Open Access Article
Open Access ArticleThe transcriptome of Euglena gracilis reveals unexpected metabolic capabilities for carbohydrate and natural product biochemistry†
Ellis C.
O'Neill
 a,
Martin
Trick
b,
Lionel
Hill
c,
Martin
Rejzek
a,
Renata G.
Dusi
d,
Chris J.
Hamilton
d,
Paul V.
Zimba
e,
Bernard
Henrissat
fgh and
Robert A.
Field
*a
a,
Martin
Trick
b,
Lionel
Hill
c,
Martin
Rejzek
a,
Renata G.
Dusi
d,
Chris J.
Hamilton
d,
Paul V.
Zimba
e,
Bernard
Henrissat
fgh and
Robert A.
Field
*a
aDepartment of Biological Chemistry, John Innes Centre, Norwich Research Park, Norwich, NR4 7UH, UK. E-mail: rob.field@jic.ac.uk
bComputational and Systems Biology, John Innes Centre, Norwich Research Park, Norwich, NR4 7UH, UK
cDepartment of Metabolic Biology, John Innes Centre, Norwich Research Park, Norwich, NR4 7UH, UK
dSchool of Pharmacy, University of East Anglia, Norwich Research Park, Norwich, NR4 7TJ, UK
eCenter for Coastal Studies, Texas A&M University-Corpus Christi, 6300 Ocean Drive, Unit 5866, Corpus Christi, Texas 78412, USA
fAix-Marseille University, Architecture et Fonction des Macromolécules Biologiques, 163 Avenue de Luminy, 13288 Marseille, France
gCentre National de la Recherche Scientifique, UMR 7257, 163 Avenue de Luminy, 13288 Marseille, France
hDepartment of Biological Sciences, King Abdulaziz University, Jeddah, Saudi Arabia
First published on 13th August 2015
Abstract
Euglena gracilis is a highly complex alga belonging to the green plant line that shows characteristics of both plants and animals, while in evolutionary terms it is most closely related to the protozoan parasites Trypanosoma and Leishmania. This well-studied organism has long been known as a rich source of vitamins A, C and E, as well as amino acids that are essential for the human diet. Here we present de novo transcriptome sequencing and preliminary analysis, providing a basis for the molecular and functional genomics studies that will be required to direct metabolic engineering efforts aimed at enhancing the quality and quantity of high value products from E. gracilis. The transcriptome contains over 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 protein-encoding genes, supporting metabolic pathways for lipids, amino acids, carbohydrates and vitamins, along with capabilities for polyketide and non-ribosomal peptide biosynthesis. The metabolic and environmental robustness of Euglena is supported by a substantial capacity for responding to biotic and abiotic stress: it has the capacity to deploy three separate pathways for vitamin C (ascorbate) production, as well as producing vitamin E (α-tocopherol) and, in addition to glutathione, the redox-active thiols nor-trypanothione and ovothiol.
000 protein-encoding genes, supporting metabolic pathways for lipids, amino acids, carbohydrates and vitamins, along with capabilities for polyketide and non-ribosomal peptide biosynthesis. The metabolic and environmental robustness of Euglena is supported by a substantial capacity for responding to biotic and abiotic stress: it has the capacity to deploy three separate pathways for vitamin C (ascorbate) production, as well as producing vitamin E (α-tocopherol) and, in addition to glutathione, the redox-active thiols nor-trypanothione and ovothiol.
Introduction
Euglenoids are abundant algae, typically found in freshwater rich in organics, where they can be so populous as to give their characteristic colour to the water, such as the verdant green Euglena viridis or blood red Euglena sanguinea. They were first documented1 by van Leeuwenhoek in his 1674 letter to the Royal Society,2 although Harris is usually credited with the first description of this species.3 These large unicellular organisms (Fig. 1A), up to 100 μm in length, possess an eye spot photoreceptor, enabling Euglena to perceive light and swim towards it using its single flagellum for motility or by a unique so-called euglenoid locomotion. Euglena exhibits mixotrophy, switching between photosynthesis, absorbing nutrients/small metabolites, and engulfing other eukaryotes and bacteria as the opportunity or need arises.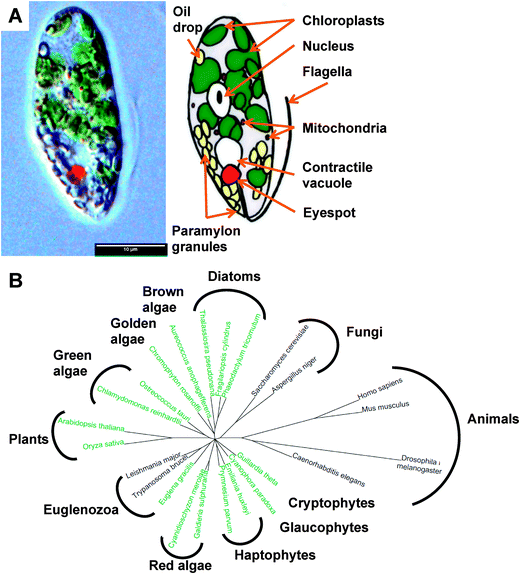 | ||
| Fig. 1 The biology of Euglena gracilis. (A) Physiology of Euglena gracillis. (B) Phylogeny showing relationship between Euglena gracilis and sequenced algae and model organisms.112 Organisms shown in green are photosynthetic. | ||
Euglena gracilis has been extensively investigated for the production of vitamins A, C, E,4 and essential amino acids and is also a good source of polyunsaturated fatty acids.5 Although a photosynthetic organism, Euglena does not produce a typical α-1,4/6-glucan, such as starch. Instead, when grown aerobically in light it produces an insoluble β-1,3-glucan, paramylon,6 contributing up to ca. 85% of its' dry weight. This glycopolymer shows immune stimulatory properties7 and is reported to have anti-HIV8 effects. In contrast, under anaerobic conditions wax esters9,10 comprise over 50% of the dry weight of some strains of Euglena. These compounds are produced even under adverse conditions such as those encountered in acid mine drains,11 which suggests potential for the use of this taxa in waste water management12 and/or the production of biodiesel. Euglenoids are physiologically very flexible, resulting in their use in many diverse scientific fields, including: as a reporter for vitamin B12 production13 and for the clinical analysis of B12 in human serum;14 to study the ecotoxicity of zinc oxide nanoparticles;15 in a neurocomputer based on their motion in a micro aquarium;16 and for the intracellular biosynthesis of ferri-hydrite17 and silver18 nanoparticles. Euglena behaviour has been studied using zero gravity flight19 and the nature of their active gravitaxis mechanisms has been investigated under varying acceleration conditions aboard the space shuttle Columbia.20
Phylogenetically, Euglena are unlike other rigorously studied algae in that their closest neighbours are the human pathogenic protozoa Trypanosoma and Leishmania (Fig. 1B). Their unique phylogenetic position and the ease with which Euglena can be cultured has made them one of the most highly studied eukaryotes, playing a pivotal role in the development of cell biology and biochemistry, a topic that is thoroughly reviewed in The Biology of Euglena by Buetow21–24 and A Color Atlas of Photosynthetic Euglenoids by Ciugulea and Triemer.25 However, despite the well-established potential for biotechnology applications for euglenoids, we know little about their genetically-encoded metabolic capacity. Sequencing the genome of Euglena has proved problematic due to its size and complexity, which has arisen from a series of endosymbiotic events during its evolution.26,27 Aside from typical eukaryotic epigenetic modifications, including DNA methylation and histone acetylation, the genome of Euglena also contains the modified nucleotide Base J (glucosylated hydroxythymidine), also found in kinetoplastids,28 which complicates DNA sequencing. Additionally Euglena has the ability to extensively process mRNA during transcription,29 altering the sequences before translation; hence the proteome would be difficult to predict from its genome. Avoiding the complications of algal genome sequencing30 and in order to begin to explore the full metabolic capability of Euglena, we have sequenced the transcriptome of Euglena gracilis var. saccharophila. Transcriptomic approaches are proving increasingly useful for illuminating the functional diversity of microbial eukaryotic life, for instance in the oceans.31 Here we present an initial analysis of Euglena transcriptomes obtained from dark and light grown cells, with an emphasis on genes encoding carbohydrate and natural products biochemistry, serving to illustrate the versatile metabolic capacity of this species.
Results and discussion
Nucleic acid extraction and transcript sequencing
To obtain a broad set of transcripts, RNA was extracted from Euglena gracilis cells grown under two radically different growth conditions. For autotrophic (photosynthetic) growth, cells were cultured under ambient illumination on low nutrient media solid agar, containing no amino acids or carbon source (referred to as “light”); high nutrient media liquid culture, containing glucose and amino acids with no light source (referred to as “dark”), was used to stimulate mixotrophic growth. Equal amounts of mRNA from these two conditions were sequenced using an Illumina HiSeq 2000 to give 26.5 Gb and 11.9 Gb of good quality reads for light and dark grown cells, respectively (Table 1). These sequences were assembled into contigs and likely coding regions were predicted using the Trinity software package.32 In this manner, 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 814 predicted protein-encoding genes were identified in light-grown cells, while 26
814 predicted protein-encoding genes were identified in light-grown cells, while 26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 738 were evident in dark-grown cells, accounting for 32
738 were evident in dark-grown cells, accounting for 32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 128 non-redundant proteins predicted overall. This indicates that, depending on growth conditions, there is a dramatic shift in metabolic capability.
128 non-redundant proteins predicted overall. This indicates that, depending on growth conditions, there is a dramatic shift in metabolic capability.
| Light-grown cells | Dark-grown cells | Combined | |
|---|---|---|---|
| a Non-redundant subsets derived with CD-HIT108 using identity threshold 0.95, word length 5. | |||
| Total number of reads | 264![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 808 808![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 150 150 |
118![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 608 608![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 486 486 |
383![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 414 414![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 636 636 |
| Total number of nucleotides | 26.5 Gb | 11.9 Gb | 38.4 Gb |
| Total number of contigs | 233![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 748 748 |
231![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 176 176 |
|
| Median length of contigs | 391 bp | 364 bp | |
| Total number of predicted ORFs longer than 100 aa | 45![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 126 126 |
47![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 607 607 |
92![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 733 733 |
| Number of unique proteins | 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 814a 814a |
26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 738a 738a |
32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 128a 128a |
| Mean length of unique proteins | 456 aa | 401 aa | |
Various eukaryotes splice a short leader sequence onto the 5′ end of the mRNA, encoded from a different region of the genome, to process the polycistronic transcripts and stabilise the produced mRNAs.33 Previously a 26 nucleotide short leader sequence had been identified in Euglena from 4 sequences,34 but this sequence was present in <1% of the transcripts presented here. However a reduced 14 base sequence, “TCTATTTTTTTTCGA”, was present at the 5′ UTR of 16.0% and 15.6% of the transcripts from light and dark grown cells, though it is unclear how many sequences without this modification are in fact full length, rather than truncated by limitations of the sequencing.
In order to annotate gene functions, the 32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 128 sequences were compared to the UNiRef 100 database using BLASTP, which produced 14
128 sequences were compared to the UNiRef 100 database using BLASTP, which produced 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 389 matches to proteins with annotated functions.‡ The majority of transcript sequences identified therefore show no match to any previously reported protein sequence from any organism. Of the sequences that can be annotated, 12
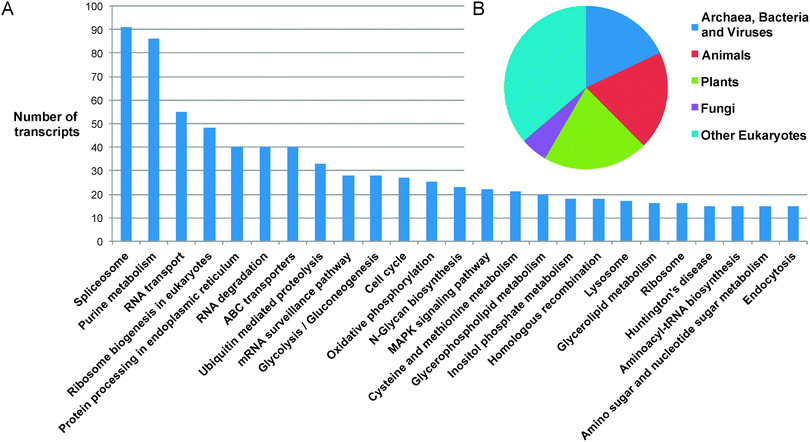
389 matches to proteins with annotated functions.‡ The majority of transcript sequences identified therefore show no match to any previously reported protein sequence from any organism. Of the sequences that can be annotated, 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 020 were classified into 157 Gene Ontology classifications, covering the full spectrum of functions that one might expect in a complex eukaryotic organism (Fig. 2). The distribution of the top BLASTP hits shows a distribution across the kingdoms of life, highlighting the diversity of possible origins of genetic material present in the Euglena genome, which have been integrated through its complex phenotypic and genotypic history.27
020 were classified into 157 Gene Ontology classifications, covering the full spectrum of functions that one might expect in a complex eukaryotic organism (Fig. 2). The distribution of the top BLASTP hits shows a distribution across the kingdoms of life, highlighting the diversity of possible origins of genetic material present in the Euglena genome, which have been integrated through its complex phenotypic and genotypic history.27
Metabolism
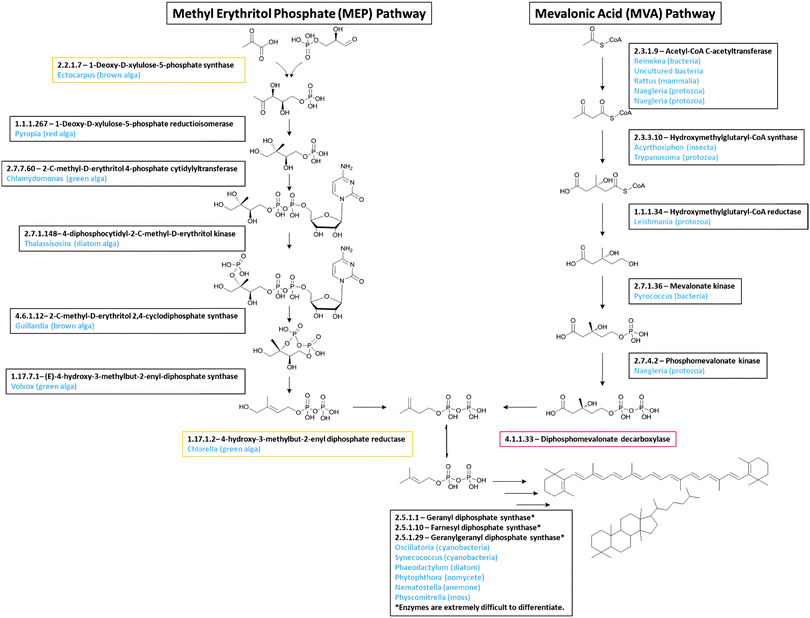
Candidate genes were identified in the Euglena transcriptome for all of the core metabolic pathways, including glycolysis and gluconeogenesis, the citric acid cycle and the pentose phosphate pathway. There are also candidates for carotenoid and thylakoid glycolipid biosynthesis, and the Calvin cycle, which are required for photosynthesis. Reassuringly, the completeness of these pathways confirms both adequate sequencing depth and accurate functional assignments.In addition to the expected candidates for fatty acid biosynthesis, a capacity to produce isoprenoids, which have functions in primary metabolism, is also evident in the Euglena transcriptome. There are two pathways for the formation of the isoprenoid precursor in plants: the cytosolic mevalonate (MVA)35 and the plastidial methyl erythritol phosphate (MEP) pathway.36 Both pathways are known in Euglena, although unusually the MEP pathway only contributes to carotenoid biosynthesis and is not involved in phytol synthesis.37 All enzymes for both pathways can readily be identified in the transcriptome, with the exception of the final decarboxylase of the MVA pathway (Fig. 3). The presence of the rest of the pathway in Euglena implies this last reaction is likely catalysed by a non-canonical enzyme. The initial and final enzymes for the chloroplast-localised MEP pathway are only apparent in the transcripts from dark grown cells, suggesting that under the conditions utilised in this study, the MEP pathway is only active in chloroplasts that are not supplying energy to the cells.
Euglena is self-contained in its amino acid requirements, producing transcripts of all the obvious genes required for the production of this class of molecule. Intriguingly, however, Euglena appears to have acquired and retained genes for amino acid biosynthesis that are reminiscent of both plant and bacterial pathways. Moreover, it appears to duplicate functionality in places: for aromatic amino acid biosynthesis, for instance, transcripts for single function enzymes are evident alongside much larger transcripts encoding several steps in the pathway (Fig. S1, ESI†).27 The ability to produce multi-functional polydomain proteins, arising from gene fusions, presents interesting opportunities from a synthetic biology perspective.
Carbohydrate-active enzymes
Euglena is not reported to have a carbohydrate-based cell wall but it is known to utilise a β-1,3-glucan carbohydrate storage polymer and it can become encased in a carbohydrate sheath, forming cysts.38 Although the exact nature of the surface glycan(s) is not known, it is thought to be highly complex, containing glucose, galactose, mannose, fucose, xylose, rhamnose and at least one hexosamine.39 The anticipated variety of glycans and the metabolic versatility displayed by Euglena make this a potentially attractive organism in which to study carbohydrate metabolism. This notion is supported by the wealth of carbohydrate-active enzymes that are evident from transcriptome data (Table 2).| Family | Proteins | Families |
|---|---|---|
| a For a full breakdown of the carbohydrate active enzymes in Euglena gracilis see http://www.cazy.org/eE.html. | ||
| Glycosyl transferases (GTs) | 229 | 34 |
| Glycoside hydrolases (GHs) | 126 | 26 |
| Carbohydrate binding modules (CBM) | 2 (+2 on GHs) | 4 |
| Carbohydrate esterases | 9 | 2 |
With around 230 glycosyltransferases (GTs) (depending upon the stringency used in annotation), Euglena is rich in GTs like Homo sapiens or Caenorhabditis elegans and much richer than the red alga Porphyridium purpureum, the green algae Micromonas sp, the basal alga Cyanophora paradoxa, the cryptophyte Guillardia theta and the chlorarachniophyte Bigelowiella natans (data from the CAZy database40). Viridiplantae (‘green plants’) have significantly more GTs than Euglena, and this is attributable to several rounds of genome duplication. A repertoire of around 130 glycosyl hydrolases, dominated by enzymes for the digestion of β-glucans (vide infra), and very few different carbohydrate-binding modules (CBMs), underlies a modest commitment to glycan digestion which may reflect the photosynthetic nature of this organism, which enables it to be largely self-sufficient in terms of carbohydrate metabolism.
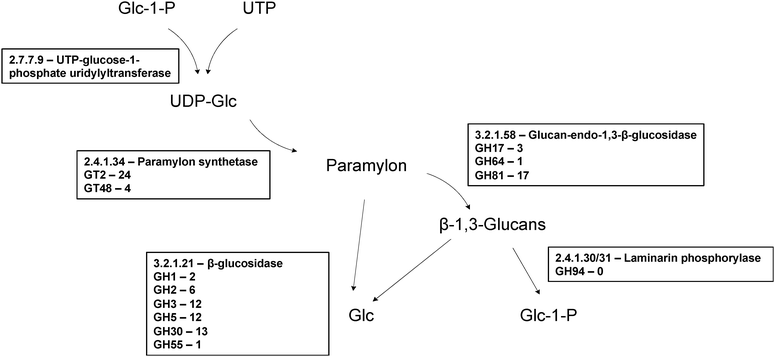 | ||
| Fig. 4 Enzymes proposed to be involved in paramylon biosynthesis. In Euglena paramylon is synthesised by glucosyl transfer from UDP-glucose, initially onto a membrane bound protein,113 by a protein complex, forming strictly linear β-1,3-glucan chains which form intra chain triple helices. In plants and fungi members of GT48 and GT2 are used to make β-1,3-glucans and members of both are present in the transcriptome. Paramylon is degraded by a series of endo- and exo-β-glucosidases to form glucose,114 or by laminarin phosphorylase to form α-glucose-1-P.115 There are many members of β-glucosidase families in the transcriptome but there is no member of GH94, the family to which the only sequenced laminaribiose phosphorylases belong,51 suggesting a novel family for this activity in Euglena. | ||
Plants and fungi synthesise the β-1,3-glucans in their cell walls using membrane-bound members of the GT48 family, which may represent candidate paramylon synthases in Euglena. Four sequences could be identified in the transcriptome which encode candidate GT48 β-1,3-glucan synthases, three of which are closely related to each other (94% identity). These proteins are predicted to be around 300 kDa in size – much larger than the proteins reported to make up the paramylon synthase complex,42 although proteolytic processing cannot be ruled out. Enzymes of the GT2 family can also participate in the biosynthesis of β-1,3- and β-1,4-glycans, such as callose, chitin, and cellulose,40 and thus also represent potential candidates for paramylon synthase. This family is heavily represented in the Euglena transcriptome, with 16 unique members, although few of these proteins has the QXXRW signature of processive polysaccharide synthases.44
Both endo- and exo-β-1,3-glucan hydrolases have been biochemically identified in Euglena,45,46 and there are many glycoside hydrolases present in the Euglena transcriptome that belong to the various GH families capable of degrading β-1,3-glucans. There are transcripts encoding 17 members of family GH81, as well as three members of family GH17 and a single member of family GH64, all families which contain endo-acting hydrolases capable of cleaving in the middle of β-1,3-glucan chains. These enzymes release short glucan chains which could then be further degraded to glucose by β-glucosidases, for which there are sequences for members of 6 families (GH1, 2, 3, 5, 30, 55) in the Euglena transcriptome. Alternatively, glucans could be degraded by phosphorylases,47 releasing Glc-1-P. β-1,3-Glucan phosphorylases have been well characterised in Euglena48,49 and other unrelated algae,50 and have since been identified in bacteria.51,52 The bacterial enzymes are in family GH94 but exclusively act upon the disaccharide, laminaribiose. No members of this family are present in the transcriptome, suggesting Euglena utilises another enzyme family for this activity.
Euglena has a full complement of genes for the biosynthesis of glycosylphosphatidylinositol (GPI) membrane anchors. This is perhaps unsurprising for an organism that is phylogenetically related to the Trypanosomes, which make extensive use of such glycolipids to attach proteins to their surface, (Table S2, ESI†). However, this class of compounds remains to be identified and characterised in Euglena species; it is unclear whether the prevalent use of GPI biochemistry in the parasitic protozoa, with a need to rapidly alter their surface coat in order to evade the host immune response, is replicated in their rather benign water-borne relatives.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 in the light sample, but in the dark no short sequence variant is detectable. This suggests that in the light the enzyme activity is required in both subcellular locations, but in the dark it is not required in the cytosol. Euglena appears to make use of alternative splicing to control subcellular targeting of a single gene product, as has been seen for many enzymes,60 including glycolytic enzymes in fungi61 and amino acid metabolic enzymes in plants.62
1 in the light sample, but in the dark no short sequence variant is detectable. This suggests that in the light the enzyme activity is required in both subcellular locations, but in the dark it is not required in the cytosol. Euglena appears to make use of alternative splicing to control subcellular targeting of a single gene product, as has been seen for many enzymes,60 including glycolytic enzymes in fungi61 and amino acid metabolic enzymes in plants.62
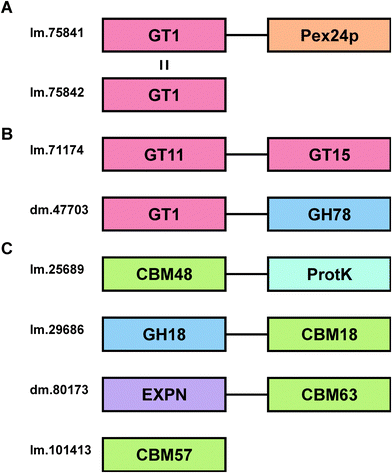
Whilst fusion of CBMs to other CAZYs is relatively common in nature, and contiguity of multiple glycoside hydrolase domains in a single protein is well known, it is much rarer to find glycosyltransferases in a protein containing other domains with carbohydrate binding or enzyme activity.63 Two transcripts were identified in the Euglena transcriptome that encode proteins with more than one carbohydrate-active enzyme domain (Fig. 5B). The first protein (lm.71174) has a putative GT11 fucosyltransferase domain, whose active site does not contain the second arginine in the usual HxRRxD consensus motif,64 and a putative GT15 mannosyltransferase domain, which contains the nucleophile and a zwitterionic ion binding motif (Fig. S2, ESI†).65 A second two-domain protein (dm.47703) appears to have a GT1 sugar transferase domain, most closely related to glucuronic acid transferases,66 linked to a C-terminal GH78 α-rhamnosidase domain.67 This protein might conceivably be involved in cleaving rhamnose from a natural product and adding a glucuronic acid moiety, which is known to facilitate sub-cellular relocalisation and xenobiotic detoxification.68 Alternatively, it might be involved in editing glycan sequences and structures. Further work is required to assess these prospects.
Carbohydrate binding modules (CBMs) are typically found as part of the same polypeptide sequence as carbohydrate-active enzymes.69 There are 4 CBMs encoded in the Euglena transcriptome (Fig. 5C), compared to 126 in Arabidopsis and 40 in humans.40 The other sequenced Euglenozoa, Trypanosoma brucei and Leishmania braziliensis, only contain one CBM each, although these organisms live in stable animal hosts and so do not require complicated carbohydrate degradation mechanisms. The common conserved Euglenozoan CBM sits in family 48, typically associated with binding to glycogen, though Euglena do not themselves produce or use glycogen. This CBM is fused to the AMP-activated protein kinase beta subunit,70 suggesting a phosphorylation-based regulatory role in carbohydrate metabolism for this bifunctional protein. Euglena also encodes a GH18 chitinase with a chitin-targeting CBM18, with a potential function in the degradation of extracellular chitin. A potential expansin for loosening plant cell wall glycan architectures,71 fused to a cellulose-binding CBM63, is also evident along with a single member of CBM57, a poorly characterised protein family thought to bind N-glycans72 (Fig. 5C).
Small molecule redox regulators
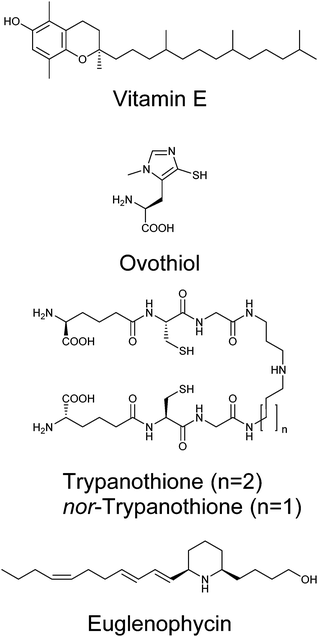
Given the genetic commitment to the production or water-soluble antioxidant vitamin C in Euglena, we were drawn to consider how much broader this commitment to handling redox stress might be. Euglena has long been known to produce high levels of vitamin E,82 a lipid-soluble antioxidant (Fig. 7).83 This vitamin is synthesised from tyrosine and phytol; genes encoding the key dioxygenase and homogentisate phytyl transferase are present in the transcriptome, along with strong candidates for the rest of the pathway (Fig. S3, ESI†).Whilst the tripeptide thiol glutathione (γ-glutamyl-cysteinyl-glycine) plays a significant redox-modulating role in most eukaryotes and microorganisms, many bacteria and protozoa use additional unusual thiol compounds.84 These include mycothiol in actinobacteria,85 bacillithiol in some bacilli,86 and trypanothione and ovothiol in trypanosomatids.87 Independent reductases with exclusive activity towards either glutathione or trypanothione, have previously been purified from E. gracilis,88 although the occurrence of trypanothione has not been demonstrated in any Euglena. The conjugation of glutathione to spermidine to form trypanothione is carried out by either a two enzyme process (e.g. in Crithidia), with the initial conjugation of glutathione to spermidine by glutathionylspermidine synthase followed by addition of another glutathione by trypanothione synthase,89 or by a single bifunctional enzyme (e.g. in Trypanosoma).90 In the Euglena transcriptome, the biosynthetic pathways for glutathione and spermidine are present (Fig. S4, ESI†), and there are two gene sequences which more closely match the trypanothione synthase than the glutathionylspermidine synthase of Crithidia, although they are most closely related to bacterial glutathionylspermidine synthases. It remains to be established whether these enzymes each carry out a single glutathione addition, or perform both conjugation steps.
With clear support for Euglena having a capacity to perform trypanothione biochemistry, we sought to demonstrate the presence of this thiol in Euglena extracts. HPLC analysis of bromobimane-derivatised cell extracts clearly show peaks for the ubiquitous glutathione and cysteine; there is also a small peak co-eluting with trypanothione, (Fig. S5, ESI†). LC-MS revealed this fraction contained both trypanothione and a compound with a molecular weight 14 Da lower (Fig. S6, ESI†). MS2 analysis revealed that this corresponds to a spermidine backbone that is one carbon shorter than is usual in trypanothione, which suggests the presence of the previously unreported nor-trypanothione (Fig. 7). Euglena is known to make the nor-spermine and nor-spermidine from 1,3-diaminopropane, which is thought to be derived from diaminobutyric acid.91 This would require some flexibility in the polyamine chain length in the trypanothione synthase, as is seen in T. cruzi, which makes homo-trypanothione when supplied with exogenous cadavarine, one carbon longer than spermidine.92
The only known enzyme for the biosynthesis of ovothiol (Fig. 7) encodes a non-heme iron dioxygenase at the N-terminus and an N-methyl transferase at the C-terminus, capable of effecting oxidative C–S bond formation, sulfoxidation and N-methylation (Fig. S4, ESI†).93 In the Euglena transcriptome, there is one sequence related to the N-terminus and one related to the C-terminus in both dark and light transcript sets, and a single transcript coding for both domains found only in the dark grown cells. A small peak eluting in the correct region of the HPLC,87 with consistent MS fragmentation, suggests that ovothiol is indeed present in Euglena (Fig. S5 and S6, ESI†).
Given the prevalence of thiol biochemistry in Euglena, and the central role that ovothiol-related compounds (ergothioneine) have recently been shown to have in biosynthesis of the antibiotic lincomycin,94 we were drawn to evaluate the capacity of E. gracilis for coenzyme A/acyl carrier protein-dependent processes, such as those that are often central to natural product biosynthesis. No polyketides or non-ribosomal peptides have been confirmed in Euglena to date, but there are transcripts apparent for the complex secondary metabolite synthases needed to make such compounds, as is evident for an increasing array of algae now that genome/transcriptome sequence data is becoming available.95
Polyketides
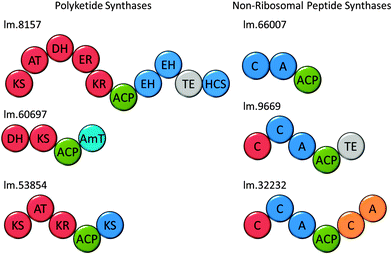
Polyketides comprise a huge range of compounds, formed by repeated condensation of acetate units, followed by variable reduction and further elaboration. Broadly speaking polyketide synthases (PKSs) can be large multidomain proteins (type I) or composed of discrete proteins with individual functions (type II), although there are other architectures possible.96 Only one possible polyketide, the alkaloid euglenophycin (Fig. 7), has been isolated and characterised from a Euglena (Euglena sanguinea), based on its toxicity to fish.97 However, using the established isolation methods,98 we could find no evidence for the production of this compound by E. gracilis.To identify polyketide synthases the proteome was searched for ketosynthases, the key catalytic domain, using BLASTP.99 Fourteen potential PKSs were identified as having this domain (Table S3, ESI†): six sequences encode multiple domains indicative of type I PKSs; three sequences encode one catalytic domain and an acyl carrier protein domain; one sequence carries two catalytic domains; four sequences only encode individual domains. The latter may be true type II PKS modules, or fragments of type I caused by failure to assemble full length transcripts. They may also be enzymes with other functions that this search technique reveals, such as fatty acid synthases. The poor Kozak consensus and lack of upstream stop codons indicate these genes could encode fragments of longer proteins.
Attempts to predict the structures of the compounds synthesised by these putative Euglena PKS enzymes, based on domain architectures, using SBSPKS100 and the PKS/NRPS Analysis Web-site,101 were not successful. This is probably due to the evolutionary distance from the bacterial and fungal species these pieces of software were designed to deal with. Analysis of the domain sequences of these enzymes using DELTA-BLAST, has allowed some assessment of the production line-like machinery and the products that could be made (Fig. 8). For example, in addition to a fully reducing PKS module, the largest polyketide synthase encoded in the Euglena transcriptome contains two enoyl hydratases and an HMGCoA synthase. As single domain proteins, these have been characterised in bacterial gene clusters as adding a β-methyl branch to polyketides such as bacillaene.102 This novel domain architecture suggests the formation of an alkane followed by addition of a β-methyl branch. Thus, although Euglena is not known to make polyketides, there is apparent capacity for the synthesis of complex natural products of this type.
Non-ribosomal peptides
Non-ribosomal peptide synthetases (NRPSs) are multi-domain proteins that join amino acids together to form small molecules with a diversity of function, including siderophores, such as enterobactin.103,104 No non-ribosomal peptides have been isolated from Euglenoids, but in the E. gracilis transcriptome, 19 putative NRPSs can be recognised based on the presence of the adenylation (A) domain and 16 based on the condensation (C) domain (Table S3, ESI†). This search strategy reveals many sequences that are most closely related to enzymes involved in fatty acid biosynthesis and two transcripts for L-aminoadipate-semialdehyde dehydrogenase, involved in lysine biosynthesis. Five transcripts encode proteins with both an A and a C domain (Fig. 8), and the remainder have either only an A or only a C domain. Notably, three adenylation domain-encoding transcripts also have ankyrin domains, potentially involved in protein–protein interactions,105 which may imply formation of non-covalent assemblies of NRPS modules. The function prediction programs, SBSPKS100 and NRPS/PKS Analysis Web-site,101 were unable to predict structure from these sequences. However, given the ability of E. gracilis to actively uptake iron from growth medium (Fig. S7, ESI†), it may be reasonable to speculate that Euglena use NRPSs to produce peptide-based siderophores, a well-known strategy amongst the bacteria.106Conclusions and outlook
Euglena gracilis has been a mainstay of 20th century biology because of its ease of culture, large cell size and metabolic diversity. It is now evident that a combination of protozoan, animal, plant, fungal and prokaryotic genes contribute to its transcriptome, highlighting the evolutionary complexity of this unicellular alga. The wide range of metabolites in Euglena is a function of this evolutionary history, while sequences for enzymes involved in biosynthetic pathways not previously identified in Euglena suggests that we have barely scratched the surface of the metabolic potential of this class of organism.Although its chloroplast genome, a prokaryote-like plasmid, was sequenced in 1993,107 Euglena has lagged behind as attention has largely focused on a limited number of model organisms. With the cost of nucleic acid sequencing decreasing rapidly, the opportunity to widen the repertoire of study organisms is considerable. The transcriptome dataset reported herein brings Euglena into the post genomic era and should facilitate the exploitation of this powerful bioresource, hopefully also stimulating wider consideration of microalgae as vehicles for natural products chemistry and synthetic biology.
Experimental
E. gracilis cell culture
Euglena gracilis var. saccharophila Klebs (strain 1224/7a) was obtained from the Culture Collection of Algae and Protozoa (http://www.ccap.ac.uk/).RNA extraction, sequencing and bioinformatics
Total RNA was isolated from 106 dark and light grown cells using RNeasy Minikit (Qiagen) and stored at −80 °C. Source Bioscience (Nottingham, UK) then performed mRNA purification, library preparation and sequencing on an Illumina HiSeq 2000 platform. A total of 313![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 205
205![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 944 and 137
944 and 137![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 689
689![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 062 100-base, paired-end reads were obtained from the light- and dark-grown samples respectively. The light-grown sequence data, down-sampled to 100 million pairs of reads, and the entire dataset of 68
062 100-base, paired-end reads were obtained from the light- and dark-grown samples respectively. The light-grown sequence data, down-sampled to 100 million pairs of reads, and the entire dataset of 68![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 844
844![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 531 read-pairs from the dark-grown sample were used for separate de novo assemblies using Trinity30 version r2013-02-25, executed in parallel on two 256 GB cluster nodes. 233
531 read-pairs from the dark-grown sample were used for separate de novo assemblies using Trinity30 version r2013-02-25, executed in parallel on two 256 GB cluster nodes. 233![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 748 and 231
748 and 231![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 176 raw assemblies were generated from light and dark respectively. Likely coding sequences were extracted from these reads using the Perl utility script supplied with the Trinity distribution, transcripts_to_best_scoring_ORFs.pl, producing 45
176 raw assemblies were generated from light and dark respectively. Likely coding sequences were extracted from these reads using the Perl utility script supplied with the Trinity distribution, transcripts_to_best_scoring_ORFs.pl, producing 45![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 126 and 47
126 and 47![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 607 candidate ORFs of lengths greater than 100 amino acids. Putative coding sequences were combined and a non-redundant set of 32
607 candidate ORFs of lengths greater than 100 amino acids. Putative coding sequences were combined and a non-redundant set of 32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 128 peptides produced using CD-HIT108 (v4.5.4) with an identity threshold of 0.95 and a word length of 5. For functional annotation, the set was queried against the UniRef 100 protein database using BLASTP with an E-value threshold of 1 × 10−10. Sequence identifiers of the best hits were harvested and used to programmatically collect from databases, via SOAP-based web services, GO terms and KEGG pathway objects, and also used to enumerate kingdom-level taxonomic classifications for the species of origin. Transcript levels for isoforms were estimated from the reads using the RSEM wrapper script supplied in the Trinity distribution to implement the RNA-Seq by Expectation Maximisation methods.109
128 peptides produced using CD-HIT108 (v4.5.4) with an identity threshold of 0.95 and a word length of 5. For functional annotation, the set was queried against the UniRef 100 protein database using BLASTP with an E-value threshold of 1 × 10−10. Sequence identifiers of the best hits were harvested and used to programmatically collect from databases, via SOAP-based web services, GO terms and KEGG pathway objects, and also used to enumerate kingdom-level taxonomic classifications for the species of origin. Transcript levels for isoforms were estimated from the reads using the RSEM wrapper script supplied in the Trinity distribution to implement the RNA-Seq by Expectation Maximisation methods.109
Annotation of carbohydrate-active enzymes (CAZys)
Putative carbohydrate-active enzymes were identified and classified using the methods used for updating the carbohydrate-active enzymes database (CAZy; http://www.cazy.org) as described previously.40 Briefly, the translated protein sequences of Euglena were compared to the full-length sequences derived from CAZy using BLAST.110 The sequences that gave an E-value < 0.1 were then subjected to a second BLAST search against a library made with the constitutive modules of glycoside hydrolases (GH), glycosyltransferases (GT), polysaccharide lyases (PL) and carbohydrate esterases (CE) and their associated carbohydrate-binding (CBM) modules. In parallel the sequences were subjected to a HMMer search using hidden Markov models built for each CAZy module family.111 A protein was considered reliably assigned when it was placed in the same family by the two methods. No particular E-value threshold was used as the E-value varies extensively with the length of the aligned sequences. Instead we relied on manual curation and examination of conserved features such as catalytic residues. Difficult cases were resolved by manual inspection of conserved features such as catalytic residues.Identification of thiols
Cells grown in the high nutrient media, at 21 °C with ambient light, were collected by centrifugation at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, flash cooled in liquid nitrogen and stored at −80 °C until analysis. 20 mg of cells were extracted into 50% acetonitrile at 60 °C and the thiols were labelled with monobromobimane (Invitrogen) and separated on an ACE AR C18 4.6 × 250 mm, 5 μm (HiChrom) by HPLC (JASCO) with fluorescence detection (JASCO FP2020 Plus) with excitation at 385 nm and emission at 460 nm. The gradient used started from 0.225% aqueous acetic acid, pH 4.0 to 100% 90% aqueous methanol over 50 min, hold for 2 min, reduced to 0% in 2 min and hold for 10 min with a flow rate of 1 ml min−1.85 Fluorescent peaks were collected and further analysed by LC-MS using a Surveyor HPLC attached to a DecaXPplus ion trap MS (Thermo). Separation was on a Kinetex XB-C18 50 × 2.1 mm, 2.6 μm column (Phenomenex) running the following gradient of acetonitrile (B) versus 0.1% formic acid in water (A) at 300 μL min−1 and 30 °C: 0 min, 2% B; 10 min, 30% B; 20 min, 95% B; 22 min, 95% B; 22.5 min, 2% B; 26 min, 2% B. Bimane-labelled thiols were detected by UV at 390 nm, and positive electrospray MS. MS spectra were collected from m/z 250–2000 and MS2 spectra of the most abundant ions were collected at an isolation width of m/z 4.0 and 35% collision energy. Spray chamber conditions were 50 units sheath gas, 5 units aux gas, 350 °C capillary temperature, and a spray voltage of 3.8 kV using a steel needle kit.
000 rpm, flash cooled in liquid nitrogen and stored at −80 °C until analysis. 20 mg of cells were extracted into 50% acetonitrile at 60 °C and the thiols were labelled with monobromobimane (Invitrogen) and separated on an ACE AR C18 4.6 × 250 mm, 5 μm (HiChrom) by HPLC (JASCO) with fluorescence detection (JASCO FP2020 Plus) with excitation at 385 nm and emission at 460 nm. The gradient used started from 0.225% aqueous acetic acid, pH 4.0 to 100% 90% aqueous methanol over 50 min, hold for 2 min, reduced to 0% in 2 min and hold for 10 min with a flow rate of 1 ml min−1.85 Fluorescent peaks were collected and further analysed by LC-MS using a Surveyor HPLC attached to a DecaXPplus ion trap MS (Thermo). Separation was on a Kinetex XB-C18 50 × 2.1 mm, 2.6 μm column (Phenomenex) running the following gradient of acetonitrile (B) versus 0.1% formic acid in water (A) at 300 μL min−1 and 30 °C: 0 min, 2% B; 10 min, 30% B; 20 min, 95% B; 22 min, 95% B; 22.5 min, 2% B; 26 min, 2% B. Bimane-labelled thiols were detected by UV at 390 nm, and positive electrospray MS. MS spectra were collected from m/z 250–2000 and MS2 spectra of the most abundant ions were collected at an isolation width of m/z 4.0 and 35% collision energy. Spray chamber conditions were 50 units sheath gas, 5 units aux gas, 350 °C capillary temperature, and a spray voltage of 3.8 kV using a steel needle kit.
Accession codes
Sequence reads were deposited in the Sequence Read Archive under submission number PRJEB10085.Acknowledgements
These studies were supported by Basic Technology Grant GR/S79268/02 from Research Councils UK, the UK BBSRC Institute Strategic Programme Grant on Understanding and Exploiting Metabolism (MET) [BB/J004561/1], the John Innes Foundation and NIES/NSF [R01 ES21968-1 for PVZ]. We thank Tom Turner for assistance with RNA preparation and Mark Field for advice on protozoan biology.References
- C. Dobell and A. van Leeuwenhoek, Antony van Leeuwenhoek and his “Little animals”: being some account of the father of protozoology and bacteriology and his multifarious discoveries in these disciplines; collected, translated, and edited, from his printed works, unpublished manuscripts, and contemporary records, Bale., 1932.
- A. Van Leeuwenhoek, Philos. Trans. R. Soc. London, 1674, 9, 178–182 Search PubMed.
- J. Harris, Philos. Trans. R. Soc. London, 1695, 19, 254–259 CrossRef.
- H. Takeyama, A. Kanamaru, Y. Yoshino, H. Kakuta, Y. Kawamura and T. Matsunaga, Biotechnol. Bioeng., 1997, 53, 185–190 CrossRef CAS.
- E. D. Korn, J. Lipid Res., 1964, 5, 352–362 CAS.
- J. S. Rodríguez-Zavala, M. A. Ortiz-Cruz, G. Mendoza-Hernández and R. Moreno-Sánchez, J. Appl. Microbiol., 2010, 109, 2160–2172 CrossRef PubMed.
- Y. Kondo, A. Kato, H. Hojo, S. Nozoe, M. Takeuchi and K. Ochi, J. Pharmacobio-Dyn., 1992, 15, 617–621 CrossRef CAS.
- N. Koizumi, H. Sakagami, A. Utsumi, S. Fujinaga, M. Takeda, K. Asano, I. Sugawara, S. Ichikawa, H. Kondo, S. Mori, K. Miyatake, Y. Nakano, H. Nakashima, T. Murakami, N. Miyano and N. Yamamoto, Antiviral Res., 1993, 21, 1–14 CrossRef CAS.
- H. Inui, K. Miyatake, Y. Nakano and S. Kitaoka, FEBS Lett., 1982, 150, 89–93 CrossRef CAS.
- S. Tucci, R. Vacula, J. Krajcovic, P. Proksch and W. Martin, J. Eukaryotic Microbiol., 2010, 57, 63–69 CrossRef CAS PubMed.
- S. Dasgupta, J. Fang, S. S. Brake, S. T. Hasiotis and L. Zhang, Chem. Geol., 2012, 306–307, 139–145 CrossRef CAS PubMed.
- K. Y. Park, B.-R. Lim and K. Lee, Water Sci. Technol., 2009, 59, 2111–2116 CrossRef CAS PubMed.
- H. N. Guttman and H. B. Funk, Nature, 1967, 213, 103–104 CrossRef CAS PubMed.
- B. B. Anderson, J. Clin. Pathol., 1964, 17, 14–26 CrossRef CAS.
- R. Brayner, S. A. Dahoumane, C. Yéprémian, C. Djediat, M. l. Meyer, A. Couté and F. Fiévet, Langmuir, 2010, 26, 6522–6528 CrossRef CAS PubMed.
- K. Ozasa, J. Lee, S. Song, M. Hara and M. Maeda, Appl. Soft Comput., 2013, 13, 527–538 CrossRef PubMed.
- R. Brayner, T. Coradin, P. Beaunier, J.-M. Grenèche, C. Djediat, C. Yéprémian, A. Couté and F. Fiévet, Colloids Surf., B, 2012, 93, 20–23 CrossRef CAS PubMed.
- Y. Li, X. Tang, W. Song, L. Zhu, X. Liu, X. Yan, C. Jin and Q. Ren, IET Nanobiotechnol., 2015, 9, 19–26 CrossRef PubMed.
- S. M. Strauch, P. Richter, M. Schuster and D. P. Häder, J. Plant Physiol., 2010, 167, 41–46 CrossRef CAS PubMed.
- D.-P. Häderi, A. Rosumi, J. Schäfer and R. Hemmersbach, J. Plant Physiol., 1995, 146, 474–480 CrossRef.
- D. E. Buetow, The biology of Euglena: general biology and ultrastructure, Academic Press, 1968 Search PubMed.
- D. E. Buetow, The biology of Euglena: biochemistry, Academic Press, 1968 Search PubMed.
- D. E. Buetow, The biology of Euglena: physiology, Academic Press, 1968 Search PubMed.
- D. E. Buetow, The biology of Euglena: subcellular biochemistry and molecular biology, Academic Press, 1989 Search PubMed.
- I. Ciugulea and R. E. Triemer, A Color Atlas of Photosynthetic Euglenoids, Michigan State University Press, 2010 Search PubMed.
- E. W. Linton, A. Karnkowska-Ishikawa, J. I. Kim, W. Shin, M. S. Bennett, J. Kwiatowski, B. Zakrys and R. E. Triemer, Protist, 2010, 161, 603–619 CrossRef CAS PubMed.
- m. i. p. An expanded discussion of these events will be published separately: E. C. O'Neill and R. A. Field.
- P. Borst and R. Sabatini, Annu. Rev. Microbiol., 2008, 62, 235–251 CrossRef CAS PubMed.
- L.-H. Tessier, R. L. Chan, M. Keller, J.-H. Weil and P. Imbault, FEBS Lett., 1992, 304, 252–255 CrossRef CAS.
- H. Rismani-Yazdi, B. Z. Haznedaroglu, K. Bibby and J. Peccia, BMC Genomics, 2011, 12, 148 CrossRef CAS PubMed.
- P. J. Keeling, F. Burki, H. M. Wilcox, B. Allam, E. E. Allen, L. A. Amaral-Zettler, E. V. Armbrust, J. M. Archibald, A. K. Bharti, C. J. Bell, B. Beszteri, K. D. Bidle, C. T. Cameron, L. Campbell, D. A. Caron, R. A. Cattolico, J. L. Collier, K. Coyne, S. K. Davy, P. Deschamps, S. T. Dyhrman, B. Edvardsen, R. D. Gates, C. J. Gobler, S. J. Greenwood, S. M. Guida, J. L. Jacobi, K. S. Jakobsen, E. R. James, B. Jenkins, U. John, M. D. Johnson, A. R. Juhl, A. Kamp, L. A. Katz, R. Kiene, A. Kudryavtsev, B. S. Leander, S. Lin, C. Lovejoy, D. Lynn, A. Marchetti, G. McManus, A. M. Nedelcu, S. Menden-Deuer, C. Miceli, T. Mock, M. Montresor, M. A. Moran, S. Murray, G. Nadathur, S. Nagai, P. B. Ngam, B. Palenik, J. Pawlowski, G. Petroni, G. Piganeau, M. C. Posewitz, K. Rengefors, G. Romano, M. E. Rumpho, T. Rynearson, K. B. Schilling, D. C. Schroeder, A. G. B. Simpson, C. H. Slamovits, D. R. Smith, G. J. Smith, S. R. Smith, H. M. Sosik, P. Stief, E. Theriot, S. N. Twary, P. E. Umale, D. Vaulot, B. Wawrik, G. L. Wheeler, W. H. Wilson, Y. Xu, A. Zingone and A. Z. Worden, PLoS Biol., 2014, 12, e1001889 Search PubMed.
- M. G. Grabherr, B. J. Haas, M. Yassour, J. Z. Levin, D. A. Thompson, I. Amit, X. Adiconis, L. Fan, R. Raychowdhury, Q. Zeng, Z. Chen, E. Mauceli, N. Hacohen, A. Gnirke, N. Rhind, F. di Palma, B. W. Birren, C. Nusbaum, K. Lindblad-Toh, N. Friedman and A. Regev, Nat. Biotechnol., 2011, 29, 644–652 CrossRef CAS PubMed.
- E. L. Lasda and T. Blumenthal, Wiley Interdiscip. Rev.: RNA, 2011, 2, 417–434 CAS.
- L. H. Tessier, M. Keller, R. L. Chan, R. Fournier, J. H. Weil and P. Imbault, EMBO J., 1991, 10, 2621–2625 CAS.
- I. Coppens and P.-J. Courtoy, Exp. Parasitol., 1996, 82, 76–85 CrossRef CAS PubMed.
- H. K. Lichtenthaler, Annu. Rev. Plant Physiol. Plant Mol. Biol., 1999, 50, 47–65 CrossRef CAS PubMed.
- D. Kim, M. R. Filtz and P. J. Proteau, J. Nat. Prod., 2004, 67, 1067–1069 CrossRef CAS PubMed.
- T. L. Jahn, Manual of phycology: an introduction to the algae and their biology, Chronica Botanica Co., 1951 Search PubMed.
- D. R. Barras and B. A. Stone, Biochem. J., 1965, 97, 14–15 Search PubMed.
- B. L. Cantarel, P. M. Coutinho, C. Rancurel, T. Bernard, V. Lombard and B. Henrissat, Nucleic Acids Res., 2009, 37, D233–D238 CrossRef CAS PubMed.
- A. K. Monfils, R. E. Triemer and E. F. Bellairs, Phycologia, 2011, 50, 156–169 CrossRef.
- D. Baumer, A. Preisfeld and H. G. Ruppel, J. Phycol., 2001, 37, 38–46 CrossRef CAS.
- J. Lomako, W. M. Lomako and W. J. Whelan, Biochim. Biophys. Acta, Gen. Subj., 2004, 1673, 45–55 CrossRef CAS PubMed.
- I. M. Saxena, R. M. Brown, M. Fevre, R. A. Geremia and B. Henrissat, J. Bacteriol., 1995, 177, 1419–1424 CAS.
- D. R. Barras and B. A. Stone, Biochim. Biophys. Acta, 1969, 191, 329–341 CrossRef CAS.
- D. R. Barras and B. A. Stone, Biochim. Biophys. Acta, 1969, 191, 342–353 CrossRef CAS.
- E. C. O’Neill and R. A. Field, Carbohydr. Res., 2015, 403, 23–37 CrossRef PubMed.
- S. H. Goldemberg, L. R. Maréchal and B. C. De Souza, J. Biol. Chem., 1966, 241, 45–50 CAS.
- M. Kitaoka, T. Sasaki and H. Taniguchi, Arch. Biochem. Biophys., 1993, 304, 508–514 CrossRef CAS PubMed.
- Y. Yamamoto, D. Kawashima, A. Hashizume, M. Hisamatsu and N. Isono, Biosci., Biotechnol., Biochem., 2013, 77, 1949–1954 CrossRef CAS PubMed.
- M. Kitaoka, Y. Matsuoka, K. Mori, M. Nishimoto and K. Hayashi, Biosci., Biotechnol., Biochem., 2012, 76, 343–348 CrossRef CAS PubMed.
- T. Nihira, Y. Saito, M. Kitaoka, M. Nishimoto, K. I. Otsubo and H. Nakai, Carbohydr. Res., 2012, 361, 49–54 CrossRef CAS PubMed.
- S. A. Longwell and D. H. Dube, Curr. Opin. Chem. Biol., 2013, 17, 41–48 CrossRef CAS PubMed.
- P. Burda and M. Aebi, Biochim. Biophys. Acta, Gen. Subj., 1999, 1426, 239–257 CrossRef CAS.
- L. de la Canal and A. J. Parodi, Comp. Biochem. Physiol., Part B: Biochem. Mol. Biol., 1985, 81, 803–805 CrossRef.
- J. Samuelson, S. Banerjee, P. Magnelli, J. Cui, D. J. Kelleher, R. Gilmore and P. W. Robbins, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 1548–1553 CrossRef CAS PubMed.
- M. Geetha-Habib and G. B. Bouck, J. Cell Biol., 1982, 93, 432–441 CrossRef CAS.
- G. B. Bouck, A. Rogalski and A. Valaitis, J. Cell Biol., 1978, 77, 805–826 CrossRef CAS.
- H. Ankel, E. Ankel, D. S. Feingold and J. S. Schutzba, Biochim. Biophys. Acta, 1967, 136, 172–175 CrossRef CAS.
- C. J. Danpure, Trends Cell Biol., 1995, 5, 230–238 CrossRef CAS.
- J. Freitag, J. Ast and M. Bolker, Nature, 2012, 485, 522–525 CrossRef CAS PubMed.
- J. S. Gebhardt, G. J. Wadsworth and B. F. Matthews, Plant Mol. Biol., 1998, 37, 99–108 CrossRef CAS.
- F. M. Medie, G. J. Davies, M. Drancourt and B. Henrissat, Nat. Rev. Microbiol., 2012, 10, 227–234 CrossRef CAS PubMed.
- T. Takahashi, Y. Ikeda, A. Tateishi, Y. Yamaguchi, M. Ishikawa and N. Taniguchi, Glycobiology, 2000, 10, 503–510 CrossRef CAS PubMed.
- Y. D. Lobsanov, P. A. Romero, B. Sleno, B. Yu, P. Yip, A. Herscovics and P. L. Howell, J. Biol. Chem., 2004, 279, 17921–17931 CrossRef CAS PubMed.
- A. M. Mulichak, W. Lu, H. C. Losey, C. T. Walsh and R. M. Garavito, Biochemistry, 2004, 43, 5170–5180 CrossRef CAS PubMed.
- Z. Cui, Y. Maruyama, B. Mikami, W. Hashimoto and K. Murata, J. Mol. Biol., 2007, 374, 384–398 CrossRef CAS PubMed.
- R. H. Tukey and C. P. Strassburg, Annu. Rev. Pharmacol. Toxicol., 2000, 40, 581–616 CrossRef CAS PubMed.
- A. B. Boraston, D. N. Bolam, H. J. Gilbert and G. J. Davies, Biochem. J., 2004, 382, 769–781 CrossRef CAS PubMed.
- G. Gao, C. S. Fernandez, D. Stapleton, A. S. Auster, J. Widmer, J. R. B. Dyck, B. E. Kemp and L. A. Witters, J. Biol. Chem., 1996, 271, 8675–8681 CrossRef CAS PubMed.
- F. Kerff, A. Amoroso, R. Herman, E. Sauvage, S. Petrella, P. Filée, P. Charlier, B. Joris, A. Tabuchi, N. Nikolaidis and D. J. Cosgrove, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 16876–16881 CrossRef CAS PubMed.
- T. Schallus, C. Jaeckh, K. Fehér, A. S. Palma, Y. Liu, J. C. Simpson, M. Mackeen, G. Stier, T. J. Gibson, T. Feizi, T. Pieler and C. Muhle-Goll, Mol. Biol. Cell, 2008, 19, 3404–3414 CrossRef CAS PubMed.
- T. Ishikawa and S. Shigeoka, Biosci., Biotechnol., Biochem., 2008, 72, 1143–1154 CrossRef CAS PubMed.
- T. Ishikawa, N. Tajima, H. Nishikawa, Y. Gao, M. Rapolu, H. Shibata, Y. Sawa and S. Shigeoka, Biochem. J., 2010, 426, 125–134 CrossRef CAS PubMed.
- F. A. Isherwood, Y. T. Chen and L. W. Mapson, Biochem. J., 1954, 56, 1–15 CrossRef CAS.
- N. Smirnoff and G. L. Wheeler, Crit. Rev. Biochem. Mol. Biol., 2000, 35, 291–314 CrossRef CAS PubMed.
- E. I. Urzica, L. N. Adler, M. D. Page, C. L. Linster, M. A. Arbing, D. Casero, M. Pellegrini, S. S. Merchant and S. G. Clarke, J. Biol. Chem., 2012, 287, 14234–14245 CrossRef CAS PubMed.
- G. Wheeler, T. Ishikawa, V. Pornsaksit and N. Smirnoff, eLife, 2015, 4, 1–25 Search PubMed.
- S. Shigeoka, Y. Nakano and S. Kitaoka, J. Nutr. Sci. Vitaminol., 1979, 25, 299–307 CrossRef CAS.
- T. Ishikawa, I. Masumoto, N. Iwasa, H. Nishikawa, Y. Sawa, H. Shibata, A. Nakamura, Y. Yabuta and S. Shigeoka, Biosci. Biotechnol., Biochem., 2006, 70, 2720–2726 CrossRef CAS PubMed.
- T. Ishikawa, H. Nishikawa, Y. Gao, Y. Sawa, H. Shibata, Y. Yabuta, T. Maruta and S. Shigeoka, J. Biol. Chem., 2008, 283, 31133–31141 CrossRef CAS PubMed.
- J. C. Ogbonna, Appl. Microbiol. Biotechnol., 2009, 84, 217–225 CrossRef CAS PubMed.
- P. B. McCay and M. M. King, in Basic and clinical nutrition, Vitamin E: a comprehensive treatise, ed. L. J. Machlin, Marcel Dekker, Inc., 1980, vol. 1, pp. P289–P317 Search PubMed.
- R. L. Krauth-Siegel and A. E. Leroux, Antioxid. Redox Signaling, 2012, 17, 583–607 CrossRef CAS PubMed.
- T. Koledin, G. Newton and R. Fahey, Arch. Microbiol., 2002, 178, 331–337 CrossRef CAS PubMed.
- G. L. Newton, M. Rawat, J. J. La Clair, V. K. Jothivasan, T. Budiarto, C. J. Hamilton, A. Claiborne, J. D. Helmann and R. C. Fahey, Nat. Chem. Biol., 2009, 5, 625–627 CrossRef CAS PubMed.
- M. R. Ariyanayagam and A. H. Fairlamb, Mol. Biochem. Parasitol., 2001, 115, 189–198 CrossRef CAS.
- F. Montrichard, F. Le Guen, D. L. Laval-Martin and E. Davioud-Charvet, FEBS Lett., 1999, 442, 29–33 CrossRef CAS.
- E. Tetaud, F. Manai, M. P. Barrett, K. Nadeau, C. T. Walsh and A. H. Fairlamb, J. Biol. Chem., 1998, 273, 19383–19390 CrossRef CAS PubMed.
- M. Comini, U. Menge and L. Flohe, Biol. Chem., 2003, 384, 653–656 CrossRef CAS PubMed.
- V. R. Villanueva, R. C. Adlakha and R. Calvayrac, Phytochemistry, 1980, 19, 787–790 CrossRef CAS.
- K. J. Hunter, S. A. L. Quesne and A. H. Fairlamb, Eur. J. Biochem., 1994, 226, 1019–1027 CrossRef CAS PubMed.
- A. Braunshausen and F. P. Seebeck, J. Am. Chem. Soc., 2011, 133, 1757–1759 CrossRef CAS PubMed.
- Q. Zhao, M. Wang, D. Xu, Q. Zhang and W. Liu, Nature, 2015, 518, 115–119 CrossRef CAS PubMed.
- S. Sasso, G. Pohnert, M. Lohr, M. Mittag and C. Hertweck, FEMS Microbiol. Rev., 2012, 36, 761–785 CrossRef CAS PubMed.
- B. Shen, Y.-Q. Cheng, D. Christenson Steven, H. Jiang, J. Ju, H.-J. Kwon, S.-K. Lim, W. Liu, K. Nonaka, J.-W. Seo, C. Smith Wyatt, S. Standage, G.-L. Tang, S. Van Lanen and J. Zhang, Polyketides, American Chemical Society, 2007, ch. 11, vol. 955, pp. 154–166 Search PubMed.
- P. V. Zimba, P. D. Moeller, K. Beauchesne, H. E. Lane and R. E. Triemer, Toxicon, 2010, 55, 100–104 CrossRef CAS PubMed.
- D. B. Gutierrez, A. Rafalski, K. Beauchesne, P. D. Moeller, R. E. Triemer and P. V. Zimba, Toxins, 2013, 5, 1587–1596 CrossRef CAS PubMed.
- M. Johnson, I. Zaretskaya, Y. Raytselis, Y. Merezhuk, S. McGinnis and T. L. Madden, Nucleic Acids Res., 2008, 36, W5–W9 CrossRef CAS PubMed.
- S. Anand, M. V. R. Prasad, G. Yadav, N. Kumar, J. Shehara, M. Z. Ansari and D. Mohanty, Nucleic Acids Res., 2010, 38, W487–W496 CrossRef CAS PubMed.
- B. O. Bachmann and J. Ravel, in Complex enzymes in Microbial natural product biosynthesis, part A: overview articles and peptides, ed. D. A. Hopwood, 2009, vol. 458, pp. 181–217 Search PubMed.
- R. A. Butcher, F. C. Schroeder, M. A. Fischbach, P. D. Straight, R. Kolter, C. T. Walsh and J. Clardy, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 1506–1509 CrossRef CAS PubMed.
- C. T. Walsh, J. Liu, F. Rusnak and M. Sakaitani, Chem. Rev., 1990, 90, 1105–1129 CrossRef CAS.
- T. Noguchi, A. Shinohara, A. Nishizawa, M. Asayama, T. Nakano, M. Hasegawa, K.-I. Harada, T. Nishizawa and M. Shirai, J. Gen. Appl. Microbiol., 2009, 55, 111–123 CrossRef CAS.
- L. K. Mosavi, T. J. Cammett, D. C. Desrosiers and Z. Y. Peng, Protein Sci., 2004, 13, 1435–1448 CrossRef CAS PubMed.
- H. Drechsel and G. Jung, J. Pept. Sci., 1998, 4, 147–181 CrossRef CAS.
- R. B. Hallick, L. Hong, R. G. Drager, M. R. Favreau, A. Monfort, B. Orsat, A. Spielmann and E. Stutz, Nucleic Acids Res., 1993, 21, 3537–3544 CrossRef CAS PubMed.
- W. Li and A. Godzik, Bioinformatics, 2006, 22, 1658–1659 CrossRef CAS PubMed.
- E. Rebuffet, A. Groisillier, A. Thompson, A. Jeudy, T. Barbeyron, M. Czjzek and G. Michel, Environ. Microbiol., 2011, 13, 1253–1270 CrossRef CAS PubMed.
- S. F. Altschul, T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller and D. J. Lipman, Nucleic Acids Res., 1997, 25, 3389–3402 CrossRef CAS PubMed.
- S. R. Eddy, Bioinformatics, 1998, 14, 755–763 CrossRef CAS PubMed.
- I. Letunic and P. Bork, Nucleic Acids Res., 2011, 39, W475–W478 CrossRef CAS PubMed.
- A. D. Tomos and D. H. Northcote, Biochem. J., 1978, 174, 283–290 CrossRef CAS.
- K. Vogel and A. A. Barber, J. Protozool., 1968, 15, 657–662 CrossRef CAS PubMed.
- L. R. Marechal and S. H. Goldemberg, Biochem. Biophys. Res. Commun., 1963, 13, 106–107 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5mb00319a |
| ‡ It should be noted that whilst some Euglena gracilis sequences already available in the databases match the transcripts presented here, many do not. This is likely due to the strains used in other studies not being related to the one used in the present study: caution should be taken when comparing sequence data from different algal isolates. |
| This journal is © The Royal Society of Chemistry 2015 |