.
- R. Ritson-Williams, M. Yotsu-Yamashita and V. J. Paul, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 3176–3179 CrossRef CAS PubMed.
- J. W. Daly, J. Nat. Prod., 2004, 67, 1211–1215 CrossRef CAS PubMed.
- C. T. Hanifin, Mar. Drugs, 2010, 8, 577–593 CrossRef CAS PubMed.
- T. Y. Magarlamov, I. A. Beleneva, A. V. Chernyshev and A. D. Kuhlevsky, Toxicon, 2014, 85, 46–51 CrossRef CAS PubMed.
- V. Pratheepa and V. Vasconcelos, Environ. Toxicol. Pharmacol., 2013, 36, 1046–1054 CrossRef CAS PubMed.
- M. Asakawa, K. Ito and H. Kajihara, Toxins, 2013, 5, 376–395 CrossRef CAS PubMed.
- D. F. Hwang, O. Arakawa, T. Saito, T. Noguchi, U. Simidu, K. Tsukamoto, Y. Shida and K. Hashimoto, Mar. Biol., 1989, 100, 327–332 CrossRef CAS.
- R. Chau, J. A. Kalaitzis and B. A. Neilan, Aquat. Toxicol., 2011, 104, 61–72 CrossRef CAS PubMed.
- S. K. Goffredi, A. Warén, V. J. Orphan, C. L. Van Dover and R. C. Vrijenhoek, Appl. Environ. Microbiol., 2004, 70, 3082–3090 CrossRef CAS.
- H. Yao, M. Dao, T. Imholt, J. Huang, K. Wheeler, A. Bonilla, S. Suresh and C. Ortiz, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 987–992 CrossRef CAS PubMed.
- Y. Suzuki, R. E. Kopp, T. Kogure, A. Suga, K. Takai, S. Tsuchida, N. Ozaki, K. Endo, J. Hashimoto, Y. Kato, C. Mizota, T. Hirata, H. Chiba, K. H. Nealson, K. Horikoshi and J. L. Kirschvink, Earth Planet. Sci. Lett., 2006, 242, 39–50 CrossRef CAS PubMed.
- O. Peraud, J. S. Biggs, R. W. Hughen, A. R. Light, G. P. Concepcion, B. M. Olivera and E. W. Schmidt, Appl. Environ. Microbiol., 2009, 75, 6820–6826 CrossRef CAS PubMed.
- Z. J. Lin, R. R. Antemano, R. W. Hughen, M. D. B. Tianero, O. Peraud, M. G. Haygood, G. P. Concepcion, B. M. Olivera, A. Light and E. W. Schmidt, J. Nat. Prod., 2010, 73, 1922–1926 CrossRef CAS PubMed.
- Z. Lin, C. A. Reilly, R. Antemano, R. W. Hughen, L. Marett, G. P. Concepcion, M. G. Haygood, B. M. Olivera, A. Light and E. W. Schmidt, J. Med. Chem., 2011, 54, 3746–3755 CrossRef CAS PubMed.
- Z. J. Lin, L. Marett, R. W. Hughen, M. Flores, I. Forteza, M. A. Ammon, G. P. Concepcion, S. Espino, B. M. Olivera, G. Rosenberg, M. G. Haygood, A. R. Light and E. W. Schmidt, Bioorg. Med. Chem. Lett., 2013, 23, 4867–4869 CrossRef CAS PubMed.
- Z. Lin, M. Koch, C. D. Pond, G. Mabeza, R. A. Seronay, G. P. Concepcion, L. R. Barrows, B. M. Olivera and E. W. Schmidt, J. Antibiot., 2014, 67, 121–126 CrossRef CAS PubMed.
- A. Marin, L. A. Alvarez, G. Cimino and A. Spinella, J. Molluscan Stud., 1999, 65, 121–131 CrossRef PubMed.
- Z. J. Lin, J. P. Torres, M. A. Ammon, L. Marett, R. W. Teichert, C. A. Reilly, J. C. Kwan, R. W. Hughen, M. Flores, M. D. Tianero, O. Peraud, J. E. Cox, A. R. Light, A. J. L. Villaraza, M. G. Haygood, G. P. Concepcion, B. M. Olivera and E. W. Schmidt, Chem. Biol., 2013, 20, 73–81 CrossRef CAS PubMed.
- D. L. Distel, D. J. Beaudoin and W. Morrill, Appl. Environ. Microbiol., 2002, 68, 6292–6299 CrossRef CAS.
- M. A. Betcher, J. M. Fung, A. W. Han, R. O'Connor, R. Seronay, G. P. Concepcion, D. L. Distel and M. G. Haygood, PLoS One, 2012, 7, e45309 CAS.
- D. L. Distel, W. Morrill, N. MacLaren-Toussaint, D. Franks and J. Waterbury, Int. J. Syst. Evol. Microbiol., 2002, 52, 2261–2269 CrossRef CAS.
- J. C. Yang, R. Madupu, A. S. Durkin, N. A. Ekborg, C. S. Pedamallu, J. B. Hostetler, D. Radune, B. S. Toms, B. Henrissat, P. M. Coutinho, S. Schwarz, L. Field, A. E. Trindade-Silva, C. A. G. Soares, S. Elshahawi, A. Hanora, E. W. Schmidt, M. G. Haygood, J. Posfai, J. Benner, C. Madinger, J. Nove, B. Anton, K. Chaudhary, J. Foster, A. Holman, S. Kumar, P. A. Lessard, Y. A. Luyten, B. Slatko, N. Wood, B. Wu, M. Teplitski, J. D. Mougous, N. Ward, J. A. Eisen, J. H. Badger and D. L. Distel, PLoS One, 2009, 4, e6085 Search PubMed.
- S. I. Elshahawi, A. E. Trindade-Silva, A. Hanora, A. W. Han, M. S. Flores, V. Vizzoni, C. G. Schrago, C. A. Soares, G. P. Concepcion, D. L. Distel, E. W. Schmidt and M. G. Haygood, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, E295–E304 CrossRef CAS PubMed.
- S. V. Nyholm and M. J. McFall-Ngai, Nat. Rev. Microbiol., 2004, 2, 632–642 CrossRef CAS PubMed.
- M. J. McFall-Ngai, Annu. Rev. Microbiol., 2014, 68, 177–194 CrossRef CAS PubMed.
- B. W. Jones and M. K. Nishiguchi, Mar. Biol., 2004, 144, 1151–1155 CrossRef PubMed.
- E. G. Ruby, Annu. Rev. Microbiol., 1996, 50, 591–624 CrossRef CAS PubMed.
- A. J. Collins, B. A. LaBarre, B. S. Wong Won, M. V. Shah, S. Heng, M. H. Choudhury, S. A. Haydar, J. Santiago and S. V. Nyholm, Appl. Environ. Microbiol., 2012, 78, 4200–4208 CrossRef CAS PubMed.
- S. Grigioni, R. Boucher-Rodoni, A. Demarta, M. Tonolla and R. Peduzzi, Mar. Biol., 2000, 136, 217–222 CrossRef CAS.
- E. Barbieri, B. J. Paster, D. Hughes, L. Zurek, D. P. Moser, A. Teske and M. L. Sogin, Environ. Microbiol., 2001, 3, 151–167 CrossRef CAS.
- W. Decleir and A. Richard, in Biologisch Jaarboek (Dodonaea), Koninklijk Natuurwetenschappelijk Genootschap Dodonaea, Gent, Belgium, 1972 Search PubMed.
- C. Van den Branden, M. Gillis and A. Richard, Comp. Biochem. Physiol., Part B: Biochem. Mol. Biol., 1980, 66, 331–334 CrossRef.
- A. Richard, C. Van den Branden and W. Decleir, in Cyclic Phenomena in Marine Plants and Animals: Proceedings of the 13th European Marine Biology Symposium, ed. E. Naylor and R. G. Hartnoll, Pergamon Press, Exeter, UK, 1979, pp. 173–180 Search PubMed.
- M. R. Kaufman, Y. Ikeda, C. Patton, G. Van Dykhuizen and D. Epel, Biol. Bull., 1998, 194, 36–43 CrossRef.
- A. Lum-Kong, J. Zool., 1992, 226, 469–490 CrossRef PubMed.
- P. Gomathi, J. R. Nair and P. M. Sherief, Indian J. Mar. Sci., 2010, 39, 100–104 CAS.
- E. Barbieri, K. Barry, A. Child and N. Wainwright, Biol. Bull., 1997, 193, 275–276 Search PubMed.
- K. Benkendorff, A. R. Davis and J. Bremner, J. Invertebr. Pathol., 2001, 78, 109–118 CrossRef CAS PubMed.
- J. R. Nair, D. Pillai, S. M. Joseph, P. Gomathi, P. V. Senan and P. M. Sherief, Indian J. Geo-Mar. Sci., 2011, 40, 13–27 CAS.
- M. S. Gil-Turnes, M. E. Hay and W. Fenical, Science, 1989, 246, 116–118 CAS.
- M. S. Gil-Turnes and W. Fenical, Biol. Bull., 1992, 182, 105–108 CrossRef.
- A. Stoessl, Biochem. Biophys. Res. Commun., 1969, 35, 186–191 CrossRef CAS.
- N. Claydon, J. F. Grove and M. Pople, Phytochemistry, 1985, 24, 937–943 CrossRef CAS.
- N. Lindquist, P. H. Barber and J. B. Weisz, Proc. R. Soc. B, 2005, 272, 1209–1216 CrossRef PubMed.
- E. W. Schmidt and M. S. Donia, Curr. Opin. Biotechnol., 2010, 21, 827–833 CrossRef CAS PubMed.
- E. W. Schmidt, M. S. Donia, J. A. McIntosh, W. F. Fricke and J. Ravel, J. Nat. Prod., 2012, 75, 295–304 CrossRef CAS PubMed.
- H. L. Sings and K. L. Rinehart, J. Ind. Microbiol. Biotechnol., 1996, 17, 385–396 CrossRef CAS.
- B. S. Davidson, Chem. Rev., 1993, 93, 1771–1791 CrossRef CAS.
- F. Lafargue and G. Duclaux, Ann. Inst. Oceanogr., 1979, 55, 163–184 Search PubMed.
- P. Kott, Mem. Queensl. Mus., 1980, 20, 1–48 Search PubMed.
- P. Kott, Micronesica, 1982, 18, 95–128 Search PubMed.
- I. Koike, M. Yamamuro and P. C. Pollard, Aust. J. Mar. Freshwater Res., 1993, 44, 173–182 Search PubMed.
- K. L. Rinehart, J. B. Gloer, J. C. Cook, S. A. Mizsak and T. A. Scahill, J. Am. Chem. Soc., 1981, 103, 1857–1859 CrossRef CAS.
- K. L. Rinehart, J. B. Gloer, R. G. Hughes, H. E. Renis, J. P. McGovren, E. B. Swynenberg, D. A. Stringfellow, S. L. Kuentzel and L. H. Li, Science, 1981, 212, 933–935 CAS.
- N. Lindquist, J. Chem. Ecol., 2002, 28, 1987–2000 CrossRef CAS.
- N. Lindquist and M. E. Hay, Ecology, 1995, 76, 1347–1358 CrossRef.
- M. Tsukimoto, M. Nagaoka, Y. Shishido, J. Fujimoto, F. Nishisaka, S. Matsumoto, E. Harunari, C. Imada and T. Matsuzaki, J. Nat. Prod., 2011, 74, 2329–2331 CrossRef CAS PubMed.
- Y. Xu, R. D. Kersten, S. J. Nam, L. Lu, A. M. Al-Suwailem, H. J. Zheng, W. Fenical, P. C. Dorrestein, B. S. Moore and P. Y. Qian, J. Am. Chem. Soc., 2012, 134, 8625–8632 CrossRef CAS PubMed.
- C. Ireland and P. J. Scheuer, J. Am. Chem. Soc., 1980, 102, 5688–5691 CrossRef CAS.
- M. S. Donia, B. J. Hathaway, S. Sudek, M. G. Haygood, M. J. Rosovitz, J. Ravel and E. W. Schmidt, Nat. Chem. Biol., 2006, 2, 729–735 CrossRef CAS PubMed.
- P. F. Long, W. C. Dunlap, C. N. Battershill and M. Jaspars, ChemBioChem, 2005, 6, 1760–1765 CrossRef CAS PubMed.
- E. W. Schmidt, J. T. Nelson, D. A. Rasko, S. Sudek, J. A. Eisen, M. G. Haygood and J. Ravel, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 7315–7320 CrossRef CAS PubMed.
- M. S. Donia, J. Ravel and E. W. Schmidt, Nat. Chem. Biol., 2008, 4, 341–343 CrossRef CAS PubMed.
- T. M. Zabriskie, C. L. Mayne and C. M. Ireland, J. Am. Chem. Soc., 1988, 110, 7919–7920 CrossRef CAS.
- A. D. Richardson, W. Aalbersberg and C. M. Ireland, Anti-Cancer Drugs, 2005, 16, 533–541 CrossRef CAS PubMed.
- M. S. Donia, W. F. Fricke, F. Partensky, J. Cox, S. I. Elshahawi, J. R. White, A. M. Phillippy, M. C. Schatz, J. Piel, M. G. Haygood, J. Ravel and E. W. Schmidt, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, E1423–E1432 CrossRef CAS PubMed.
- J. C. Kwan, M. S. Donia, A. W. Han, E. Hirose, M. G. Haygood and E. W. Schmidt, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 20655–20660 CrossRef CAS PubMed.
- A. Nakabachi, R. Ueoka, K. Oshima, R. Teta, A. Mangoni, M. Gurgui, N. J. Oldham, G. van Echten-Deckert, K. Okamura, K. Yamamoto, H. Inoue, M. Ohkuma, Y. Hongoh, S.-y. Y. Miyagishima, M. Hattori, J. Piel and T. Fukatsu, Curr. Biol., 2013, 23, 1478–1484 CrossRef CAS PubMed.
- J. F. Biard, C. Grivois, J. F. Verbist, C. Debitus and J. B. Carre, J. Mar. Biol. Assoc. U. K., 1990, 70, 741–746 CrossRef.
- J. F. Biard, C. Roussakis, J. M. Kornprobst, D. Gouiffesbarbin, J. F. Verbist, P. Cotelle, M. P. Foster, C. M. Ireland and C. Debitus, J. Nat. Prod., 1994, 57, 1336–1345 CrossRef CAS.
- D. Gouiffes, M. Juge, N. Grimaud, L. Welin, M. P. Sauviat, Y. Barbin, D. Laurent, C. Roussakis, J. P. Henichart and J. F. Verbist, Toxicon, 1988, 26, 1129–1136 CrossRef CAS.
- B. M. Degnan, C. J. Hawkins, M. F. Lavin, E. J. McCaffrey, D. L. Parry and D. J. Watters, J. Med. Chem., 1989, 32, 1354–1359 CrossRef CAS.
- C. S. Riesenfeld, A. E. Murray and B. J. Baker, J. Nat. Prod., 2008, 71, 1812–1818 CrossRef CAS PubMed.
- M. S. Donia, W. F. Fricke, J. Ravel and E. W. Schmidt, PLoS One, 2011, 6, e17897 CAS.
- C. Moss, D. H. Green, B. Perez, A. Velasco, R. Henriquez and J. D. McKenzie, Mar. Biol., 2003, 143, 99–110 CrossRef CAS PubMed.
- A. E. Perez-Matos, W. Rosado and N. S. Govind, Antonie van Leeuwenhoek, 2007, 92, 155–164 CrossRef PubMed.
- C. M. Rath, B. Janto, J. Earl, A. Ahmed, F. Z. Hu, L. Hiller, M. Dahlgren, R. Kreft, F. A. Yu, J. J. Wolff, H. K. Kweon, M. A. Christiansen, K. Hakansson, R. M. Williams, G. D. Ehrlich and D. H. Sherman, ACS Chem. Biol., 2011, 6, 1244–1256 CrossRef CAS PubMed.
- R. Gaugler, Entomopathogenic Nematology, CABI Publishing, New York, NY, USA, 2002 Search PubMed.
- S. Forst and K. Nealson, Microbiol. Rev., 1996, 60, 21–43 CAS.
- J. Li, K. Hu and J. M. Webster, Chem. Heterocycl. Compd., 1998, 34, 1331–1339 CrossRef CAS.
- S. Forst, B. Dowds, N. Boemare and E. Stackebrandt, Annu. Rev. Microbiol., 1997, 51, 47–72 CrossRef CAS PubMed.
- S. R. Dutky, Adv. Appl. Microbiol., 1959, 1, 175–200 CAS.
- V. J. Paul, S. Frautschy, W. Fenical and K. H. Nealson, J. Chem. Ecol., 1981, 7, 589–597 CrossRef CAS PubMed.
- S. Forst and D. Clarke, in Entomopathogenic Nematology, ed. R. Gaugler, CABI Publishing, New York, NY, USA, 2002, pp. 57–77 Search PubMed.
- E. E. Lewis, in Entomopathogenic Nematology, ed. R. Gaugler, CABI Publishing, New York, NY, USA, 2002, pp. 205–223 Search PubMed.
- G. O. Poinar, Fundam. Appl. Nematol., 1993, 16, 333–338 Search PubMed.
- G. Chen, G. B. Dunphy and J. M. Webster, Biol. Control, 1994, 4, 157–162 CrossRef.
- R. J. Akhurst, J. Gen. Microbiol., 1982, 128, 3061–3065 CAS.
- J. M. Webster, G. Chen, K. Hu and J. Li, in Entomopathogenic Nematology, ed. R. Gaugler, CABI Publishing, New York, NY, USA, 2002, pp. 99–114 Search PubMed.
- S. Sharma, N. Waterfield, D. Bowen, T. Rocheleau, L. Holland, R. James and R. ffrench-Constant, FEMS Microbiol. Lett., 2002, 214, 241–249 CrossRef CAS PubMed.
- R. ffrench-Constant, N. Waterfield, P. Daborn, S. Joyce, H. Bennett, C. Au, A. Dowling, S. Boundy, S. Reynolds and D. Clarke, FEMS Microbiol. Rev., 2003, 26, 433–456 CrossRef CAS PubMed.
- H. Hawlena, F. Bashey and C. M. Lively, Ecol. Evol., 2012, 2, 2516–2521 Search PubMed.
- N. E. Boemare, M. H. Boyergiglio, J. O. Thaler, R. J. Akhurst and M. Brehelin, Appl. Environ. Microbiol., 1992, 58, 3032–3037 CAS.
- J. O. Thaler, S. Baghdiguian and N. Boemare, Appl. Environ. Microbiol., 1995, 61, 2049–2052 CAS.
- L. Lango-Scholey, A. O. Brachmann, H. B. Bode and D. J. Clarke, PLoS One, 2013, 8, e82152 Search PubMed.
- J. Li, G. Chen, H. Wu and J. M. Webster, Appl. Environ. Microbiol., 1995, 61, 4329–4333 CAS.
- F. Sztaricskai, Z. Dinya, G. Y. Batta, E. Szallas, A. Szentirmai and A. Fodor, ACH - Models Chem., 1992, 129, 697–707 CAS.
- T. A. Ciche, M. Blackburn, J. R. Carney and J. C. Ensign, Appl. Environ. Microbiol., 2003, 69, 4706–4713 CrossRef CAS.
- S. W. Fuchs, F. Grundmann, M. Kurz, M. Kaiser and H. B. Bode, ChemBioChem, 2014, 15, 512–516 CrossRef CAS PubMed.
- B. V. McInerney, R. P. Gregson, M. J. Lacey, R. J. Akhurst, G. R. Lyons, S. H. Rhodes, D. R. Smith, L. M. Engelhardt and A. H. White, J. Nat. Prod., 1991, 54, 774–784 CrossRef CAS.
- J. Li, G. Chen, J. M. Webster and E. Czyzewska, J. Nat. Prod., 1995, 58, 1081–1086 CrossRef CAS.
- J. Li, G. Chen and J. M. Webster, Can. J. Microbiol., 1997, 43, 770–773 CrossRef CAS.
- Q. Zhou, F. Grundmann, M. Kaiser, M. Schiell, S. Gaudriault, A. Batzer, M. Kurz and H. B. Bode, Chem.–Eur. J., 2013, 19, 16772–16779 CrossRef CAS PubMed.
- G. Lang, T. Kalvelage, A. Peters, J. Wiese and J. F. Imhoff, J. Nat. Prod., 2008, 71, 1074–1077 CrossRef CAS PubMed.
- B. V. McInerney, W. C. Taylor, M. J. Lacey, R. J. Akhurst and R. P. Gregson, J. Nat. Prod., 1991, 54, 785–795 CrossRef CAS.
- D. Ji, Y. Yi, G. H. Kang, Y. H. Choi, P. Kim, N. I. Baek and Y. Kim, FEMS Microbiol. Lett., 2004, 239, 241–248 CrossRef CAS PubMed.
- M. Kronenwerth, K. A. J. Bozhüyük, A. S. Kahnt, D. Steinhilber, S. Gaudriault, M. Kaiser and H. B. Bode, Chem.–Eur. J., 2014, 20, 17478–17487 CrossRef CAS PubMed.
- M. Kronenwerth, C. Dauth, M. Kaiser, I. Pemberton and H. B. Bode, Eur. J. Org. Chem., 2014, 2014, 8026–8028 CrossRef CAS PubMed.
- F. Grundmann, M. Kaiser, M. Schiell, A. Batzer, M. Kurz, A. Thanwisai, N. Chantratita and H. B. Bode, J. Nat. Prod., 2014, 77, 779–783 CrossRef CAS PubMed.
- F. I. Nollmann, A. Dowling, M. Kaiser, K. Deckmann, S. Grosch, R. Ffrench-Constant and H. B. Bode, Beilstein J. Org. Chem., 2012, 8, 528–533 CrossRef CAS PubMed.
- B. Ohlendorf, S. Simon, J. Wiese and J. F. Imhoff, Nat. Prod. Commun., 2011, 6, 1247–1250 CAS.
- J. M. Crawford, C. Portmann, X. Zhang, M. B. J. Roeffaers and J. Clardy, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 10821–10826 CrossRef CAS PubMed.
- G. Chen, Y. Zhang, J. Li, G. B. Dunphy, Z. K. Punja and J. M. Webster, J. Invertebr. Pathol., 1996, 68, 101–108 CrossRef CAS PubMed.
- P. J. Isaacson and J. M. Webster, J. Invertebr. Pathol., 2002, 79, 146–153 CrossRef CAS.
- S. Derzelle, E. Duchaud, F. Kunst, A. Danchin and P. Bertin, Appl. Environ. Microbiol., 2002, 68, 3780–3789 CrossRef CAS.
- W. H. Richardson, T. M. Schmidt and K. H. Nealson, Appl. Environ. Microbiol., 1988, 54, 1602–1605 CAS.
- I. Eleftherianos, S. Boundy, S. A. Joyce, S. Aslam, J. W. Marshall, R. J. Cox, T. J. Simpson, D. J. Clarke, R. H. ffrench-Constant and S. E. Reynolds, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 2419–2424 CrossRef CAS PubMed.
- K. Hu and J. M. Webster, FEMS Microbiol. Lett., 2000, 189, 219–223 CAS.
- A. O. Brachmann, S. A. Joyce, H. Jenke-Kodama, G. Schwar, D. J. Clarke and H. B. Bode, ChemBioChem, 2007, 8, 1721–1728 CrossRef CAS PubMed.
- K. Hu, J. Li, W. Wang, H. Wu, H. Lin and J. M. Webster, Can. J. Microbiol., 1998, 44, 1072–1077 CrossRef CAS.
- S. Paik, Y. H. Park, S. I. Suh, H. S. Kim, I. S. Lee, M. K. Park, C. S. Lee and S. H. Park, Bull. Korean Chem. Soc., 2001, 22, 372–374 CAS.
- M. E. Baur, H. K. Kaya and D. R. Strong, Biol. Control, 1998, 12, 231–236 CrossRef.
- X. S. Zhou, H. K. Kaya, K. Heungens and H. Goodrich-Blair, Appl. Environ. Microbiol., 2002, 68, 6202–6209 CrossRef CAS.
- A. E. Douglas, Cell Host Microbe, 2011, 10, 359–367 CAS.
- R. L. L. Kellner, Insect Biochem. Mol. Biol., 2002, 32, 389–395 CrossRef CAS.
- R. L. L. Kellner and K. Dettner, Oecologia, 1996, 107, 293–300 CrossRef.
- J. Piel, I. Hofer and D. Q. Hui, J. Bacteriol., 2004, 186, 1280–1286 CrossRef CAS.
- K. M. Oliver, J. A. Russell, N. A. Moran and M. S. Hunter, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 1803–1807 CrossRef CAS PubMed.
- P. Łukasik, M. A. Dawid, J. Ferrari and H. C. Godfray, Oecologia, 2013, 173, 985–996 CrossRef PubMed.
- J. Xie, I. Vilchez and M. Mateos, PLoS One, 2010, 5, e12149 Search PubMed.
- A. K. Hansen, G. Jeong, T. D. Paine and R. Stouthamer, Appl. Environ. Microbiol., 2007, 73, 7531–7535 CrossRef CAS PubMed.
- T. H. Hsiao, in The Ecology of Agricultural Pests: Biochemical Approaches, ed. E. O. C. Symondson and J. E. Liddell, Chapman and Hall, London, UK, 1996, p. 517 Search PubMed.
- P. Łukasik, M. van Asch, H. Guo and J. Ferrari, Ecol. Lett., 2013, 16, 214–218 CrossRef PubMed.
- C. L. Scarborough, J. Ferrari and H. C. J. Godfray, Science, 2005, 310, 1781 CrossRef CAS PubMed.
- P. T. Hamilton, J. S. Leong, B. F. Koop and S. J. Perlman, Mol. Ecol., 2014, 23, 1558–1570 CrossRef CAS PubMed.
- J. Jaenike, R. Unckless, S. N. Cockburn, L. M. Boelio and S. J. Perlman, Science, 2010, 329, 212–215 CrossRef CAS PubMed.
- G. Yim, H. Huimi Wang and J. Davies Frs, Philos. Trans. R. Soc. London, Ser. B, 2007, 362, 1195–1200 CrossRef CAS PubMed.
- J. Clardy, M. A. Fischbach and C. R. Currie, Curr. Biol., 2009, 19, R437–R441 CrossRef CAS PubMed.
- M. Kaltenpoth, W. Goettler, C. Dale, J. W. Stubblefield, G. Herzner, K. Roeser-Mueller and E. Strohm, Int. J. Syst. Evol. Microbiol., 2006, 56, 1403–1411 CrossRef CAS PubMed.
- M. Kaltenpoth, W. Gottler, G. Herzner and E. Strohm, Curr. Biol., 2005, 15, 475–479 CrossRef CAS PubMed.
- M. Kaltenpoth, K. Roeser-Mueller, S. Koehler, A. Peterson, T. Nechitaylo, J. W. Stubblefield, G. Herzner, J. Seger and E. Strohm, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 6359–6364 CrossRef CAS PubMed.
- M. Kaltenpoth, T. Schmitt, C. Polidori, D. Koedam and E. Strohm, Physiol. Entomol., 2010, 35, 196–200 CrossRef CAS PubMed.
- M. Kaltenpoth, E. Yildirim, M. F. Gürbüz, G. Herzner and E. Strohm, Appl. Environ. Microbiol., 2012, 78, 822–827 CrossRef CAS PubMed.
- M. Kaltenpoth, W. Goettler, S. Koehler and E. Strohm, Evol. Ecol., 2010, 24, 463–477 CrossRef.
- S. Koehler, J. Doubsky and M. Kaltenpoth, Front. Zool., 2013, 10, 13 CrossRef PubMed.
- S. Koehler and M. Kaltenpoth, J. Chem. Ecol., 2013, 39, 978–988 CrossRef CAS PubMed.
- J. Kroiss, M. Kaltenpoth, B. Schneider, M.-G. G. Schwinger, C. Hertweck, R. K. Maddula, E. Strohm and A. Svatos, Nat. Chem. Biol., 2010, 6, 261–263 CrossRef CAS PubMed.
- C. R. Currie, J. A. Scott, R. C. Summerbell and D. Malloch, Nature, 1999, 398, 701–704 CrossRef CAS.
- A. S. Vieira, E. D. Morgan, F. P. Drijfhout and M. I. Camargo-Mathias, J. Chem. Ecol., 2012, 38, 1289–1297 CrossRef CAS PubMed.
- C. R. Currie, U. G. Mueller and D. Malloch, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 7998–8002 CrossRef CAS.
- C. R. Currie and A. E. Stuart, Proc. R. Soc. B, 2001, 268, 1033–1039 CrossRef CAS PubMed.
- M. J. Cafaro, M. Poulsen, A. E. F. Little, S. L. Price, N. M. Gerardo, B. Wong, A. E. Stuart, B. Larget, P. Abbot and C. R. Currie, Proc. R. Soc. B, 2011, 278, 1814–1822 CrossRef PubMed.
- U. G. Mueller, D. Dash, C. Rabeling and A. Rodrigues, Evolution, 2008, 62, 2894–2912 CrossRef CAS PubMed.
- M. Poulsen, M. Cafaro, J. J. Boomsma and C. R. Currie, Mol. Ecol., 2005, 14, 3597–3604 CrossRef CAS PubMed.
- J. Barke, R. F. Seipke, S. Gruschow, D. Heavens, N. Drou, M. J. Bibb, R. J. Goss, D. W. Yu and M. I. Hutchings, BMC Biol., 2010, 8, 10 CrossRef PubMed.
- R. Sen, H. D. Ishak, D. Estrada, S. E. Dowd, E. Hong and U. G. Mueller, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 17805–17810 CrossRef CAS PubMed.
- C. Kost, T. Lakatos, I. Bottcher, W. R. Arendholz, M. Redenbach and R. Wirth, Naturwissenschaften, 2007, 94, 821–828 CrossRef CAS PubMed.
- G. Carr, E. R. Derbyshire, E. Caldera, C. R. Currie and J. Clardy, J. Nat. Prod., 2012, 75, 1806–1809 CrossRef CAS PubMed.
- D. C. Oh, M. Poulsen, C. R. Currie and J. Clardy, Nat. Chem. Biol., 2009, 5, 391–393 CrossRef CAS PubMed.
- S. Haeder, R. Wirth, H. Herz and D. Spiteller, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 4742–4746 CrossRef CAS PubMed.
- I. Schoenian, M. Spiteller, M. Ghaste, R. Wirth, H. Herz and D. Spiteller, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 1955–1960 CrossRef PubMed.
- T. D. Zucchi, A. S. Guidolin and F. L. Consoli, Microbiol. Res., 2011, 166, 68–76 CrossRef CAS PubMed.
- M. X. Ruiz-González, P.-J. G. Malé, C. Leroy, A. Dejean, H. Gryta, P. Jargeat, A. Quilichini and J. Orivel, Biol. Lett., 2011, 7, 475–479 CrossRef PubMed.
- A. Dejean, P. J. Solano, J. Ayroles, B. Corbara and J. Orivel, Nature, 2005, 434, 973 CrossRef CAS PubMed.
- R. F. Seipke, J. Barke, M. X. Ruiz-Gonzalez, J. Orivel, D. W. Yu and M. I. Hutchings, Antonie van Leeuwenhoek, 2012, 101, 443–447 CrossRef CAS PubMed.
- T. R. Ramadhar, C. Beemelmanns, C. R. Currie and J. Clardy, J. Antibiot., 2014, 67, 53–58 CrossRef CAS PubMed.
- U. G. Mueller, N. M. Gerardo, D. K. Aanen, D. L. Six and T. R. Schultz, Annu. Rev. Ecol. Evol. Syst., 2005, 36, 563–595 CrossRef.
- A. A. Visser, T. Nobre, C. R. Currie, D. K. Aanen and M. Poulsen, Microb. Ecol., 2012, 63, 975–985 CrossRef CAS PubMed.
- G. Carr, M. Poulsen, J. L. Klassen, Y. Hou, T. P. Wyche, T. S. Bugni, C. R. Currie and J. Clardy, Org. Lett., 2012, 14, 2822–2825 CrossRef CAS PubMed.
- S. Um, A. Fraimout, P. Sapountzis, D.-C. Oh and M. Poulsen, Sci. Rep., 2013, 3, 3250 Search PubMed.
- T. C. Harrington, in Ecological and evolutionary advances in insect-fungal associations, ed. F. E. Vega and M. Blackwell, Oxford University Press, New York, NY, USA, 2005, pp. 257–291 Search PubMed.
- H. Francke-Grosmann, in Symbiosis, ed. S. M. Henry, Academic Press, New York, NY, USA, 1967, pp. 141–205 Search PubMed.
- J. J. Scott, D. C. Oh, M. C. Yuceer, K. D. Klepzig, J. Clardy and C. R. Currie, Science, 2008, 322, 63 CrossRef CAS PubMed.
- D. C. Oh, J. J. Scott, C. R. Currie and J. Clardy, Org. Lett., 2009, 11, 633–636 CrossRef CAS PubMed.
- J. A. V. Blodgett, D. C. Oh, S. G. Cao, C. R. Currie, R. Kolter and J. Clardy, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 11692–11697 CrossRef CAS PubMed.
- J. Hulcr, A. Adams, K. Raffa, R. Hofstetter, K. Klepzig and C. Currie, Microb. Ecol., 2011, 61, 759–768 CrossRef PubMed.
- R. J. Dillon and A. K. Charnley, J. Invertebr. Pathol., 1995, 66, 72–75 CrossRef CAS.
- R. Dillon and K. Charnley, Res. Microbiol., 2002, 153, 503–509 CrossRef CAS.
- R. J. Dillon, C. T. Vennard, A. Buckling and A. K. Charnley, Ecol. Lett., 2005, 8, 1291–1298 CrossRef PubMed.
- T. C. Olofsson, E. Butler, P. Markowicz, C. Lindholm, L. Larsson and A. Vásquez, Int. Wound J., 2014 DOI:10.1111/iwj.12345.
- E. Forsgren, T. C. Olofsson, A. Vásquez and I. Fries, Apidologie, 2010, 41, 99–108 CrossRef.
- A. Vásquez, E. Forsgren, I. Fries, R. J. Paxton, E. Flaberg, L. Szekely and T. C. Olofsson, PLoS One, 2012, 7, e33188 Search PubMed.
- H. Koch and P. Schmid-Hempel, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 19288–19292 CrossRef CAS PubMed.
- D. P. Cariveau, J. Elijah Powell, H. Koch, R. Winfree and N. A. Moran, ISME J., 2014, 8, 2369–2379 CrossRef CAS PubMed.
- K. Lam, K. Thu, M. Tsang, M. Moore and G. Gries, Naturwissenschaften, 2009, 96, 1127–1132 CrossRef CAS PubMed.
- L. M. Hedges, J. C. Brownlie, S. L. O'Neill and K. N. Johnson, Science, 2008, 332, 702 CrossRef PubMed.
- L. Teixeira, Á. Ferreira and M. Ashburner, PLoS Biol., 2008, 6, e1000002 CrossRef PubMed.
- P. T. Hamilton and S. J. Perlman, PLoS Pathog., 2013, 9, e1003808 Search PubMed.
- R. L. Glaser and M. A. Meola, PLoS One, 2010, 5, e11977 Search PubMed.
- L. A. Moreira, I. Iturbe-Ormaetxe, J. A. Jeffery, G. Lu, A. T. Pyke, L. M. Hedges, B. C. Rocha, S. Hall-Mendelin, A. Day, M. Riegler, L. E. Hugo, K. N. Johnson, B. H. Kay, E. A. McGraw, A. F. van den Hurk, P. A. Ryan and S. L. O'Neill, Cell, 2009, 139, 1268–1278 CrossRef PubMed.
- E. Rances, H. Y. Yixin, M. Woolfit and E. A. McGraw, PLoS Pathog., 2012, 8, e1002548 CAS.
- S. C. Carreiro, F. C. Pagnocca, M. Bacci, O. C. Bueno, M. J. A. Hebling and W. J. Middelhoven, Folia Microbiol., 2002, 47, 259–262 CrossRef CAS.
- A. Rodrigues, R. Cable, U. Mueller, M. Bacci Jr and F. Pagnocca, Antonie van Leeuwenhoek, 2009, 96, 331–342 CrossRef PubMed.
- T. S. Davis, R. W. Hofstetter, J. T. Foster, N. E. Foote and P. Keim, Microb. Ecol., 2011, 61, 626–634 CrossRef PubMed.
- T. Nakashima, T. Iizuka, K. Ogura, M. Maeda and T. Tanaka, J. Fac. Agric., Hokkaido Univ., 1982, 61, 60–72 CAS.
- A. Hervey and M. S. R. Nair, Mycologia, 1979, 71, 1064–1066 CrossRef CAS.
- M. S. R. Nair and A. Hervey, Phytochemistry, 1979, 18, 326–327 CrossRef CAS.
- Y. Wang, U. Mueller and J. Clardy, J. Chem. Ecol., 1999, 25, 935–941 CrossRef CAS.
- S. A. Van Bael, H. Fernandez-Marin, M. C. Valencia, E. I. Rojas, W. T. Wcislo and E. A. Herre, Proc. R. Soc. B, 2009, 276, 2419–2426 CrossRef PubMed.
- S. A. O. Armitage, H. Fernandez-Marin, W. T. Wcislo and J. J. Boomsma, Evolution, 2012, 66, 1966–1975 CrossRef PubMed.
- U. G. Mueller, A. Ortiz and M. Bacci Jr, Insectes Soc., 2010, 57, 209–215 CrossRef.
- D. K. Aanen, H. H. D. Licht, A. J. M. Debets, N. A. G. Kerstes, R. F. Hoekstra and J. J. Boomsma, Science, 2009, 326, 1103–1106 CrossRef CAS PubMed.
- L. Wang, Y. Feng, J. Tian, M. Xiang, J. Sun, J. Ding, W.-B. Yin, M. Stadler, Y. Che and X. Liu, ISME J., 2015 DOI:10.1038/ismej.2014.26.
- X. Li, G. Wheeler and J. Ding, Arthropod Plant Interact., 2012, vol. 6, pp. 417–424 Search PubMed.
- C. Kobayashi, Y. Fukasawa, D. Hirose and M. Kato, Evol. Ecol., 2008, 22, 711–722 Search PubMed.
- M. Gilliam, S. Taber III, B. J. Lorenz and D. B. Prest, J. Invertebr. Pathol., 1988, 52, 314–325 CrossRef.
- J. Frisvad, U. Thrane, R. Samson and J. Pitt, in Advances in Food Mycology, ed. A. D. Hocking, J. I. Pitt, R. Samson and U. Thrane, Springer, New York, NY, USA, 2006, pp. 3–31 Search PubMed.
- M. Rohlfs and L. Kurschner, J. Appl. Entomol., 2010, 134, 667–671 Search PubMed.
- P. Daszak, A. A. Cunningham and A. D. Hyatt, Divers. Distrib., 2003, 9, 141–150 CrossRef.
- J. M. Myers, J. P. Ramsey, A. L. Blackman, A. E. Nichols, K. P. C. Minbiole and R. N. Harris, J. Chem. Ecol., 2012, 38, 958–965 CrossRef PubMed.
- R. M. Brucker, R. N. Harris, C. R. Schwantes, T. N. Gallaher, D. C. Flaherty, B. A. Lam and K. P. C. Minbiole, J. Chem. Ecol., 2008, 34, 1422–1429 CrossRef CAS PubMed.
- P. J. Wiggins, J. M. Smith, R. N. Harris and K. P. C. Minbiole, J. Herpetol., 2011, 45, 329–332 CrossRef.
- M. R. Brucker, C. M. Baylor, R. L. Walters, A. Lauer, R. N. Harris and K. P. C. Minbiole, J. Chem. Ecol., 2008, 34, 39–43 CrossRef PubMed.
- M. H. Becker, R. M. Brucker, C. R. Schwantes, R. N. Harris and K. P. Minbiole, Appl. Environ. Microbiol., 2009, 75, 6635–6638 CrossRef CAS PubMed.
- B. A. Lam, J. B. Walke, V. T. Vredenburg and R. N. Harris, Biol Conservat, 2010, 143, 529–531 CrossRef PubMed.
- R. N. Harris, R. M. Brucker, J. B. Walke, M. H. Becker, C. R. Schwantes, D. C. Flaherty, B. A. Lam, D. C. Woodhams, C. J. Briggs, V. T. Vredenburg and K. P. C. Minbiole, ISME J., 2009, 3, 818–824 CrossRef CAS PubMed.
- A. H. Loudon, J. A. Holland, T. P. Umile, E. A. Burzynski, K. P. Minbiole and R. N. Harris, Front. Microbiol., 2014, 5, e00441 Search PubMed.
- M. C. Bastos, H. Ceotto, M. L. Coelho and J. S. Nascimento, Curr. Pharm. Biotechnol., 2009, 10, 38–61 CAS.
- R. L. Gallo and T. Nakatsuji, J. Invest. Dermatol., 2011, 131, 1974–1980 CrossRef CAS PubMed.
- H. Muellner, M. A. Folger, A. Werner, U. Gierlich, K. Eyer, K. Heinzmann, N. M. Shaw and F. Wyer, Germany Pat., WO/2007/093548, 2008.
- A. L. Cogen, K. Yamasaki, K. M. Sanchez, R. A. Dorschner, Y. Lai, D. T. MacLeod, J. W. Torpey, M. Otto, V. Nizet, J. E. Kim and R. L. Gallo, J. Invest. Dermatol., 2010, 130, 192–200 CrossRef CAS PubMed.
- A. L. Cogen, K. Yamasaki, J. Muto, K. M. Sanchez, L. C. Alexander, J. Tanios, Y. Lai, J. E. Kim, V. Nizet and R. L. Gallo, PLoS One, 2010, 5, e8557 Search PubMed.
- T. Iwase, Y. Uehara, H. Shinji, A. Tajima, H. Seo, K. Takada, T. Agata and Y. Mizunoe, Nature, 2010, 465, 346–349 CrossRef CAS PubMed.
- M. Otto, R. Süssmuth, C. Vuong, G. Jung and F. Götz, FEBS Lett., 1999, 450, 257–262 CrossRef CAS.
- J. H. Daskin and R. A. Alford, Proc. R. Soc. B, 2012, 279, 1457–1465 CrossRef PubMed.
- H. M. Wexler, Clin. Microbiol. Rev., 2007, 20, 593–621 CrossRef CAS PubMed.
- S. K. Mazmanian, J. L. Round and D. L. Kasper, Nature, 2008, 453, 620–625 CrossRef CAS PubMed.
- S. Yurist-Doutsch, M.-C. C. Arrieta, S. L. Vogt and B. B. Finlay, Annu. Rev. Genet., 2014, 48, 361–382 CrossRef CAS PubMed.
- L. Rizzetto, C. De Filippo and D. Cavalieri, Eur. J. Immunol., 2014, 44, 3166–3181 CrossRef CAS PubMed.
- J. C. Clemente, L. K. Ursell, L. Wegener Parfrey and R. Knight, Cell, 2012, 148, 1258–1270 CrossRef CAS PubMed.
- F. J. Gatesoupe, J. Mol. Microbiol. Biotechnol., 2008, 14, 107–114 CrossRef CAS PubMed.
- S. Ghosh, E. Ringø, A. D. G. Selvam, M. Rahinam, N. Sathyan, N. John and A. A. M. Hatha, Int. J. Fish. Aquacult., 2014, 4, 1–11 Search PubMed.
- E. Ringø, U. Schillinger and W. Holzapfel, in Biology of Growing Animals, ed. W. H. Holzapfel and P. J. Naughton, Elsevier, Edinburgh, UK, 2005, pp. 416–453 Search PubMed.
- E. Ringø, L. Løvmo, M. Kristiansen and Y. Bakken, Aquacult. Res., 2010, 41, 451–467 CrossRef PubMed.
- J. J. Soler, M. Martín-Vivaldi, M. Ruiz-Rodriguez, E. Valdivia, A. M. Martín-Platero, M. Martínez-Bueno, J. M. Peralta-Sanchez and M. Méndez, Funct. Ecol., 2008, 22, 864–871 CrossRef PubMed.
- M. Martín-Vivaldi, J. J. Soler, J. M. Peralta-Sánchez, L. Arco, A. M. Martín-Platero, M. Martínez-Bueno, M. Ruiz-Rodríguez and E. Valdivia, J. Anim. Ecol., 2014, 83, 1289–1301 CrossRef PubMed.
- M. Ruiz-Rodriguez, E. Valdivia, J. J. Soler, M. Martín-Vivaldi, A. M. Martín-Platero and M. Martínez-Bueno, J. Exp. Biol., 2009, 212, 3621–3626 CrossRef CAS PubMed.
- A. M. Martín-Platero, E. Valdivia, M. Ruiz-Rodriguez, J. J. Soler, M. Martin-Vivaldi, M. Maqueda and M. Martínez-Bueno, Appl. Environ. Microbiol., 2006, 72, 4245–4249 CrossRef PubMed.
- M. Ruiz-Rodriguez, M. Martínez-Bueno, M. Martín-Vivaldi, E. Valdivia and J. J. Soler, FEMS Microbiol. Ecol., 2013, 85, 495–502 CrossRef CAS PubMed.
- M. Martin-Vivaldi, A. Pena, J. M. Peralta-Sanchez, L. Sanchez, S. Ananou, M. Ruiz-Rodriguez and J. J. Soler, Proc. R. Soc. B, 2010, 277, 123–130 CrossRef CAS PubMed.
- T. Nosenko, F. Schreiber, M. Adamska, M. Adamski, M. Eitel, J. Hammel, M. Maldonado, W. E. Müller, M. Nickel, B. Schierwater, J. Vacelet, M. Wiens and G. Wörheide, Mol. Phylogenet. Evol., 2013, 67, 223–233 CrossRef PubMed.
- W. Wheeler, Cladistics, 2001, 17, 113–169 CrossRef PubMed.
- G. D. Edgecombe, G. Giribet, C. W. Dunn, A. Hejnol, R. M. Kristensen, R. C. Neves, G. W. Rouse, K. Worsaae and M. V. Sørensen, Org. Divers. Evol., 2011, 11, 151–172 CrossRef PubMed.
- G. Giribet and G. D. Edgecombe, Annu. Rev. Entomol., 2012, 57, 167–186 CrossRef CAS PubMed.
- S. Cremer, S. A. O. Armitage and P. Schmid-Hempel, Curr. Biol., 2007, 17, R693–R702 CrossRef CAS PubMed.
- W. D. Hamilton, in Animal societies: Theories and Facts, ed. Y. Ito, J. L. Brown and J. Kikkawa, Japan Scientific Society Press, Tokyo, Japan, 1987, pp. 81–102 Search PubMed.
- M. P. Lombardo, Behav. Ecol. Sociobiol., 2007, 62, 479–497 CrossRef.
- S. E. Marsh, M. Poulsen, A. Pinto-Tomás and C. R. Currie, PLoS One, 2014, 9, e103269 Search PubMed.
- V. G. Martinson, J. Moy and N. A. Moran, Appl. Environ. Microbiol., 2012, 78, 2830–2840 CrossRef CAS PubMed.
- F. Romero, F. Espliego, J. P. Baz, T. G. DeQuesada, D. Gravalos, F. DelaCalle and J. L. FernadezPuertes, J. Antibiot., 1997, 50, 734–737 CrossRef CAS.
- R. Sakai, K. Yoshida, A. Kimura, K. Koike, M. Jimbo, K. Koike, A. Kobiyama and H. Kamiya, ChemBioChem, 2008, 9, 543–551 CrossRef CAS PubMed.
- C. Vorburger and A. Gouskov, J. Evol. Biol., 2011, 24, 1611–1617 CrossRef CAS PubMed.
- N. A. Moran, P. Tran and N. M. Gerardo, Appl. Environ. Microbiol., 2005, 71, 8802–8810 CrossRef CAS PubMed.
- H. Salem, L. Flórez, N. M. Gerardo and M. Kaltenpoth, Proc. R. Soc. B, 2015, 282 DOI:10.1098/rspb.2014.2957.
- M. J. Cafaro and C. R. Currie, Can. J. Microbiol., 2005, 51, 441–446 CrossRef CAS PubMed.
- L. M. Henry, J. Peccoud, J.-C. C. Simon, J. D. Hadfield, M. J. Maiden, J. Ferrari and H. C. Godfray, Curr. Biol., 2013, 23, 1713–1717 CrossRef CAS PubMed.
- P. H. W. Biedermann and M. Kaltenpoth, J. Chem. Ecol., 2014, 40, 99 CrossRef CAS PubMed.
- J. Piel, D. Butzke, N. Fusetani, D. Q. Hui, M. Platzer, G. P. Wen and S. Matsunaga, J. Nat. Prod., 2005, 68, 472–479 CrossRef CAS PubMed.
- R. J. Worthington and C. Melander, Trends Biotechnol., 2013, 31, 177–184 CrossRef CAS PubMed.
- A. Svatoš, Trends Biotechnol., 2010, 28, 425–434 CrossRef PubMed.
- A. Svatoš, Anal. Chem., 2011, 83, 5037–5044 CrossRef PubMed.
- C. M. Gibson and M. S. Hunter, Ecol. Lett., 2010, 13, 223–234 CrossRef PubMed.
- W. B. Turner and D. C. Aldridge, Fungal metabolites, Academic Press, London, UK, 1971 Search PubMed.
- P. Steyn, The Biosynthesis of Mycotoxins: A study in secondary Metabolism, Elsevier Science, New York, NY, USA, 2012 Search PubMed.
- N. E. Beckage and J.-M. Drezen, Parasitoid viruses: symbionts and pathogens, Academic Press, San Diego, CA, USA, 2012 Search PubMed.
- H. Thoetkiattikul, M. H. Beck and M. R. Strand, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 11426–11431 CrossRef CAS PubMed.
- N. M. Dheilly, F. Maure, M. Ravallec, R. Galinier, J. Doyon, D. Duval, L. Leger, A.-N. Volkoff, D. Misse, S. Nidelet, V. Demolombe, J. Brodeur, B. Gourbal, F. Thomas and G. Mitta, Proc. R. Soc. B, 2015, 282, 20142773 CrossRef PubMed.
- J. J. Barr, R. Auro, M. Furlan, K. L. Whiteson, M. L. Erb, J. Pogliano, A. Stotland, R. Wolkowicz, A. S. Cutting, K. S. Doran, P. Salamon, M. Youle and F. Rohwer, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 10771–10776 CrossRef CAS PubMed.
- E. W. Schmidt, Nat. Chem. Biol., 2008, 4, 466–473 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5np00010f |
| ‡ All authors contributed equally to this work. |
| § Present address: Johannes Gutenberg University Mainz, Institute for Zoology, Department for Evolutionary Ecology, Johann-Joachim-Becher-Weg 13, 55128 Mainz, Germany. Email: mkaltenp@uni-mainz.de |
| This journal is © The Royal Society of Chemistry 2015 |