Open Access Article
Open Access ArticleMetal binding spectrum and model structure of the Bacillus anthracis virulence determinant MntA†
Elena
Vigonsky‡
a,
Inbar
Fish‡
b,
Nurit
Livnat-Levanon
a,
Elena
Ovcharenko
a,
Nir
Ben-Tal
b and
Oded
Lewinson
*a
aDepartment of Biochemistry, The Bruce and Ruth Rappaport Faculty of Medicine Technion-Israel Institute of Technology, Haifa, Israel. E-mail: lewinson@tx.technion.ac.il; Fax: +972-4-8295205; Tel: +972-4-8295428
bDepartment of Biochemistry and Molecular Biology, Tel-Aviv University, Tel-Aviv, Israel
First published on 16th June 2015
Abstract
The potentially lethal human pathogen Bacillus anthracis expresses a putative metal import system, MntBCA, which belongs to the large family of ABC transporters. MntBCA is essential for virulence of Bacillus anthracis: deletion of MntA, the system's substrate binding protein, yields a completely non-virulent strain. Here we determined the metal binding spectrum of MntA. In contrast to what can be inferred from growth complementation studies we find no evidence that MntA binds Fe2+ or Fe3+. Rather, MntA binds a variety of other metal ions, including Mn2+, Zn2+, Cd2+, Co2+, and Ni2+ with affinities ranging from 10−6 to 10−8 M. Binding of Zn2+ and Co2+ have a pronounced thermo-stabilizing effect on MntA, with Mn2+ having a milder effect. The thermodynamic stability of MntA, competition experiments, and metal binding and release experiments all suggest that Mn2+ is the metal that is likely transported by MntBCA and is therefore the limiting factor for virulence of Bacillus anthracis. A homology-model of MntA shows a single, highly conserved metal binding site, with four residues that participate in metal coordination: two histidines, a glutamate, and an aspartate. The metals bind to this site in a mutually exclusive manner, yet surprisingly, mutational analysis shows that for proper coordination each metal requires a different subset of these four residues. ConSurf evolutionary analysis and structural comparison of MntA and its homologues suggest that substrate binding proteins (SBPs) of metal ions use a pair of highly conserved prolines to interact with their cognate ABC transporters. This proline pair is found exclusively in ABC import systems of metal ions.
Introduction
Trace metals such as Cu1/2+, Zn2+, Mn2+, and Fe2/3+ are essential cofactors to the structure and function of many enzymes and structural proteins, and it is estimated that metalloproteins comprise 30–40% of most proteomes.1,2 However, since metal ions are toxic at high concentrations, all organisms must maintain a delicate balance between essential uptake and toxic overload.3 This necessity places trace metals at the crossroads of host–pathogen interactions: to fight infections, hosts employ both metal sequestering/depravation and bombardment.4–6 In turn, to maintain and control their intracellular metal quote, bacterial pathogens evolved elaborate uptake and extrusion systems. It is therefore not surprising that bacterial genes involved in metal homeostasis have been broadly demonstrated to be key virulence determinants.7–10Bacterial pathogens primarily rely on their ATP Binding Cassette (ABC) transporters for the high-affinity acquisition of essential metals. This dependence is mirrored in the pivotal role of ABC transporters in bacterial virulence and pathogenesis. In many important human pathogens such as Mycobacterium tuberculosis,11Staphylococcus aureus,12Streptococcus pneumoniae,13–15Salmonella typhi,16,17Streptococcus mutans,18 and Yersinia pestis19 ABC importers of Zn2+, Mn2+, and Fe2/3+ are essential to bacterial virulence and pathogenesis.
ABC transporters comprise a very large super-family of proteins.20,21 They couple the energy of ATP hydrolysis to the energetically uphill translocation of molecules across biological membranes. These ubiquitous enzymes are present in all extant phyla with 79 genes in E. coli,22 ∼120 in higher plants,23 and 48 in human.24 The transported molecules are extremely diverse, ranging from small molecules (such as metal ions, sugars, amino acids) to large and bulky compounds (peptides, proteins, organo-metal complexes, and antibiotics).
ABC transporters consist minimally of four domains: two transmembrane domains (TMDs) and two cytoplasmic nucleotide-binding domains (NBDs).25 Binding and hydrolysis of ATP at the NBDs power the translocation of substrates through the permeation pathway formed by the TMDs. A given ABC transporter may function either as an exporter or an importer, with importers present only in prokaryotes. ABC transporters that function as importers depend on a high-affinity substrate binding protein (SBP) that delivers the substrate to the cognate transporter.26–28
The mntBCA operon of the potentially lethal human pathogen Bacillus anthracis encodes an ABC import system, putatively of manganese and/or iron. MntA (hereafter baMntA), the system's high-affinity substrate binding protein (SBP) is one of this pathogen's most important virulence determinants: its genetic deletion yields a completely non-virulent strain, less virulent than the Sterne strain used for vaccination.29 Many other SBPs of ABC importers of iron, zinc, and manganese play significant roles in bacterial virulence,8,11–13,15–19,30,31 but the effect of the baMntA deletion is the most dramatic, leading to a 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000-fold increase in the LD50 of Bacillus anthracis. Remarkably, beyond its crucial role as a virulence determinant, very little is known of the structure and function of baMntA.
000-fold increase in the LD50 of Bacillus anthracis. Remarkably, beyond its crucial role as a virulence determinant, very little is known of the structure and function of baMntA.
Here, we present the first structure–function analysis of baMntA. We determined its metal binding specificity and affinities, identified residues that participate in metal coordination, and describe a novel structural element that is likely required for the interaction of baMntA with its cognate ABC transporter.
Results
Overexpression and purification of baMntA
baMntA was cloned without its first 21 amino acids that code for the protein's membrane anchoring domain. To not interfere with subsequent metal binding measurements the protein was expressed without an affinity tag. E. coli were transformed with plasmid p-mntA and extracts were prepared before or after addition of IPTG. A prominent band of ∼35 kDa (corresponding to the expected molecular weight of baMntA) was observed in SDS-PAGE of extracts prepared from induced cells (ESI,† Fig. S1A). For purification, cell extracts were subjected to ion exchange chromatography and gel filtration. This procedure yielded a preparation that was 80–90% pure (ESI,† Fig. S1B). The protein could be readily concentrated to 30 mg mL−1 (a process that improved its purity, ESI,† Fig. S1B) and was highly mono-disperse as judged by size exclusion chromatography (ESI,† Fig. S1C). Comparison of the elution volume of baMntA to the elution volumes (using the same column) of known molecular weight standards suggests that baMntA is a monomer (ESI,† Fig. S1D).Metal binding selectivity of baMntA
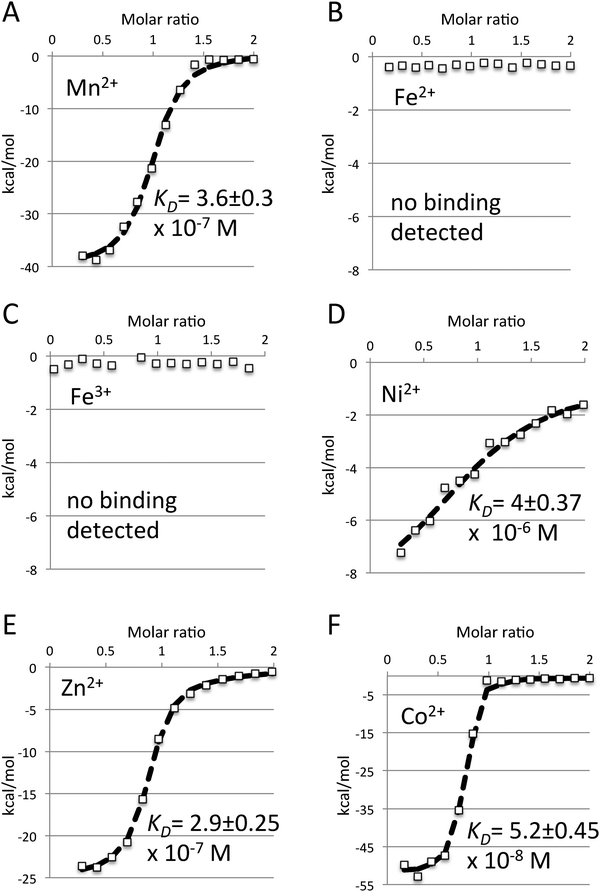
Growth complementation studies implied that baMntA is the substrate binding protein (SBP) of a manganese/iron ABC metal uptake system.29 We therefore began by characterizing the interaction of baMntA with Mn2+and Fe2/3+. Using Isothermal Titration Calorimetry (ITC) we readily detected a high affinity (KD = 3.6 × 10−7 M) binding of Mn2+ by baMntA. The reaction is exothermic, and the data could be readily fitted using a simple 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 interaction model (Fig. 1A, see Methods for model fitting). In contrast to the conclusions that may be drawn from the previously reported growth studies,29 we could not detect binding of iron in either of its oxidation states (Fe2/3+, Fig. 1B and C). Binding of iron was also not observed in the presence of reducing agent 2-mercaptoethanol. We next tested baMntA for its ability to bind a variety of other metals. We observed that in addition to Mn2+ baMntA also binds Cd2+, Zn2+, Co2+, and Ni2+. The metal binding affinities of baMntA ranged over nearly two orders of magnitude (Fig. 1D–F, ESI,† Table S1). The highest affinity was to Co2+ (KD = 5.2 ×10−8 M) and the lowest to Ni2+ (KD = 4 × 10−6 M). The binding affinities of baMntA to Cd2+ and Zn2+ were of intermediate values at 5.8 × 10−7 M and 2.9 × 10−7 M, respectively. In addition to Fe2+/3+, we also did not detect binding of Mg2+ or Cu2+ (ESI,† Table S1). Rukhman et al. demonstrated that binding of Mn2+ by MntC of the cyanobacterium Synechocystis PCC is redox dependent, and is negatively affected by reducing agents.32 This effect was attributed to the reduction of a disulfide bond that is unique to cyanobacterial SBPs of manganese. The two cysteines that form this bond are missing in baMntA and indeed binding of Mn2+ was unaffected by reducing agents (not shown).
1 interaction model (Fig. 1A, see Methods for model fitting). In contrast to the conclusions that may be drawn from the previously reported growth studies,29 we could not detect binding of iron in either of its oxidation states (Fe2/3+, Fig. 1B and C). Binding of iron was also not observed in the presence of reducing agent 2-mercaptoethanol. We next tested baMntA for its ability to bind a variety of other metals. We observed that in addition to Mn2+ baMntA also binds Cd2+, Zn2+, Co2+, and Ni2+. The metal binding affinities of baMntA ranged over nearly two orders of magnitude (Fig. 1D–F, ESI,† Table S1). The highest affinity was to Co2+ (KD = 5.2 ×10−8 M) and the lowest to Ni2+ (KD = 4 × 10−6 M). The binding affinities of baMntA to Cd2+ and Zn2+ were of intermediate values at 5.8 × 10−7 M and 2.9 × 10−7 M, respectively. In addition to Fe2+/3+, we also did not detect binding of Mg2+ or Cu2+ (ESI,† Table S1). Rukhman et al. demonstrated that binding of Mn2+ by MntC of the cyanobacterium Synechocystis PCC is redox dependent, and is negatively affected by reducing agents.32 This effect was attributed to the reduction of a disulfide bond that is unique to cyanobacterial SBPs of manganese. The two cysteines that form this bond are missing in baMntA and indeed binding of Mn2+ was unaffected by reducing agents (not shown).
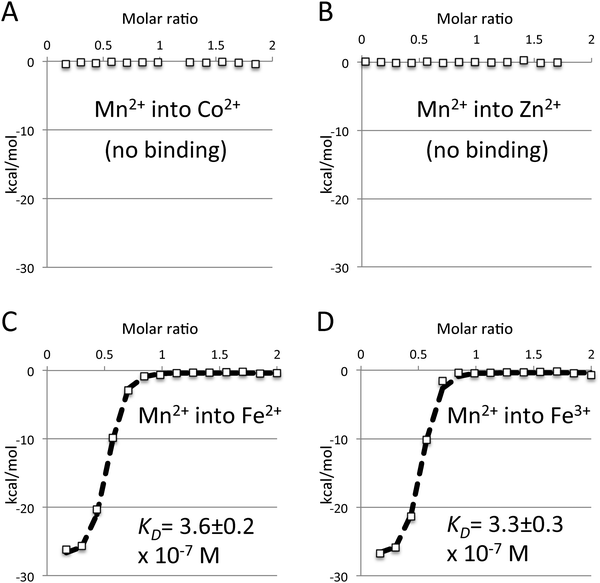
Iron does not compete with manganese for binding to baMntA
The contradiction between the previously performed growth studies29 and our inability to detect binding of either Fe2+ or Fe3+ prompted us to further investigate this issue. Lack of detection of iron binding may stem from limitations of the Isothermal Titration Calorimetry (ITC) approach. ITC measures heat released or absorbed during a reaction (enthalpy). It is therefore possible that if an association is driven mainly (or exclusively) by entropy it will not be detected by ITC. This is highly unlikely in the case of metal binding by baMntA, as metal binding by SBPs is strongly driven by enthalpy.33–35 This is also specifically the case with baMntA, as binding of Mn2+, Cd2+, Zn2+, Co2+, and Ni2+ all have a strong enthalpic signature. To experimentally test whether iron binding occurs but is undetected in the ITC experiments we conducted binding competition experiments. Metal SBPs of ABC transporters such as baMntA have a single metal binding site (see also later) that binds a single metal ion at a time. Indeed, pre-incubation of baMntA with a molar excess of either Zn2+ or Co2+ completely abolished binding of Mn2+ (Fig. 2A and B), and identical results were obtained when Co2+ was injected into baMntA pre-incubated with Zn2+. In contrast, pre-incubation of baMntA with a 10-fold molar excess of either Fe2+ or Fe3+ had no effect on Mn2+ binding, and the affinity of Mn2+ binding remained unchanged (Fig. 2C and D). Inhibition of Mn2+ binding by iron was also not observed in the presence of reducing agent 2-mercaptoethanol. Taken together, these results show that the metals that bind to baMntA do so in a mutually exclusive manner and that baMntA does not bind Fe2+ or Fe3+.A 3-D structural model of baMntA
To date, our efforts to determine the structure of baMntA by X-ray crystallography have failed. We therefore used comparative (homology) modeling to predict the structure of baMntA. A BLAST search of the protein data bank (PDB) identified MtsA (PDB ID 3HH8) from Streptococcus pyogenes36 as the most suitable template. The high sequence similarity between baMntA and spMtsA (53% identity, 90% similarity) and the similar functions30 are indicative of the appropriateness of the template.Three modeling tools were used to build a homology model of baMntA: MODELLER (version number 9.8),37 I-TASSER,38 and SWISS-MODEL.39 Practically identical results were obtained using the three different algorithms (Fig. 3A), with RMSDs of 0.85–1.02 Å (backbone atoms) and 1.5–1.7 Å (whole atom comparison) between the three models. The model that was constructed with MODELLER was evaluated using three quality control programs. The stereo-chemical quality of the model was evaluated with PROCHECK,40 the compatibility of the atomic model (3D) with its own amino acid sequence (1D) was evaluated with Verify3D,41 and the statistical (pseudo) energy potential of the model was computed with ProSA.42 Highly similar Verify3D scores were obtained for the template and the model: 95.71% of the residues of the template had an average 3D-to-1D score >0.2, whereas 96.79% of the residues in the model had an average score >0.2. Ramachandran plot analysis shows that most of the residues are in allowed areas of the plot (ESI,† Fig. S2A), and the ProSA score of the model falls well within the allowed area and is highly similar to that of template (ESI,† Fig. S2B).
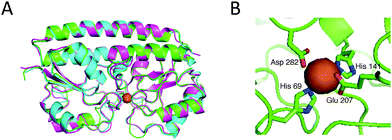 | ||
| Fig. 3 Model structure of baMntA. (A) Structural alignment of the baMntA models created with MODELLER (green), SWISS-MODEL (cyan), or I-TASSER (magenta). MtsA (PDB ID 3HH8) was specified as a template for MODELLER. No template was specified for either SWISS-MODEL or I-TASSER. The protein is shown as a cartoon representation and the metal (here Zn2+) as an orange sphere. (B) The proposed metal binding site of MntA (using MODELLER). His69, His141, Glu207, and Asp282 are intimately positioned to interact with the metal (Zn2+, orange sphere). | ||
It is widely observed that the interior of proteins is much more evolutionarily conserved than their periphery.43 We used the ConSurf webserver (http://consurf.tau.ac.il/) to validate that the model adheres to this anticipation (wrong models often do not43). As shown in ESI,† Fig. S2C, the ConSurf conservation analysis reveals a highly conserved core and an overall variable surface. The highest degree of conservation surrounds a central cavity, which is characteristic of binding sites. An extremely similar distribution of conserved and variable residues is also observed in the experimentally determined structures of the closely related SBPs MntC, PsaA, and MtsA (see later in Discussion and Fig. 5). Not surprisingly, the overall predicted fold of baMntA is very similar to other metal ion SBPs.32,36,44
The model suggests the existence of a single metal binding site that is positioned in the interface between the N- and C-terminal domains of baMntA. Extensive loops surround the metal binding site and leave little unoccupied space. Four residues intimately interact with the metal: His69, His141, Glu207, and Asp282 (Fig. 3B). These four residues are invariably conserved in homologues of baMntA (ESI,† Fig. S3, and ref. 32, 36, 45 and 46). Three of these residues (His69, His141, Asp282) are conserved both in SBPs of Mn2+/Fe2+ and in SBPs of Zn2+. The fourth residue is glutamate in SBPs of Mn2+/Fe2+ or histidine in SBPs of Zn2+ (ESI,† Fig. S3). In this respect, it is noteworthy that although this residue in baMntA is a glutamate (Glu207) it binds Mn2+, Cd2+, Zn2+ and Co2+ with high affinity but does not bind Fe2+.
Role of conserved metal binding residues of baMntA
To test the involvement of the predicted residues in metal binding we generated single alanine substitutions. The mutant variants were overexpressed and purified as wild type baMntA and the Zn2+, Co2+, and Mn2+ binding affinities were measured in ITC experiments.Quite surprisingly, the most dramatic effect was of the Glu207 substitution. This mutant (Glu207A) completely lost the ability to bind any of the tested metals (Table 1). Mutating His69 had the mildest effect: the mutant could still bind all three metals, albeit with reduced affinities (∼10-fold reductions, Table 1). Substituting either His141 or D282 with alanine led to a selective loss of metal recognition: both mutants bound Zn2+ with near wild type affinities yet lost the ability to bind Co2+ or Mn2+. Taken together, the simplest interpretation of these results is that although the metals bind to the same binding site, each of them binds in a different manner, and is differentially coordinated by residues of baMntA (see also below). Relative to its homologues, the metal binding site of baMntA is highly intolerant to substitutions. In both MtsA of Streptococcus pyogenes36 and PsaA of Streptococcus pneumoniae46 much milder effects were observed for similar substitutions.
| K D (M) | |||
|---|---|---|---|
| CoSO4 | MnCl2 | ZnCl2 | |
| Wild type | 5.2 ± 0.45 × 10−8 | 3.6 ± 0.3 × 10−7 | 2.9 ± 0.25 × 10−7 |
| His69Ala | 4.7 ± 0.2 × 10−7 | 4.2 ± 0.12 × 10−6 | 3.6 ± 0.33 × 10−6 |
| His141Ala | No binding | No binding | 3.4 ± 0.37 × 10−7 |
| Glu207Ala | No binding | No binding | No binding |
| Asp282Ala | Affinity too low to be determined | No binding | 3.1 ± 0.28 × 10−7 |
Co2+, Zn2+, and Mn2+ bind differently to baMntA
The binding competition experiments shown in Fig. 2 show that metal binding to baMntA is mutually exclusive. In these experiments baMntA was first saturated with a molar excess of one metal and then injected with another. To further investigate the competition of metal binding we conducted experiments where we pre-incubated baMntA with one metal at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio and then gradually injected a second metal to concentrations that surpass those of the first one. As shown, when baMntA was incubated with Mn2+ at a 1
1 molar ratio and then gradually injected a second metal to concentrations that surpass those of the first one. As shown, when baMntA was incubated with Mn2+ at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
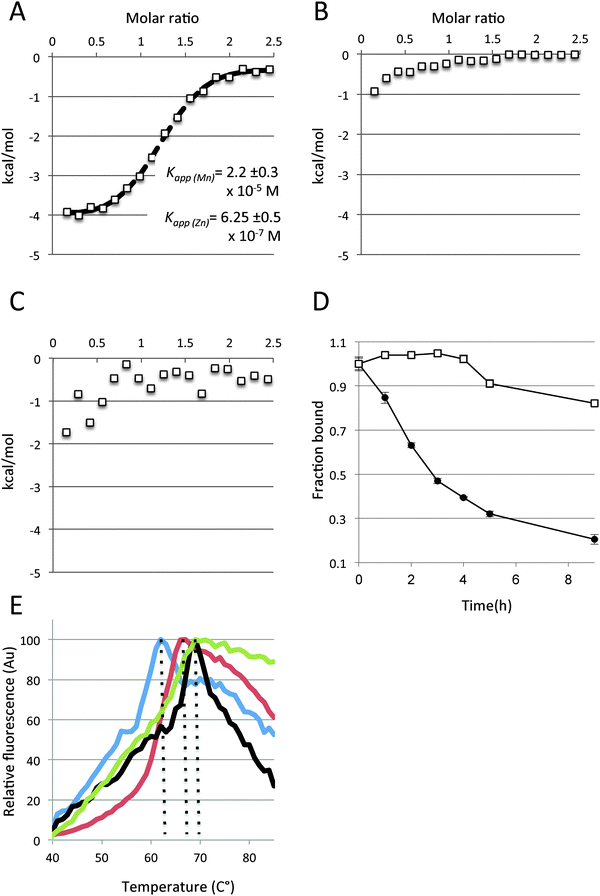
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio and then Zn2+ was gradually added, we still observed robust binding of Zn2+ (Fig. 4A). Analysis of the data using a simple competition model (see Methods for models) suggests that Zn2+ displaces pre-bound Mn2+ and that the Zn2+KD(apparent) only mildly increases (∼2-fold) in the presence of 15 μM Mn2+ (concentration of Mn2+ used in this experiment). Interestingly, analyzing the same data to determine the Mn2+KD(apparent) suggests that Mn2+ is much more affected by Zn2+ than vice versa, its KD(apparent) increasing by ∼100-fold. This is surprising since individually both these metals bind to baMntA with very similar affinities. This asymmetry between Mn2+ and Zn2+ is further displayed by conducting the reverse experiment: titrating Mn2+ into baMntA that was pre-incubated with Zn2+ at a 1
1 molar ratio and then Zn2+ was gradually added, we still observed robust binding of Zn2+ (Fig. 4A). Analysis of the data using a simple competition model (see Methods for models) suggests that Zn2+ displaces pre-bound Mn2+ and that the Zn2+KD(apparent) only mildly increases (∼2-fold) in the presence of 15 μM Mn2+ (concentration of Mn2+ used in this experiment). Interestingly, analyzing the same data to determine the Mn2+KD(apparent) suggests that Mn2+ is much more affected by Zn2+ than vice versa, its KD(apparent) increasing by ∼100-fold. This is surprising since individually both these metals bind to baMntA with very similar affinities. This asymmetry between Mn2+ and Zn2+ is further displayed by conducting the reverse experiment: titrating Mn2+ into baMntA that was pre-incubated with Zn2+ at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio. As shown (Fig. 4B), under these conditions binding of Mn2+ was hardly observed, suggesting that once Zn2+ has bound it cannot be displaced by Mn2+. Like Mn2+, Co2+ hardly displaced pre-bound Zn2+, despite the fact that it binds to baMntA with higher affinity than Zn2+ (Fig. 4C). This suggests that Zn2+ and Mn2+, despite having similar binding affinities, bind differently to baMntA and that Mn2+ is released much more readily than Zn2+. The unfavorable release of Zn2+ was also observed in experiments where baMntA was loaded with either Zn2+ or Mn2+ and then dialyzed against an EDTA-containing buffer. At intervals we withdrew samples and used inductively coupled plasma phase mass spectroscopy (ICP-MS) to measure the fraction of metal that remain bound. As shown (Fig. 4D), the two metals were released from baMntA at very different rates, with Mn2+ dissociating much faster than Zn2+. Given that the two metals bind to baMntA with similar affinities (Fig. 1, ESI,† Table S1) this means that the association rate of Mn2+ to baMntA is much faster than that of Zn2+. Kinetic experiments demonstrated that binding of Mn2+ by PsaA is a fast first-order reaction, while that of Zn2+ is a slow second-order reaction.46 It seems that PsaA and baMntA that employ an identical set of metal-ligating residues share also this feature.
1 molar ratio. As shown (Fig. 4B), under these conditions binding of Mn2+ was hardly observed, suggesting that once Zn2+ has bound it cannot be displaced by Mn2+. Like Mn2+, Co2+ hardly displaced pre-bound Zn2+, despite the fact that it binds to baMntA with higher affinity than Zn2+ (Fig. 4C). This suggests that Zn2+ and Mn2+, despite having similar binding affinities, bind differently to baMntA and that Mn2+ is released much more readily than Zn2+. The unfavorable release of Zn2+ was also observed in experiments where baMntA was loaded with either Zn2+ or Mn2+ and then dialyzed against an EDTA-containing buffer. At intervals we withdrew samples and used inductively coupled plasma phase mass spectroscopy (ICP-MS) to measure the fraction of metal that remain bound. As shown (Fig. 4D), the two metals were released from baMntA at very different rates, with Mn2+ dissociating much faster than Zn2+. Given that the two metals bind to baMntA with similar affinities (Fig. 1, ESI,† Table S1) this means that the association rate of Mn2+ to baMntA is much faster than that of Zn2+. Kinetic experiments demonstrated that binding of Mn2+ by PsaA is a fast first-order reaction, while that of Zn2+ is a slow second-order reaction.46 It seems that PsaA and baMntA that employ an identical set of metal-ligating residues share also this feature.
The differential interaction between baMntA and the metals was also studied by examining the effects of metal binding on thermostability. For this we used a fluorescence-based assay (SYPRO-orange) that has been extensively used to measure protein unfolding in general and specifically of substrate binding proteins of ABC transporters.31,45,47–50 SYPRO-orange binds to hydrophobic protein patches that are gradually exposed during heat denaturation and peak fluorescence is observed when the protein is fully denatured. Subsequently, as the protein begins to aggregate less hydrophobic surfaces are available for binding and the SYPRO-orange fluorescence declines.
For these thermo stability assays we first dialyzed baMntA against a 1000-fold excess of buffer containing 20 mM EDTA to remove any metals that might have been co-purified with the protein. In the absence of exogenously added metals, SYPRO-orange binding to EDTA treated baMntA displayed peak fluorescence at 62 °C. The temperature of peak fluorescence was shifted to 66 °C in the presence of Mn2+ (Fig. 4E), and a larger shift (to 69 °C) was observed in the presence of either Zn2+ or Co2+. The irregularity of the slopes precluded the use of the first derivative to quantitate the mid-denaturation temperatures of apo and holo baMntA, as has been performed with PsaA from Streptococcus pneumoniae.31,45 Nevertheless, despite the fact that absolute fluorescence values were used (rather than first derivatives) the effects of the different metals on the thermostability of baMntA are very similar to their effects on PsaA.31,45 Taken together, the mutational analysis (Table 1) and the experiments shown in Fig. 4 suggest that although baMntA binds Mn2+, Zn2+, and Co2+ with similar affinities the geometry, chemistry, and stabilizing effects of the interactions are different.
A conserved protein:protein interaction surface in baMntA
Using the ConSurf algorithm we performed a structural conservation analysis of baMntA and compared it to that of three other SBPs of metal ions. A top view of baMntA reveals variability (not shown), as expected from a protein surface that is not engaged in functional interactions. In contrast, extensive patches of conserved structural elements are present at the bottom (cytoplasmic facing) of baMntA and its homologues (Fig. 5A). This conservation is partly due to the conserved metal binding site, but also to conserved elements that are positioned in the protein:protein interaction surface of the SBP with its transporter. Such extensive conservation on the face of the SBP that interacts with the transporter is also apparent in SBPs of Type-II ABC transporters such as BtuF, HmuT, and ShuT (Fig. 5B). However, it is largely absent in SBPs of Type-I ABC transporters such as MalE and HisJ (Fig. 5C). A pair of prolines stands out, (baMntA Pro44 and Pro204) and this proline pair is absolutely conserved in baMntA and its homologues, yet absent in SBPs of Type-I and Type-II transporters. The proline side chains point away from the SBPs and towards the transporter (Fig. 5D). Their location is reminiscent of that of the conserved glutamates of SBPs of Type-II ABC transporters (Fig. 5E), which are responsible for the high affinity interaction of the SBP with the transporter.35,51,52 The central role of prolines in mediating protein:protein interactions is well-documented.53 In this context, the orientation, location, and conservation of this pair of prolines suggests that they likely have a role in docking to the cognate ABC transporter(s).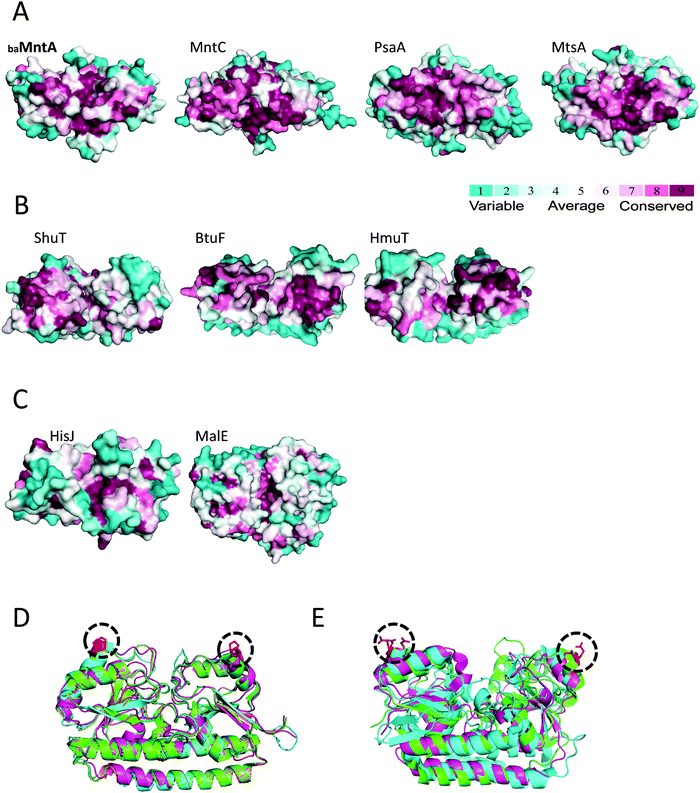 | ||
| Fig. 5 A conserved interaction surface in baMntA. (A) Surface representation (bottom view) of a ConSurf analysis of the model structure of baMntA, and the crystal structures of MntC (PDB ID 1XVL), PsaA (PDB ID 1PSZ), and MtsA (PDB ID 3HH8). The least conserved residues are colored cyan, the most conserved are in purple. (B) Same as in A, but shown are the SBPs of Type-II ABC transporters ShuT (PDB ID 2R7A), BtuF (PDB ID 1N4A), and HmuT (PDB ID 3MD9). (C) Same as in A, but shown are the SBPs of Type-I ABC transporters HisJ (PDB ID 1HSL), and MalE (PDB ID 1ANF). (D) A cartoon representation of the structural alignment of baMntA (orange), MntC (magenta), PsaA (cyan), MtsA (green). The conserved prolines are colored red and circled. (E) Same as in D, shown is the structural alignment of HmuT (green), BtuF (magenta), and MolA (cyan). The conserved glutamates are colored red and circled. | ||
Discussion
In recent years it is becoming increasingly clear that ABC transporters (importers) of d-block metals are crucial for bacterial virulence and pathogenesis.3,11–19,29,30 This is perhaps not surprising as these trace elements are essential for bacterial growth yet their availability is very scarce in the host.4 MntA of Bacillus anthracis presents an outstanding example of the great reliance of bacterial pathogens on their metal import systems. Deletion of mntA from the chromosome of Bacillus anthracis leads to complete arrest of Anthrax progression.29 In fact, the resulting strain is less virulent than the one that is currently used for vaccination. It is very rare to find a single gene (that does not code for a virulence factor) with such a great contribution to virulence. We thus deemed it important to learn more of this metal import system.baMntA is absolutely conserved (95–99% identity) in pathogenic strains of Bacillus anthracis and Bacillus cereus, yet a close homologue is absent from the non-pathogenic Bacillus subtilis. It is also absent from the chromosome of non-pathogenic E. coli strains, but present on a pathogenicity plasmid of the avian pathogenic E. coli (APEC). Closely related homologues (more than 50% identity) are also found in the pathogens Enterococcus faecalis, Enterococcus faecium, and Listeria monocytogenes. It thus seems that mntA homologues have been preserved in pathogenic bacteria.
Three metals are most commonly found at the crossroads of host–pathogen interactions: iron, zinc, and manganese. Iron and zinc are essential for countless bacterial physiological processes, including respiration, replication, translation, signaling, and cell division. Manganese is needed mainly for protection against oxidative stress, as a prosthetic group for superoxide dismutase (SOD) or directly conferring protection from oxidants. Oxidative stress is especially relevant for intracellular pathogens such as Bacillus anthracis as reactive oxygen species (ROS) are broadly used by the host as a defense mechanism. Additionally, the mammalian NRAMP transporters function to deprive pathogens of manganese by exporting this metal from the phagosomal lumen.54 This may be especially relevant for Anthrax progression as spores of Bacillus anthracis are taken up by macrophages and the phagosome is the site of exit from spore state and initial germination.
One of the main goals of this work was to assess which metal is likely transported by MntBCA and thus presents the limiting factor for virulence of Bacillus anthracis. Previous studies showed that Bacillus anthracis ΔmntA fails to grow in defined media unless supplemented with either Mn2+ or Fe2+. The complementation with Mn2+ was 5–10 times more efficient leading the authors to suggest that Mn2+ is the preferred substrate and that Fe2+ is transported with lower affinity.29 We found no evidence for binding of Fe2+ or Fe3+ by baMntA. Moreover, high concentrations of Fe2+ or Fe3+ did not inhibit binding of Mn2+. We therefore conclude at this point that Fe2/3+ is most likely not transported by MntBCA. The genome of Bacillus anthracis encodes 7–10 ABC import systems for iron, heme, iron complexes, and iron-siderophores. This redundancy highlights the central role of iron in Bacillus anthracis metabolism and ensures iron availability in changing environments. The large repertoire of iron import systems, the dramatic effect of the mntA deletion, and our inability to detect iron binding all suggest that the ΔmntA strain did not turn a-virulent because it cannot acquire iron.
baMntA binds both Mn2+ and Zn2+ with similarly high affinities and therefore both metals are possible transport substrates and potential contributors to virulence. Despite their similar binding affinities their binding by baMntA is not equivalent. Once Zn2+ has bound it cannot be replaced by Mn2+, while the reciprocal displacement readily occurs. In addition, Mn2+ associates and dissociates much more readily than Zn2+, and on the time-scale of transport reactions Zn2+ binding seems almost irreversible. Furthermore, Zn2+ has a greater thermo-stabilizing effect on the protein than does Mn2+. For a substrate to be efficiently transported (by any transport system) both its binding and release need to be reasonably fast. This does not seem to be the case for Zn2+binding by baMntA. Very similar observations have been made with PsaA from Streptococcus pneumoniae45,46 and SitA from Staphylococcus pseudintermedius.55 Like these authors we suggest that Mn2+ and not Zn2+ is the transported metal. If this is correct, the direct conclusion is that the high affinity acquisition of Mn2+ presents a major barrier to the development of Anthrax. However, this proposal is currently speculative until verified by direct transport assays.
Unfortunately, we have so far failed in conducting biochemical studies of the complete transport system MntBCA (transporter + SBP). In fact, despite their clear importance, none of these ABC metal transporters have ever been characterized. From a mechanistic perspective we know very little about ABC transporters (importers) of metal ions. ABC importers display a diversity of transport mechanisms and are roughly divided to two mechanistic groups: Type I importers (of sugars, amino acids, peptides) and Type II importers (of vitamin B12, heme, iron-siderophores).25,51,56 However, this picture appears incomplete as recent reports of the ribose importer suggest the existence of a different, novel mechanism of action.57 With respect to ABC importers of metal ions we have no knowledge if they operate by a mechanism that is similar to or different from those previously observed. Clues about their mechanism may be drawn from the structures of their substrate binding proteins (SBPs). SBPs of metal ions have the same fold as SBPs of vitamin B12, heme, or iron-siderophores, a fold that is very different from that of SBPs of sugars, amino acids, or peptides.3,34,58 The structures of the cognate ABC transporters are shaped according to the SBPs,25,59–61 and therefore one may infer that the structure of ABC transporters of metal ions is similar to that of Type II importers. This hypothesis is supported to an extent by the model structure of MntBC (Fig. 6A). We used 5 different programs to predict the structure of MntBC: MODELLER, SWISS-MODEL, HHPred, FFAS03, and I-TASSER. Practically identical structures were predicted by all 5 programs, and the whole atom RMSDs for the membrane domain was between 0.28 and 1.65 Å. In further support of the MntBC model, a ConSurf analysis shows the presence of a highly conserved membrane permeation pathway surrounded by variable residues that face the membrane (Fig. 6B).
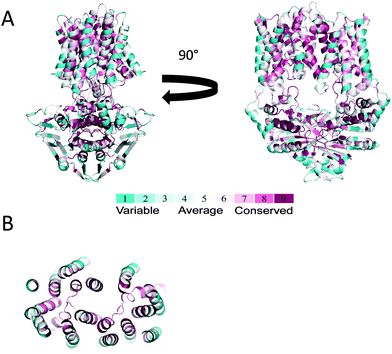 | ||
| Fig. 6 Model structure of baMntBC. (A) The transporter complex, consisting of the nucleotide-binding domain (MntB) and the transmembrane domain (MntC) was modeled with MODELLER using the structure of MolBC (PDB ID 2NQ2) as a template. Left and right panels are side views rotated by 90°. The protein backbone is shown as a cartoon representation colored according to the ConSurf scale (most variable residues in cyan, most conserved in purple). (B) A top view of the trans-membrane domain formed by the MntC dimer. Shown is a slab view at approximately the middle of the membrane plane, with MntB (the nucleotide binding domain) removed for clarity. | ||
The model predicts the presence of 8 transmembrane helices per MntC monomer (16 helices in the assembled transporter), and the same result was obtained by using the TMHMM prediction tool.62 The presence of 8 transmembrane helices per monomer is intermediate between 10 transmembrane helices of Type-II transporters such as BtuCD63 and 5–6 transmembrane helices in Type-I transporters such as MetNI.64 With respect to the number of amino acids that compose the TMDs, ABC transporters of metal ions are right between the ‘big’ Type-II transporters and the ‘small’ Type-I transporters: e.g., BtuC (326 amino acids), MntC (288 amino acids), and MetI (217 amino acids).
A structural element that is plainly missing from ABC transporters of metal ions is the “knobs and pockets” feature of Type II importers. Invariant glutamates (knobs) on the SBPs of Type II systems snugly fit into an arginine pocket of the transporter.52,60 This interaction is responsible for the high affinity complexes formed between Type II importers and their cognate SBPs.35,51 The high affinity and stability of the transporter–SBP complex is the mechanistic hallmark of Type II importers, and the stability of this complex dictates the transport mechanism. Type I importers completely lack theses “knobs and pockets” and indeed do not form high affinity transporter–SBP complexes, and thus operate by a very distinct mechanism.35,65–70 In this context, ABC transporters of metal ions may present a middle ground: they lack the glutamates–arginines interaction but instead have a pair of prolines that is perfectly positioned to mediate protein:protein interactions. Clearly, mechanistic studies of this important yet uncharacterized group of ABC transporters are much needed.
The results we report here, and the identification of mutants of baMntA with a selective (or full) loss of metal binding provide an important stepping stone for future studies of Bacillus anthracis virulence and Anthrax progression in animal models.
Materials and methods
MntA cloning, expression and purification
The mntA gene from Bacillus anthracis was cloned into pET-19b (Novagen) vector for expression without its N-terminal lipid-anchoring domain (first 21 amino acids). baMntA was expressed in E.coli BL21-Gold(DE3) cells (Stratagene) using either M9 minimal media or Terrific Broth, as indicated in the text. Protein expression was induced at mid log phase by addition of 1 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG) at 37 °C for 2 h. Cells were harvested by centrifugation (13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 600 × g, 20 min, 4 °C) and the pellet was stored at −80 °C until use.
600 × g, 20 min, 4 °C) and the pellet was stored at −80 °C until use.
For baMntA purification, cells were homogenized in 20 mM Tris-HCl pH 8, 10 mM NaCl, complete EDTA-free protease inhibitor (Roche), 30 μg mL−1 DNase (Worthington), 1 mM CaCl2 and 1 mM MgCl2 and ruptured by three passages in an EmulsiFlex-C3 homogenizer (Avestin). The lysate was ultracentrifuged at 350![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g, 30 min, 4 °C and the supernatant was loaded onto anion-exchange column (Resource Q 1 mL, GE Healthcare). The unbound fraction, containing baMntA, was collected and further purified by size-exclusion chromatography (HiLoad 16/600, GE Healthcare). Protein purification was monitored by coomassie staining of SDS-PAGE and size-exclusion chromatography (SEC).
000 × g, 30 min, 4 °C and the supernatant was loaded onto anion-exchange column (Resource Q 1 mL, GE Healthcare). The unbound fraction, containing baMntA, was collected and further purified by size-exclusion chromatography (HiLoad 16/600, GE Healthcare). Protein purification was monitored by coomassie staining of SDS-PAGE and size-exclusion chromatography (SEC).
Structural modeling
We searched the baMntA sequence against the PDB database using blastp (protein–protein BLAST). The structure of MtsA (PDB ID 3HH8) was the most relevant template with high sequence identity (53%). We constructed several multiple sequence alignments (MSAs) using different techniques and parameters in order to get the best alignment. This included collecting homologous sequences via an HMMER search71 against the UniProt and SwissProt databases. In addition we conducted PSI-BLAST72 searches against the UniProt, SwissProt, and clean_UniProt with one, two or three iterations. The collected sequences in all cases were aligned using the MUSCLE algorithm.73 We then extracted the aligned sequences of baMntA and MtsA from the different MSAs to obtain additional pairwise alignments. Reassuringly, all the pairwise alignments were indeed similar, as anticipated owing to the high sequence identity. We then used this pairwise alignment to build a model using MODELLER. Modeling of baMntA with SWISS-MODEL39 and I-TASSER38 was performed in automated mode, without specifying a template, using the default parameters of both programs.We used the same procedures as the modeling of baMntA to model MntB and MntC. For MntB we used PDB ID 2NQ2, chain C (30% seq id, 89% query cover, e-value 1e-32). For MntC we used PDB ID 2NQ2, chain A (14% seq id, 97% query cover, e-value 2e-15).
Construction and purification of MntA mutants
Point mutations (H69A, H141A, E207A, D282A) were introduced into wild type baMntA by using QuikChange Lightning site-directed mutagenesis kit (Agilent Technologies). Mutations were confirmed by sequence analysis. Mutant proteins were overexpressed and purified as the wild type baMntA.Isothermal titration calorimetry
Calorimetric measurements were performed with MicroCal iTC200 System (GE Healthcare). Metals stocks were dissolved in double-distilled water (except for Fe2+ in 1% H2SO4 and Fe3+ in 2% HCl) and diluted into the buffer of the experiment (25 mM Tris-HCl pH 8, 150 mM NaCl). All measurements were carried out at 25 °C. 2 μL aliquots of metal (0.2–0.6 mM, as indicated) were added by a rotating syringe to the reaction well containing 200 μL of 15–60 μM (as indicated) baMntA. Data fitting was performed with the AFFINImeter server (http://www.affinimeter.com). Unless stated otherwise the experimental data was fit using a simple 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 binding model, where the metal free form of the protein (FS) is in equilibrium with the bound species (MA): FS ⇔ MA. For the competition experiments shown in Fig. 4 the data was fit using the Competitive model of AFFINImeter, where the metal free form of the protein (FS) is in equilibrium with the bound species (MA) and a second bound species (MB): MA FS ⇔ MB.
1 binding model, where the metal free form of the protein (FS) is in equilibrium with the bound species (MA): FS ⇔ MA. For the competition experiments shown in Fig. 4 the data was fit using the Competitive model of AFFINImeter, where the metal free form of the protein (FS) is in equilibrium with the bound species (MA) and a second bound species (MB): MA FS ⇔ MB.
EDTA treatment of baMntA
For the metal release experiments 1.4 mL of baMntA (75 μM) were incubated with 50 μM of the indicated metal for 10 minutes at room temperature. The mixture was then placed in a dialysis tube (D-tube Dialyzer, MWCO 6–8 kDa, Novagen) and dialyzed against 1.4 L of 25 mM Tris-HCl pH 8, 150 mM NaCl and 20 mM EDTA at 4 °C. At the indicated intervals 0.2 mL samples were withdrawn and diluted 15-fold with double distilled water. The metal content of baMntA was determined by ICP-MS.Thermal shift assay
15 μM baMntA were mixed with metal ion (Mn+2, Zn+2 or Co+2) in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio and SYPRO Orange (Molecular probes) was added at 0.5× of the manufacturer's guidelines. 20 μL were subjected to thermal unfolding from 25 °C to 85 °C at a heating rate of 1 °C per min, using a Corbett real-time cycler (Rotor Gene 6000). Fluorescence was recorded by excitation at 470 and emission at 610. Data were also collected for buffer only, protein without metal ions and for each of the metal ions without protein. After subtraction of the buffer fluorescence from that of the protein containing samples, the relative fluorescence was plotted against the temperature.
1 molar ratio and SYPRO Orange (Molecular probes) was added at 0.5× of the manufacturer's guidelines. 20 μL were subjected to thermal unfolding from 25 °C to 85 °C at a heating rate of 1 °C per min, using a Corbett real-time cycler (Rotor Gene 6000). Fluorescence was recorded by excitation at 470 and emission at 610. Data were also collected for buffer only, protein without metal ions and for each of the metal ions without protein. After subtraction of the buffer fluorescence from that of the protein containing samples, the relative fluorescence was plotted against the temperature.
Inductively coupled plasma mass spectrometry (ICP-MS)
Metal ions content was determined by ICP-MS, at the Earth Science institute (Hebrew university of Jerusalem). Samples were diluted to a final concentration of 5 μM in 1% HNO3 and analyzed by ICP-MS using Agilent 7500cx ICP-MS. Each sample was measured three times and averaged.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
This work was supported by the Meriuex Research Foundation (OL), NATO's Science for Peace and Security Programme (SPS Project G4622, NBT and OL), the Israeli Academy of Sciences (OL), the European Research Council FP-7 IRG (OL), and the Rappaport Family Institute for biomedical research (OL).References
- C. Andreini, et al., MetalPDB: a database of metal sites in biological macromolecular structures, Nucleic Acids Res., 2013, 41(D1), D312–D319 CrossRef CAS PubMed.
- C. Andreini, et al., Metal ions in biological catalysis: from enzyme databases to general principles, JBIC, J. Biol. Inorg. Chem., 2008, 13(8), 1205–1218 CrossRef CAS PubMed.
- J. S. Klein and O. Lewinson, Bacterial ATP-driven transporters of transition metals: physiological roles, mechanisms of action, and roles in bacterial virulence, Metallomics, 2011, 3(11), 1098–1108 RSC.
- K. W. Becker and E. P. Skaar, Metal limitation and toxicity at the interface between host and pathogen, FEMS Microbiol. Rev., 2014, 38(6), 1235–1249 CrossRef CAS PubMed.
- T. E. Kehl-Fie and E. P. Skaar, Nutritional immunity beyond iron: a role for manganese and zinc, Curr. Opin. Chem. Biol., 2010, 14(2), 218–224 CrossRef CAS PubMed.
- V. E. Diaz-Ochoa, et al., Transition metal ions at the crossroads of mucosal immunity and microbial pathogenesis, Front. Cell. Infect. Microbiol., 2014, 4, 2 Search PubMed.
- K. M. Papp-Wallace and M. E. Maguire, Manganese Transport and the Role of Manganese in Virulence, Annu. Rev. Microbiol., 2006, 60(1), 187–209 CrossRef CAS PubMed.
- M. I. Hood, et al., Identification of an Acinetobacter baumannii Zinc Acquisition System that Facilitates Resistance to Calprotectin-mediated Zinc Sequestration, PLoS Pathog., 2012, 8(12), e1003068 CAS.
- G. Porcheron, et al., Iron, copper, zinc, and manganese transport and regulation in pathogenic Enterobacteria: correlations between strains, site of infection and the relative importance of the different metal transport systems for virulence, Front. Cell. Infect. Microbiol., 2013, 3, 90 Search PubMed.
- C. Guilhen, M.-K. Taha and F. J. Veyrier, Role of transition metal exporters in virulence: the example of Neisseria meningitidis, Front. Cell. Infect. Microbiol., 2013, 3, 102 Search PubMed.
- G. M. Rodriguez and I. Smith, Identification of an ABC Transporter Required for Iron Acquisition and Virulence in Mycobacterium tuberculosis, J. Bacteriol., 2006, 188(2), 424–430 CrossRef CAS PubMed.
- B. A. Diep, et al., Identifying potential therapeutic targets of methicillin-resistant Staphylococcus aureus through in vivo proteomic analysis, J. Infect. Dis., 2014, 209(10), 1533–1541 CrossRef CAS PubMed.
- J. S. Brown, et al., Characterization of Pit, a Streptococcus pneumoniae Iron Uptake ABC Transporter, Infect. Immun., 2002, 70(8), 4389–4398 CrossRef CAS.
- A. Marra, et al., In vivo characterization of the Psa genes from Streptococcus pneumoniae in multiple models of infection, Microbiology, 2002, 148(5), 1483–1491 CAS.
- A. Dintilhac and J. P. Claverys, The ADC locus, which affects competence for genetic transformation in Streptococcus pneumoniae, encodes an ABC transporter with a putative lipoprotein homologous to a family of streptococcal adhesins, Res. Microbiol., 1997, 148(2), 119–131 CrossRef CAS.
- S. Ammendola, et al., High-Affinity Zn2+ Uptake System ZnuABC Is Required for Bacterial Zinc Homeostasis in Intracellular Environments and Contributes to the Virulence of Salmonella enterica, Infect. Immun., 2007, 75(12), 5867–5876 CrossRef CAS PubMed.
- T. Pattery, J. P. Hernalsteens and H. De Greve, Identification and molecular characterization of a novel Salmonella enteritidis pathogenicity islet encoding an ABC transporter, Mol. Microbiol., 1999, 33(4), 791–805 CrossRef CAS.
- S. Paik, et al., The sloABCR Operon of Streptococcus Mutans Encodes an Mn and Fe Transport System Required for Endocarditis Virulence and its Mn-dependent Repressor, J. Bacteriol., 2003, 185(20), 5967–5975 CrossRef CAS.
- S. W. Bearden and R. D. Perry, The Yfe system of Yersinia pestis transports iron and manganese and is required for full virulence of plague, Mol. Microbiol., 1999, 32(2), 403–414 CrossRef CAS.
- C. F. Higgins, ABC transporters: from microorganisms to man, Annu. Rev. Cell Biol., 1992, 8, 67–113 CrossRef CAS PubMed.
- I. B. Holland, et al., in ABC Proteins: From Bacteria to Man, ed. I. B. Holland, Academic Press, London, 2003, p. 647 Search PubMed.
- K. J. Linton and C. F. Higgins, The Escherichia coli ATP-Binding Cassette (ABC) proteins, Mol. Microbiol., 1998, 28(1), 5–13 CrossRef CAS.
- R. o. Sánchez-Fernández, et al., The Arabidopsis thaliana ABC Protein Superfamily, A Complete Inventory, J. Biol. Chem., 2001, 276(32), 30231–30244 CrossRef PubMed.
- M. Dean, Y. Hamon and G. Chimini, The human ATP-binding cassette (ABC) transporter superfamily, J. Lipid Res., 2001, 42(7), 1007–1017 CAS.
- D. C. Rees, E. Johnson and O. Lewinson, ABC transporters: the power to change, Nat. Rev. Mol. Cell Biol., 2009, 10(3), 218–227 CrossRef CAS PubMed.
- G. F. Ames, et al., Traffic ATPases: a superfamily of transport proteins operating from Escherichia coli to humans, Adv. Enzymol. Relat. Areas Mol. Biol., 1992, 65, 1–47 CAS.
- E. L. Borths, et al., In vitro functional characterization of BtuCD-F, the Escherichia coli ABC transporters for vitamin B12 uptake, Biochemistry, 2005, 44, 16301–16309 CrossRef CAS PubMed.
- N. Cadieux, et al., Identification of the Periplasmic Cobalamin-Binding Protein BtuF of Escherichia coli, J. Bacteriol., 2002, 184(3), 706–717 CrossRef CAS.
- O. Gat, et al., The solute-binding component of a putative Mn(II) ABC transporter (MntA) is a novel Bacillus anthracis virulence determinant, Mol. Microbiol., 2005, 58(2), 533–551 CrossRef CAS PubMed.
- R. Janulczyk, S. Ricci and L. Bjorck, MtsABC is Important for Manganese and Iron Transport, Oxidative Stress Resistance and Virulence of Streptococcus pyogenes, Infect. Immun., 2003, 71(5), 2656–2664 CrossRef CAS.
- C. A. McDevitt, et al., A Molecular Mechanism for Bacterial Susceptibility to Zinc, PLoS Pathog., 2011, 7(11), e1002357 CAS.
- V. Rukhman, et al., The MntC Crystal Structure Suggests that Import of Mn2+ in Cyanobacteria is Redox Controlled, J. Mol. Biol., 2005, 348(4), 961–969 CrossRef CAS PubMed.
- D. C. Desrosiers, et al., The general transition metal (Tro) and Zn2+ (Znu) transporters in Treponema pallidum: analysis of metal specificities and expression profiles, Mol. Microbiol., 2007, 65(1), 137–152 CrossRef CAS PubMed.
- L. Tirado-Lee, et al., Classification of a Haemophilus influenzae ABC Transporter HI1470/71 through its Cognate Molybdate Periplasmic Binding Protein, J. Mol. Struct., 2011, 19(11), 1701–1710 CAS.
- E. Vigonsky, E. Ovcharenko and O. Lewinson, Two molybdate/tungstate ABC transporters that interact very differently with their substrate binding proteins, Proc. Natl. Acad. Sci. U. S. A., 2013, 110(14), 5440–5445 CrossRef CAS PubMed.
- X. Sun, et al., Crystal Structure and Metal Binding Properties of the Lipoprotein MtsA, Responsible for Iron Transport in Streptococcus pyogenes, Biochemistry, 2009, 48(26), 6184–6190 CrossRef CAS PubMed.
- N. Eswar, et al., in Protein Structure Modeling with MODELLER, in Structural Proteomics, ed. B. Kobe, M. Guss and T. Huber, Humana Press, 2008, pp. 145–159 Search PubMed.
- A. Roy, A. Kucukural and Y. Zhang, I-TASSER: a unified platform for automated protein structure and function prediction, Nat. Protoc., 2010, 5(4), 725–738 CrossRef CAS PubMed.
- T. Schwede, et al., SWISS-MODEL: an automated protein homology-modeling server, Nucleic Acids Res., 2003, 31(13), 3381–3385 CrossRef CAS PubMed.
- R. A. Laskowski, et al., {PROCHECK}: a program to check the stereochemical quality of protein structures, J. Appl. Crystallogr., 1993, 26, 283–291 CrossRef CAS.
- R. Luthy, J. U. Bowie and D. Eisenberg, Assessment of protein models with three-dimensional profiles, Nature, 1992, 356(6364), 83–85 CrossRef CAS PubMed.
- M. Wiederstein and M. J. Sippl, ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins, Nucleic Acids Res., 2007, 35(web server issue), W407–W410 CrossRef PubMed.
- M. Kalman and N. Ben-Tal, Quality assessment of protein model-structures using evolutionary conservation, Bioinformatics, 2010, 26(10), 1299–1307 CrossRef CAS PubMed.
- M. C. Lawrence, et al., The crystal structure of pneumococcal surface antigen PsaA reveals a metal-binding site and a novel structure for a putative ABC-type binding protein, Structure, 1998, 6(12), 1553–1561 CrossRef CAS.
- R. M. Couñago, et al., Imperfect coordination chemistry facilitates metal ion release in the Psa permease, Nat. Chem. Biol., 2014, 10(1), 35–41 CrossRef PubMed.
- N. Li, et al., Varied metal-binding properties of lipoprotein PsaA in Streptococcus pneumoniae, JBIC, J. Biol. Inorg. Chem., 2014, 19(6), 829–838 CrossRef CAS PubMed.
- C. J. Layton and H. W. Hellinga, Quantitation of protein–protein interactions by thermal stability shift analysis, Protein Sci., 2011, 20(8), 1439–1450 CrossRef CAS PubMed.
- M. D. Cummings, M. A. Farnum and M. I. Nelen, Universal Screening Methods and Applications of ThermoFluor®, J. Biomol. Screening, 2006, 11(7), 854–863 CrossRef CAS PubMed.
- C. J. Layton and H. W. Hellinga, Thermodynamic Analysis of Ligand-induced Changes in Protein Thermal Unfolding Applied to High-throughput Determination of Ligand Affinities with Extrinsic Fluorescent Dyes, Biochemistry, 2010, 49(51), 10831–10841 CrossRef CAS PubMed.
- A. Niedziela-Majka, et al., High-throughput Screening of Formulations to Optimize the Thermal Stability of a Therapeutic Monoclonal Antibody, J. Biomol. Screening, 2015, 20(4), 552–559 CrossRef CAS PubMed.
- O. Lewinson, et al., A distinct mechanism for the ABC transporter BtuCD-BtuF revealed by the dynamics of complex formation, Nat. Struct. Mol. Biol., 2010, 17(3), 332–338 CAS.
- J.-S. Woo, et al., X-ray structure of the Yersinia pestis heme transporter HmuUV, Nat. Struct. Mol. Biol., 2012, 19(12), 1310–1315 CAS.
- B. K. Kay, M. P. Williamson and M. Sudol, The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains, FASEB J., 2000, 14(2), 231–241 CAS.
- Y. Nevo and N. Nelson, The NRAMP family of metal-ion transporters, Biochim. Biophys. Acta, Mol. Cell Res., 2006, 1763(7), 609–620 CrossRef CAS PubMed.
- F. Abate, et al., Apo, Zn(2+)-bound and Mn(2+)-bound structures reveal ligand-binding properties of SitA from the pathogen Staphylococcus pseudintermedius, Biosci. Rep., 2014, 34(6), e00154 CrossRef PubMed.
- A. J. Rice, A. Park and H. W. Pinkett, Diversity in ABC transporters: Type I, II and III importers, Crit. Rev. Biochem. Mol. Biol., 2014, 49(5), 426–437 CrossRef CAS PubMed.
- M. C. Clifton, et al., In vitro Reassembly of the Ribose ATP-binding Cassette Transporter Reveals a Distinct Set of Transport Complexes, J. Biol. Chem., 2015, 290(9), 5555–5565 CrossRef CAS PubMed.
- P. A. B. Ronnie, et al., A structural classification of substrate-binding proteins, FEBS Lett., 2010, 584(12), 2606–2617 CrossRef PubMed.
- K. Hollenstein, D. C. Frei and K. P. Locher, Structure of an ABC transporter in complex with its binding protein, Nature, 2007, 446(7132), 213–216 CrossRef CAS PubMed.
- R. N. Hvorup, et al., Asymmetry in the structure of the ABC transporter binding protein complex BtuCD-BtuF, Science, 2007, 317, 1387–1390 CrossRef CAS PubMed.
- M. L. Oldham and J. Chen, Crystal Structure of the Maltose Transporter in a Pretranslocation Intermediate State, Science, 2011, 332(6034), 1202–1205 CrossRef CAS PubMed.
- A. Krogh, et al., Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes1, J. Mol. Biol., 2001, 305(3), 567–580 CrossRef CAS PubMed.
- K. P. Locher, A. T. Lee and D. C. Rees, The E. coli BtuCD structure: a framework for ABC transporter architecture and mechanism, Science, 2002, 296(5570), 1091–1098 CrossRef CAS PubMed.
- N. S. Kadaba, et al., The high-affinity E. coli methionine ABC transporter: structure and allosteric regulation, Science, 2008, 321, 250–253 CrossRef CAS PubMed.
- G. F. Ames, et al., Liganded and unliganded receptors interact with equal affinity with the membrane complex of periplasmic permeases, a subfamily of traffic ATPases, J. Biol. Chem., 1996, 271(24), 14264–14270 CrossRef CAS PubMed.
- C. E. Liu, P.-Q. Liu and G. F.-L. Ames, Characterization of the Adenosine Triphosphatase Activity of the Periplasmic Histidine Permease, a Traffic ATPase (ABC Transporter), J. Biol. Chem., 1997, 272(35), 21883–21891 CrossRef CAS PubMed.
- K. Nikaido and G. F.-L. Ames, One Intact ATP-binding Subunit is Sufficient to Support ATP Hydrolysis and Translocation in an ABC Transporter, the Histidine Permease, J. Biol. Chem., 1999, 274(38), 26727–26735 CrossRef CAS PubMed.
- J. Chen, et al., Trapping the transition state of an ATP-binding cassette transporter: evidence for a concerted mechanism of maltose transport, Proc. Natl. Acad. Sci. U. S. A., 2001, 98(4), 1525–1530 CrossRef CAS.
- A. L. Davidson, H. A. Shuman and H. Nikaido, Mechanism of maltose transport in Escherichia coli: transmembrane signaling by periplasmic binding proteins, Proc. Natl. Acad. Sci. U. S. A., 1992, 89(6), 2360–2364 CrossRef CAS.
- N. Tal, E. Ovcharenko and O. Lewinson, A single intact ATPase site of the ABC transporter BtuCD drives 5% transport activity yet supports full in vivo vitamin B12 utilization, Proc. Natl. Acad. Sci. U. S. A., 2013, 110(14), 5434–5439 CrossRef CAS PubMed.
- S. R. Eddy, Profile hidden Markov models, Bioinformatics, 1998, 14(9), 755–763 CrossRef CAS PubMed.
- S. F. Altschul, et al., Gapped BLAST and PSI-BLAST: a new generation of protein database search programs, Nucleic Acids Res., 1997, 25(17), 3389–3402 CrossRef CAS PubMed.
- R. C. Edgar, MUSCLE: a multiple sequence alignment method with reduced time and space complexity, BMC Bioinf., 2004, 5, 113 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5mt00100e |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2015 |