Fatty acid biosynthesis revisited: structure elucidation and metabolic engineering
Joris
Beld
,
D. John
Lee
and
Michael D.
Burkart
*
Department of Chemistry and Biochemistry, University of California San Diego, 9500 Gilman Drive, La Jolla, CA 92093-0358, USA. E-mail: mburkart@ucsd.edu; Web: http://burkartlab.ucsd.edu
First published on 20th October 2014
Abstract
Fatty acids are primary metabolites synthesized by complex, elegant, and essential biosynthetic machinery. Fatty acid synthases resemble an iterative assembly line, with an acyl carrier protein conveying the growing fatty acid to necessary enzymatic domains for modification. Each catalytic domain is a unique enzyme spanning a wide range of folds and structures. Although they harbor the same enzymatic activities, two different types of fatty acid synthase architectures are observed in nature. During recent years, strained petroleum supplies have driven interest in engineering organisms to either produce more fatty acids or specific high value products. Such efforts require a fundamental understanding of the enzymatic activities and regulation of fatty acid synthases. Despite more than one hundred years of research, we continue to learn new lessons about fatty acid synthases' many intricate structural and regulatory elements. In this review, we summarize each enzymatic domain and discuss efforts to engineer fatty acid synthases, providing some clues to important challenges and opportunities in the field.
Introduction
Sparked by renewed interest in biofuels, green chemicals, and antibiotic research, the past decade has seen new research on fatty acid synthases (FASs). Although innovative technologies in petroleum discovery have temporarily met some of our demand for oil,1 these appear to provide short term solutions for increasing hydrocarbon demands. Our future requires alternative energy sources not only to replace liquid fuels, but also to provide renewable feedstocks for chemicals and consumables.2 With current capital investments focused on petrochemical processes, a direct petroleum replacement, or “drop-in” solution, would prove most attractive. Crude oil is a fatty-acid rich mixture of hydrocarbons formed over millions of years from ancient biomass. One critical challenge of our time is to harness the power of photosynthesis to produce large quantities of fatty acids in a much shorter timeframe. In order to improve yield or design tailored products, we must understand the underlying metabolic process of fatty acid biosynthesis and its peripheral partners.Nature produces fatty acids for biological scaffolding, such as cell walls, membranes, and protein modification; as energy storage, such as triacylglycerides; and as building blocks for primary and secondary metabolites including biotin and lipoic acid. All organisms make fatty acids using highly conserved chemistries. Typically, fatty acid biosynthesis begins with acetyl-CoA, carboxylation produces the malonyl-CoA building blocks that are subsequently condensed and reduced in an iterative fashion until the fatty acid chain matures for use by the cell. In bacteria, plants, and algae the different enzymes that catalyze this cycle are expressed as discrete proteins (type II), whereas non-plant eukaryotes use a FAS in which all functionality is supplied by multi-domain megasynthases (type I) (Fig. 1). In fungi, the synthase is encoded on two genes and assembles as a heterododecamer of 2.6 MDa, whereas the human FAS is encoded on one gene and forms a homodimer of 540 kDa. It is believed that type I synthases derive from an ancient gene fusion event of type II enzymes.3 Most eukaryotes harbor an additional type II FAS in their mitochondria, presumably dedicated to the biosynthesis of lipoic acid.4
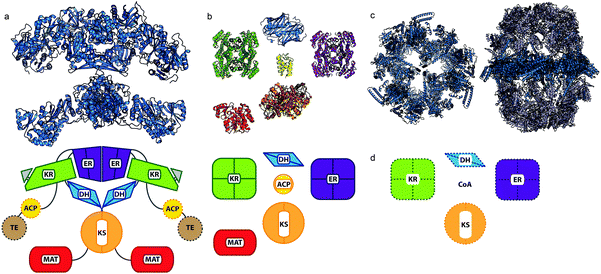 | ||
| Fig. 1 Comparison of FAS domain organizations. (a) Mammalian type I synthase (PDB: 2VZ8) structure with a diagram below of the domain organization. Domains that were not observed in the crystal structure are denoted by hashed borders. (b) E. coli type II discrete structures with a comparative diagram below denoting multimeric states of the proteins. (PDB: KR, 1Q7B; DH, 1MKB; ER, 1DFI; ACP, 2FAD; KS, 2VB9; MAT, 2G2Z) (c) Two fungal type I synthases demonstrating increased scaffolding elements and organization compared to the mammalian type I synthase in (a). At left, top-view cross-section of FAS from yeast Saccharomyces cerevisiae (PDB: 2UV8). At right, thermophilic fungus Thermomyces lanuginosus (PDB: 2UVB, 2UVC). (d) Domain organization for the ACP-independent type II-like FAS discovered in archaea, from which no structures are currently available. | ||
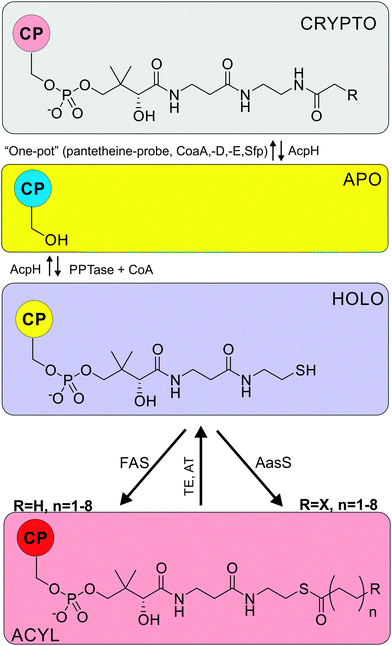
Both type I and type II FAS rely on a small protein, the acyl carrier protein (ACP, Table 1), to shuttle the fatty acid cargo from enzyme to enzyme. Apo-ACP must be activated via post-translational modification by a phosphopantetheinyl transferase (PPTase) to install a coenzyme A (CoA) derived 4′-phosphopantetheine arm (PPant) (see Fig. 3), forming holo-ACP. The terminal thiol forms the tether upon which the growing fatty acid chain is iteratively extended by ketoacyl synthases (KS, Table 1), reduced by ketoacyl ACP reductases (KR), dehydrated by dehydratases (DH), and further reduced by enoyl ACP reductases (ER), until being offloaded by dedicated thioesterases (TE) or acyltransferases (AT) (Fig. 10).
| Abbrv. 1 | Abbrv. 2 | Name | E.c. | Ref. |
|---|---|---|---|---|
| ACP | AcpP | Acyl carrier protein | — | 7–9 |
| AcpH | AcpH | ACP phosphodiesterase, ACP hydrolase | 3.1.4.14 | 10 |
| AcpS | AcpS | Holo-ACP synthase, phosphopantetheinyl transferase | 2.7.8.7 | 11 |
| DH | FabA | β-Hydroxydecanoyl-ACP dehydratase II | 5.3.3.14 | 12 |
| KS1 | FabB | β-Ketoacyl-ACP synthase I | 4.2.3.14 | 13 and 14 |
| — | FabC | Acyl carrier protein (old name, found to be AcpP) | — | — |
| AT | FabD | Malonyl-CoA:ACP transacylase | 2.3.1.39 | 15 |
| AccB | FabE | Biotin carboxyl carrier protein of ACCase | — | 16 |
| KS2 | FabF | β-Ketoacyl-ACP synthase II | 2.3.1.179 | 14 |
| KR | FabG | β-Ketoacyl-ACP reductase | 1.1.1.100 | 17 |
| KS3 | FabH | β-Ketoacyl-ACP synthase III | 2.3.1.180 | 18 and 19 |
| ER | FabI | Enoyl-ACP reductase I | 1.3.1.9 | 20 |
| — | FabJ | Ketoacyl synthase (found to be FabF) | — | 21 |
| ER | FabK | Enoyl-ACP reductase II (e.g. Streptococci) | 1.3.1.9 | 22 |
| ER | FabL | Enoyl-ACP reductase III (e.g. Bacilli) | 1.3.1.104 | 23 |
| — | FabM | cis-3-decenoyl-ACP isomerase | 5.3.3.14 | 24 |
| — | FabQ | Dehydratase/isomerase | n/a | 25 |
| — | FabR | Transcriptional repressor | — | 26 |
| — | FabT | Transcriptional regulator | — | 27 |
| ER | FabV | Enoyl-ACP reductase IV (e.g. Vibrii) | 1.3.1.9 | 28 |
| DH | FabZ | β-Hydroxyacyl-ACP dehydratase I | 4.2.1.59 | 12 |
| TE | FatA/B | Acyl-ACP thioesterase | 3.1.2.14 | 29–32 |
Archaea are well known to lack traditional fatty acid-based lipids so it was long assumed that they did not have a FAS. However, a non-typical FAS was recently discovered in archaea that appears to be ACP independent (Fig. 1).5,6 This discovery may offer an opportunity to study FAS in a unique regulatory and metabolic environment.
In this review we focus on what is known about the different enzymatic domains, their structure and function, and protein-level engineering efforts. Unless otherwise stated discussion will focus on the FAS pathway from E. coli, the most well studied system. Finally, we briefly discuss metabolic engineering of the fatty acid synthase for the production of fuel or other products.
FAS transport: ACP
Central to fatty acid biosynthesis, ACPs transport and present the growing acyl chain to appropriate reaction partners for elongation and fatty acid production. These relatively small proteins exhibit α-helical bundle topography and are dynamic in nature, making them challenging to study by crystallography.ACPs require dynamic movement because they are acted upon by many enzymes, each of which is a binding partner. ACPs from type I synthases are tethered to the megasynthase by flexible linkers in the peptide chain, allowing the ACP to sample enzymatic domains for biosynthesis. ACPs from type II synthases, however, are discrete proteins that must deliver intermediates to independent catalytic partners. Due to its central metabolic role and vital interactions with many partner proteins, ACP is both a prime target and an important consideration for metabolic engineering efforts.
Because type I FASs are so large, much structural information is based on individual domains cloned out of the full synthase. A few such truncated ACP structures are available, such as the rat ACP (PDB: 2PNG)33 and the human ACP in complex with its PPTase (PDB: 2CG5).34 One 3.2 Å crystal structure of the mammalian type I FAS is available with five domains resolved (PDB: 2VZ8), but the ACP and TE remained unresolved.35 This structure demonstrates a global “ginger-bread man” quaternary structure facilitating the interaction of the ACP with the partner domains (Fig. 1).
An organizationally different approach is observed in the fungal type I FAS, acquired at 3.1 Å resolution forming a large “soccer-ball” heterododecamer (α6β6) (PDB: 2UVB).36 In this structure three ACPs are tethered in each of two chambers within the oligomer, with three sets of enzymatic domains lining the chambers. The distances between reactive domains require that ACPs travel up to 130 Å, which is facilitated by flexible linkers.37 Extensive computer simulation work found that each ACP bounces stochastically within this chamber, but with asymmetric probability observed for multiple ACPs to simultaneously interact with equivalent catalytic sites.38 The authors conclude this to be an entropic phenomenon and hesitate to suggest cooperativity between subunits. The yeast type I FAS (PDB: 3HMJ)39 offers a similar barrel shape with a different domain arrangement, further studied by electron microscopy (EM).40 A larger reaction chamber than the previously published fungal FAS was observed by crystallography, with variable domain occupancy by the ACPs suggesting a stochastic role for the ACP within type I complexes. An excellent current review by Grininger discusses type I structure and the evolution of these megasynthases.41
Available structures of ACPs from type II synthases demonstrate a different approach to the challenge of processivity. Higher copy numbers, up to 0.25% of soluble protein,42,43 are observed. In order to transport reactive intermediates through the cytosol, type II ACPs protect their cargo by sequestration within a central hydrophobic cleft. Sequestration satisfies multiple roles: protecting the thioester linkage from hydrolysis and premature product release; shielding reactive intermediates from side-reactions; providing a limiting “ruler” to control final metabolite size; and inducing conformational changes in the ACP that trigger downstream catalytic steps.44
The nature of sequestration has been studied from many different approaches. Looking into the hydrophobic pocket, crystallographic studies of acylated ACPs yielded several structures showing an expandable hydrophobic pocket within the helical bundle accommodating the growing chain (PDB: 1LOH, 2FAC, 2FAD, 2FAE).45 This was also observed by mutagenesis, which found a chain length dependence on ACP fatty acid stabilization.46 Solution-state NMR studies of a spinach ACP loaded with decanoate and stearate found that the hydrophobic pocket could comfortably sequester ten carbons, but no conformational changes allowed it to successfully sequester the full stearate.47 The 18 carbon chain forms a flexible hairpin structure, which may represent a recognition motif for thioesterase activity. In contrast with these type II systems, the type I mammalian ACPs do not sequester the acyl chain as demonstrated by NMR.33 Acyl states of the ACP showed no chemical shift perturbations when compared to the holo state, and Nuclear Overhauser Effects (NOEs, spin–spin coupling through space observed by NMR) between the ACP and the acyl chain were not observed.
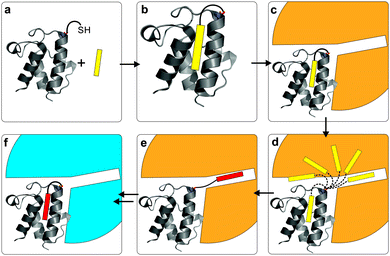
Recent work from our group provides the first mechanism-based structural glimpse of how a sequestered intermediate in the hydrophobic pocket is presented to a partner protein (PDB: 4KEH).48 The Escherichia coli ACP was covalently crosslinked to a DH, FabA (Table 1), by a mechanism-based PPant probe that mimics the natural chemistry. In the structure, the long helix II of the ACP is anchored by electrostatic interactions with the DH, while an arginine patch pulls helix III away. This allows the translocation of the acyl chain from the hydrophobic pocket of the ACP into the active site of the DH. This translocation (see Fig. 2) has been called the ‘switchblade mechanism’, a mnemonic introduced by Ban and co-workers in 2006.49 Recently, Cronan argued against this nomenclature, indicating that a ‘switchblade’ suggests an active expulsion of cargo from the inner core of the ACP for which there is no evidence.44 Thus, ‘chain-flipping’ has been adopted as a more accurate representation of this unique carrier protein behavior.
There are seven other structures of ACPs interacting with partner proteins available (PDB: 3EJB, 3EJD, 3EJE; 4IHF, 4IHG, 4IHH; 4ETW; 3NY7; 2XZ0; 2FHS; 1F80; 2CG5). Remarkably, all show strong interactions with helix II and significant structural perturbation of helix III. Indeed, in the crystal structure of E. coli ACP in complex with ER (PDB: 2FHS) much of the ACP is too dynamic to be resolved, with the exception of helix II.50 The binding interaction of ACP with its partners appears to be very consistent, based on currently available structural data (see Fig. 8).
A key question of processivity thus arises: does the ACP in type II FAS communicate the status of its cargo to prevent unproductive interactions or enhance productive ones? NMR studies of ACPs loaded with short biosynthetic intermediates found that helix II was conformationally consistent regardless of the intermediate, but significant changes were observed at the C-terminus of helix III.51 The authors conclude that while helix II must be used for docking, close interactions with helix III and the PPant arm may be used to communicate the cargo's status.
Foundational work on the biochemical properties of ACPs46 identified differences between the Vibrio harveyi ACP and the E. coli ACP. Divalent cations were critical for only the V. harveyi ACP structure at neutral pH. Later, NMR studies comparing the V. harveyi and E. coli ACPs confirmed both the importance of divalent cation binding and implicating a histidine near the C-terminus.52 Mutation studies found that acyl-ACP synthetase (see “Other enzymes” section of this review) functionality is dependent on the ACP structure and the presence of a hydrophobic pocket.
The first successful chimeric ACP produced was used to functionally replace the root nodulation protein NodF in Rhizobium,53 a carrier protein unrelated to the primary FAS. The chimera was composed of both the E. coli ACP and the NodF gene, with large portions of the proteins requiring changes to achieve functionality. This study clearly demonstrated that protein–protein recognition is necessary for these systems to function, and that such protein–protein recognition will need to be considered in FAS engineering efforts.
Work generating peptidyl carrier protein (PCP) ACP chimeras produced a Bacillus subtilis PCP carrying the recognition helix from the B. subtilis ACP.54 This work benefitted from co-crystal structures between the carrier protein and the PPTase, which clearly showed the arrangement of residues involved in recognition. Functionality was established through the ability of this PPTase to convert the apo-chimera to the holo-form. Although tempting to attempt to generate a “universal” carrier protein, structural research on the carrier protein55 suggests that mutagenizing partner proteins will be more successful.
Another approach is to create a “minimal” carrier protein with which to study truly essential protein–protein interactions. Found in a B. subtilis open reading frame, the 11 amino acid sequence dubbed “ybbR” maintains an α-helical structure in solution and offers one of the shortest polypeptide tag identified that can be acted on by PPTases.56,57 YbbR and similar domains are widespread in other species and may be involved in cell cycle regulation.58 Further studies are needed to explore ybbR as a true “minimal” ACP and not just a PPTase substrate.
In E. coli, ACP interacts with SpoT during carbon starvation, triggering (p)ppGpp accumulation and the stress response of the organism.59,60 SpoT is a bifunctional enzyme that can both synthesize and degrade the alarmone guanosine penta- or tetraphosphate (p)ppGpp, which is involved in the stringent response in bacteria causing the inhibition of RNA synthesis and conservation of amino acids. The interaction between SpoT and ACP only occurred with functional ACP suggesting that the ratio of acyl-ACP to holo-ACP may be sensed by the bacteria. Studying the ACP as a regulatory target complements earlier work demonstrating that apo-ACP would inhibit cell growth by stopping acyltransferase activity in lipid metabolism.61
ACP is an essential protein central to metabolism and organism growth, which presents a challenge for studying modifications in vivo. A temperature-sensitive ACP mutant and an ACP knockout carrying an arabinose-controlled supplementary ACP were generated.62 Interestingly, three of the four residues identified as critical when generating these strains were also identified by Burkart and Tsai in the ACP–DH structure as critical to ACP–DH interaction.48 The ACP knockout strain allows for complementation studies in vivo. V. harveyi ACP can complement the absence of endogenous ACP in E. coli, while both spinach63 and algal ACP cannot.32 The algal ACP interacts efficiently in vitro with an E. coli KS, suggesting that a different protein–protein interaction in its fatty acid biosynthesis is non-productive. Whether this is determined by the ACP dynamics or the partner protein recognition remains a question.
Early NMR studies of a spinach ACP observed a slow exchange between two structures.64 This was confirmed by later studies that observed an exchange between a folded and an unfolded state that was altered by the acylation state of the ACP.47 It might well be that these subtle differences in the ACP structure govern the processivity of the fatty acid synthase. The defining feature of an ACP is flexibility; in substrate, structure, and partner. The subtle communication in this flexibility cannot be overlooked, but offers opportunity and challenge, expanding both the potential of successful engineering and the strict requirements for any engineering to be successful.
Several detailed reviews of the ACP are available. A modern and exhaustive review was published by Crosby and Crump in 2012,9 a broader review was published by Chan and Vogel in 2010,7 and an excellent comparative review was published by Byers and Gong in 2007 looking at sequence and structural relationships.8
Modification of ACP: PPTases and AcpHs
ACPs transport the growing fatty acid chain by a covalent thioester tether. This thioester linkage is at the end of a PPant arm, which is post-translationally attached to the ACP (Fig. 3).11 A dedicated PPTase is responsible for this modification, transferring PPant from CoA to a conserved serine residue on apo-ACP, forming holo-ACP. The PPTase is thus essential for “activation” of carrier proteins and biosynthesis to occur.Almost every organism maintains at least one PPTase, and often multiple PPTases are found. PPTases are classified in three different subfamilies. The AcpS-type PPTases, which show limited promiscuity to their carrier protein and CoA substrates, are always involved in activating carrier proteins of type II FAS. The Sfp-type PPTases, named for the archetypical surfactin synthase activator in B. subtilis, show highly promiscuous behavior and are therefore used in many heterologous synthase expression studies. Finally, in some type I FAS megasynthases (e.g. from fungi) the PPTase is part of the synthase. The use of the PPant arm is not limited to FAS, but is also observed in secondary metabolism including polyketide- and non-ribosomal peptide synthases. We have recently published a comprehensive review on the superfamily of PPTases, encompassing many challenges and opportunities.11
In the field of metabolic engineering, PPTases have not surfaced as a prime target. This might be because the endogenous PPTase activity is sufficient, and thus not rate limiting. For example, E. coli has three PPTases: “AcpS” for FAS, the Sfp-type “EntD” for the siderophore enterobactin synthase, and the Sfp-type “AcpT” with currently unknown function. Endogenous apo-ACP is toxic to E. coli,61 so the ACP pool is almost completely in holo- or acyl-form. Heterologous overexpression of many ACPs in E. coli typically results in a mixture of apo- and holo-carrier protein, suggesting that AcpS, EntD or AcpT often manage to post-translationally modify these foreign carrier proteins.
In some organisms, primarily bacteria, an enzyme is found that facilitates the reverse transformation of a PPTase. The ACP hydrolase (AcpH), or phosphodiesterase, removes the PPant arm from holo-ACP, regenerating apo-ACP. Although this enzyme was discovered in the 1960s, difficulties with expression in E. coli have prohibited its study and use in the laboratory. The native E. coli enzyme is very active in vitro but unstable and difficult to work with.10 The Pseudomonas aeruginosa AcpH is much more amenable in vitro,65 facilitating its use in the laboratory to detach labeled cargo or recycling previously labeled carrier proteins.66–68
AcpH is able to efficiently remove the PPant from holo-ACP, inactivating ACP. Cronan and co-workers observe that if AcpH is as active in vivo as in vitro there would be no holo-ACP present, since AcpH is much more active than the PPTase AcpS.70 Thus, it seems likely that there is tight regulation of AcpH activity. In the context of metabolic engineering this enzyme has been overlooked and opens up some interesting possibilities for tricking species into up-regulating FAB.
Chain initiation: MCAT
Malonyl-CoA ACP transacylase (MCAT) catalyzes the initiation of fatty acid biosynthesis by conversion of malonyl-CoA to malonyl-ACP (Fig. 4), the key building block in fatty acid synthesis for all three ketoacyl synthase (KS) enzymes.71,72 MCAT is transiently malonylated at an active site serine residue (Ser92 in E. coli). E. coli MCAT can then transfer the malonyl group to a variety of substrates in vitro, including CoA, ACP, pantetheine, N-(N-acetyl-β-alanyl)-cysteamine or N-acetylcysteamine.71 Interestingly, the enzyme does not catalyze loading of acetyl-CoA.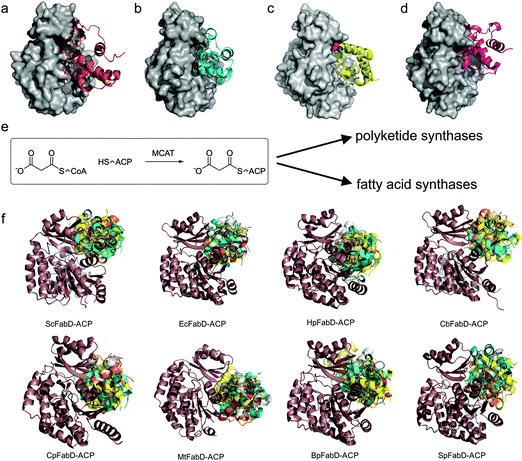 | ||
| Fig. 4 Malonyl-CoA acyltransferases (MCAT/FabD). (a–d) Similar to Arthur et al.89 we used several protein–protein docking servers to visualize the tentative interaction between ACP and MCAT. Using Patchdock,106 Grammx107 and Cluspro,108E. coli ACP (PDB: 1T8K) was docked onto E. coli FabD (PDB: 2G1H) and compared with the modeled structure of ScACP and ScFabD, PDB: 1NNZ, shown in A.88 (b) Low energy state observed by all three methods, while (c and d) were observed by two. (e) reaction catalyzed by MCAT/FabD. (f) Published structures of malonyl-CoA acyltransferases (MCAT/FabD) show very close homology. Here, we docked using Cluspro108 to various MCAT X-ray crystal structures (in dark brown) E. coli ACP, showing the top 5 hits (protein bundles in multicolor). From top left to bottom right: 1NM2 (from Streptomyces coelicolor), 2G1H (from E. coli), 2H1Y (from Helicobacter pylori), 3TQE (from Coxiella burnetii), 3PTW (from Clostridium perfringens), 2QC3 (from Mycobacterium tuberculosis), 3EZO (from Burkholderia pseudomallei) and 3IM8 (from Streptococcus pneumoniae). | ||
MCAT was first isolated from spinach in 1982, quickly followed by avocado and the cyanobacterium Anabaeana variabilis.73–75 Soybean harbors two isoforms of MCAT, presumably from gene duplication or alternative splicing.76 These two similar proteins demonstrate different activity and inhibition profiles, and both accept a variety of CoA analogs, with preference for malonyl-CoA. Only a single isoform of MCAT is found in the genome of Cuphea lanceolota77 and Brassica napus, two vascular plants with well-studied FAS pathways.78 The proteins show respectively 25% and 47% sequence homology to E. coli FabD, and overexpression of the B. napus enzyme in E. coli can complement an E. coli FabD mutant. Recently, the structure of MCAT from the cyanobacteria Synechocystis sp. PCC 6803 was solved, showing a 40% sequence identity with FabD from E. coli.79 Also, MCAT from S. aureus and Streptococcus pneumoniae were recently described, suggesting subtle differences in substrate specificity.80 Interestingly, the identification and characterization of a type II homologous MCAT in human mitochondria helped decipher the presence of a type II FAS in a type I FAS utilizing organism.81
To assay MCAT activity in vitro, a cumbersome radioactive assay was originally used.71 A new assay was more recently developed, coupling MCAT activity with α-ketoglutarate dehydrogenase monitoring by NAD+ reduction.82 This assay was used for screening inhibitors against MCAT from the apicomplexan parasites Eimeria tenella83 and P. falciparum.84,85
The structure of E. coli MCAT86 reveals the roles of conserved residues: Gln11/Leu93 (stabilization oxyanion hole), Ser92 (active site), Arg117 (binding of substrate) and His201 (activating Ser92).15 MCAT from Heliobacter pylori has been crystallized and its interaction with its ACP studied by computation.87 Also, the structure of MCAT from Streptomyces coelicolor and its binding to ACP was studied in detail by computation.88,89 Interestingly, detailed mutagenesis of the active site residues of S. coelicolor MCAT suggests that only Ser97 is required for activity and not two implicated nucleophiles (Ser and His) as previously published.90,91
M. tuberculosis has two MCATs at its disposal, FabD92 and FabD2, which show little sequence similarity or clear phylogeny.93,94 The structure of M. tuberculosis FabD has been reported by two labs.94,95 In one (PDB: 2QC3),94 the enzyme shows a slightly different active site, with the serine side chain rotated away 100°, which would suggest that the enzyme has a different active site topology than E. coli or Saccharomyces cerevisiae MCAT homologs. However, overlaying both MtMCAT structures (PDB: 2QC3 and 2QJ3) shows that only the orientation of the serine residue differs significantly between these structures.
It has been suggested that MCAT sits at the crossroads of the FAS and polyketide synthase (PKS), since both metabolic machines require conversion of their ACPs into malonyl-ACP by malonyl-CoA. However, the affinity of MCAT towards PKS ACPs is lower than FAS ACPs, and mutagenesis studies suggest that the binding mode of PKS ACP is different than FAS ACP.89 For example, the modeled FAS ACP–MCAT interaction from Xanthomonas sp. differs substantially from that of PKS ACP–MCAT in S. coelicolor.96
Interestingly, when assembling a small PKS in vitro it was found MCAT was not essential for product formation, suggesting that holo-ACP can directly load malonyl-CoA.97 It appeared that self-malonylation ability was an intrinsic property of polyketide carrier proteins.98 This phenomenon had also been observed in P. falciparum FAS holo-ACP.99 Subsequent work showed that PKS ACPs can catalyze the malonylation of FAS ACPs via trans-thioesterification of malonyl-CoA, malonyl-ACP (PKS) and malonyl-ACP (FAS).100 However, the same group later showed that this catalytic effect was in fact an artifact presumably due to a small contamination of the (PKS and FAS) carrier proteins with FabD itself.101 This topic is still controversial.
When E. coli MCAT was expressed in rapeseed and tobacco,102 no significant changes in fatty acid content or profile were observed suggesting that this enzyme does not catalyze a rate limiting step. However, overexpression of the protist Schizochytrium sp. MCAT in the yeast S. cerevisiae led to substantial increases in fatty acid production and biomass.103 Recently, the first microalgal MCAT was identified and expressed.104 Although transcription of MCAT increased when Nannochloropsis sp. was nitrogen starved to trigger lipid production, no correlation could be found between MCAT expression and fatty acid profile or quantity, similar to that seen in other plants.102 When the MCAT FabD and general purpose thioesterase TesA were overexpressed in E. coli, ∼11% increase in fatty acid content was observed over the TesA-only strain.105
Chain extension: ketoacyl synthases
KSs catalyze the chain extension step in FAS, via Claisen condensation to form the carbon–carbon bond (Fig. 5). Typically two or three KSs are found in type II FAS, whereas type I only contains one. The three subfamilies of initiation and elongation enzymes are all derived from a thiolase precursor enzyme.14 In this class of enzymes, the nucleophilic α-anion of the acyl-thioester is generated in either a non-decarboxylative or decarboxylative fashion, with KSs employing the latter.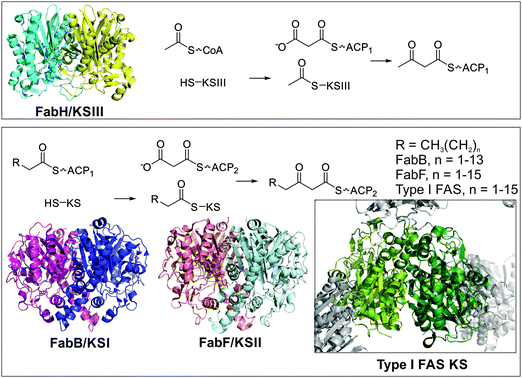 | ||
| Fig. 5 Ketoacyl synthases. The ketoacyl synthase dimer is highly conserved throughout all branches of life. Top: FabH (KSIII, PDB: 1EBL) and FabD (MCAT) are responsible for initiating fatty acid biosynthesis. The acetyl group of acetyl-CoA is transferred to the active site cysteine residue of KSIII. Malonyl-ACP (the product of FabD/MCAT) binds to KSIII and a new carbon–carbon bond is formed, while CO2 is released. Bottom: FabB (KSI, PDB: 1G5X) and FabF (KSII, PDB: 2GFW) extend the chain further by first transfer of a fatty acid from an alkyl-ACP species to a conserved cysteine residue on the ketoacyl synthase. A different carrier protein loaded with a malonyl moiety binds to the loaded KS and a new carbon-bond is formed, while CO2 is released. The type I FAS KS from pig is shown as insert, showcasing the highly conserved fold and dimer structure of these enzymes. Note that R can also be an unsaturated alkyl chain, further discussed in the ‘FAS dehydration’ section of this review. | ||
Along with MCAT, the KSIII “FabH” is responsible for the initiation of FAS. Here acetyl-CoA is loaded onto a cysteine active site residue, and malonyl-ACP is extended by two carbon units via decarboxylative addition, releasing CO2. Some organisms harbor a suite of FabHs, to facilitate the biosynthesis of various unusual fatty acids. FabH is most active on substrates with less than four carbons and is inactive with acyl-ACPs.
Overexpression of FabH leads to a general increase in shorter chain fatty acids,19 but other interesting findings have also been observed. Overexpression of FabH from B. subtilis in E. coli led to branched C15 and C17, metabolites not found in wild-type E. coli.109 Polyhydroxyalkanoate synthase, responsible for the production of polyesters of 3-hydroxyalkanoic acids as intracellular granules in many bacteria, has an important reliance on FabH. Overexpression of PhaC and FabH in E. coli led to the production of P(3HB) homopolymers in the presence of glucose.110 In the same system, mutagenesis of FabH resulted in the production of various unusual polymers.111 Most likely, FabH overexpression leads to a larger pool of acetoacetyl-ACP, which can be converted into 3-hydroxybutyryl-CoA, the monomer of PHB, or more frequent initiation of fatty acid biosynthesis, leading to higher C14 and C16, and lower C18, levels.112 Deletion of FabH hampers growth significantly and leads to enhanced C18 levels.
The two other ketoacyl synthases in E. coli are KSI FabB and KSII FabF. Both FabB and FabF show activity with saturated C4 to C14 fatty acids. FabB catalyzes the condensation of cis-3-decenoyl-ACP, cis-5-dodecenoyl-ACP and cis-7-tetradodecenoyl-ACP, with malonyl-ACP. Interestingly, strains lacking FabB are unsaturated fatty acid auxotrophs and seem to be unable to elongate cis-3-decenoyl-ACP.113 This implicates FabB in de novo production of unsaturated fatty acids in bacteria, as these organisms generally do not possess desaturases. However, overexpression of FabB does not result in an increase in unsaturated fatty acids.
In E. coli FabF has been found to be mainly responsible for the elongation of C14:1 (Cn:x, in which n = number of carbons and x = number of unsaturations in the chain). The last step to vaccenic acid (C18:1) has been demonstrated to be catalyzed by FabF and not FabB.114 Deletion of FabF leads to a temperature-sensitive mutant, whereas overexpression shows lethality. When FabF was overexpressed, a 4-fold increase in malonyl-CoA was observed due to arrest of fatty acid biosynthesis.115,116 Coexpression of FabD alleviates this effect, and it has been hypothesized that FabD forms a complex with FabF, FabH, or FabB. FabF is involved in thermal regulation of fatty acid composition of the cell membrane in E. coli.114 Interestingly, FabF and FabH are co-transcribed within the Fab cluster, whereas FabB is modulated by the FadR and FabR repressors.
Lactococcus lactis has only FabF and no FabB. Expression of this FabF can replace both FabB and FabF of E. coli, although the strain loses its thermal regulation of fatty acid unsaturation.117 Also the FabF of Clostridium acetobutylicium (which shows the same fatty acid profile as E. coli) can replace both FabB and FabF in E. coli.118 The FAS inhibitor cerulenin, which targets KSs, was found to increase the production of medium chain fatty acids in vivo. Based on this finding, FabF was mutagenized for octanoyl-ACP specificity and FabB put under inducible degradation, yielding a 12% theoretical yield of octanoate production.119
The Khosla laboratory recently reconstituted the complete E. coli FAS in vitro.120 Increasing the concentration of FabF or FabH enhances FAS activity at low concentrations but inhibits at high concentrations. On the other hand, the amount of FabB does not seem to influence activity. It is hypothesized that at high concentrations holo-ACP is removed from the pool of free holo-ACP by binding with FabF and FabH, thus lowering overall FAS activity. Supporting this conclusion are observations of acyl-ACP inhibition by FabH in vitro.121,122
Reconstituting the FAS of a cyanobacterium allows for a comparison of these bacterial synthases. Surprisingly, where FabI (ER) and FabZ (DH) are the rate limiting enzymes in E. coli, in Synechococcus sp. PCC 7002, FabH was found to be solely rate limiting.18 Additionally, FabB is not essential in Synechococcus, whereas FabF is required.
A few KSII enzymes from plants have been cloned and characterized. A mutant of Arabidopsis thaliana KSII shows an increase in palmitic acid (C16:0) and a decrease in C18 fatty acids. Similarly RNAi down-regulation of this enzyme results in a 7-fold increase in C16:0. Overexpression of CwKSII in A. thaliana reduces the amount of C16:0 and enhances the amount of C18 fatty acids. However, only a slight reduction in C16 was observed when a KSII was overexpressed in B. napus. When Jatropha curcas KSII was overexpressed in A. thaliana, the ratio of C18/C16 increased, albeit slightly.123
KSs that show promiscuity for fatty acid substrates must be able to initiate with different acyl-CoAs, as in the case of FabH; or to extend from C4 to C16, as in the case of FabB. Since these enzymes are essential to FAS, there seems to be some redundancy – e.g. the E. coli KSI can supplement the absence of KSII to a certain extent. Protein–protein interactions between KS and ACP are required for catalysis, and unrelated carrier proteins from non-ribosomal peptide synthases are not substrates, as showcased by the absence of mechanistic crosslinking in vitro.124 A chloroplastic algal FAS ACP successfully crosslinked with E. coli KSII, suggesting that the protein–protein interactions between ACP and KS are relatively permissive.32 Engineering a plant KSII into green algae resulted in minimal effects on lipid and fatty acid accumulation. Similarly, when spinach KSIII was overexpressed in tobacco, only a 5% increase in fatty acid content was observed, despite the 50-fold increase in enzymatic activity.125 Overexpression of Cuphea KSIII resulted in a 9% decrease in fatty acid content. Expression of E. coli KSIII in B. napus resulted in shorter fatty acids and growth arrest, suggesting complex regulation on fatty acid biosynthesis.126
Fungi and animals utilize a type I FAS megasynthase for fatty acid production, which contains only one KS. Recently, it has been found that this KS is responsible for the C16/C18 ratio.127S. cerevisiae has a 2.0–2.5 ratio of C16/C18 whereas Hansenula polymorpha has a ratio of 0.2–0.3, although their FAS I and FAS II subunits have 62 and 67% sequence identity. Swapping PPTase, ACP, KR and KS domains between these two fungi showed that only KS-swapping resulted in a change in C16/C18 ratio.127
All three ketoacyl synthases from E. coli have been crystallized (KSI,128 KSII,129 KSIII130,131), and demonstrate a thiolase fold that is similar to all condensing enzymes (Fig. 5).14,132 The important active site residues are Cys112, His244 and Asn274 in E. coli FabH. A Cys112Ser mutant cannot transacylate, but both His244Ala and Asn274Ala mutants show increased activity for the half-reaction. Similarly, decarboxylation is much increased in the Cys112Ser mutant but abolished in the His244Ala and Asn274Ala mutants, showing the necessity of all three residues for catalysis. The docking site of ACP onto FabH has been elucidated.133 Despite this work, there is no systematic study on how structure or active site composition controls specificity and product formation.
Reduction of ketoacyl intermediates: 3-ketoacyl-ACP reductases
In contrast to enoyl-ACP reductases (see below), surprisingly little has been published on FAS KRs, the enzymes that are responsible for reducing 3-ketoacyl-ACP to 3-hydroxyacyl-ACP, using NAD(P)H. The E. coli KR FabG was isolated, purified, and characterized in 1966.134 The enzyme appeared to be active on a range of 3-ketoacyl-ACPs with chain lengths varying between 4 and 10 carbons. FabG can function on acyl-CoAs, albeit with a large activity penalty.134 The promiscuity of KRs is also demonstrated by the development of an activity assay based on ethylacetoacetate.135Later, Cronan and co-workers showed that although there are many NAD(P)H Rossman-fold KRs annotated in the genome of E. coli, FabG is essential.136 Since the family of short-chain alcohol dehydrogenase/reductases is very large, annotation of FabGs is difficult. For example, in the genome of L. lactis two FabGs were found, but only one can complement an E. coli FabG sensitive strain.137 The difference in FabG substrate specificity has been utilized in the engineering of polyhydroxyalkanoate (PHA) biosynthesis. PHAs are biological polymers that derive from the polymerization of R-3-hydroxy fatty acid analogs of coenzyme A. When E. coli is engineered with a PHA polymerase, no medium chain length PHA was formed. However, when E. coli was co-overexpressed with P. aeruginosa FabG accumulation did occur, suggesting that this FabG shows enhanced activity on 3-ketoacyl-CoAs.110,138
The first structure of a KR was the KR of the plant B. napus.139 Each monomer of the homotetrameric enzyme harbors a Ser-Tyr-Lys triad. A comparison of the structure of E. coli FabG140 with the KR of B. napus, which was co-crystallized with NADP+, showed large conformational changes corresponding to cofactor binding.141 The tetramer from B. napus KR shows negative cooperativity in binding NADPH, which is further enhanced by the presence of ACP.140
Generation of temperature-sensitive FabG mutants in E. coli and Salmonella sp. revealed point-mutations on the twofold-axes of symmetry at the dimer interfaces, suggesting that these mutations destabilize the tetramer.17 Recently, it was shown that FabG is also essential in P. aeruginosa, and a virtual inhibitor screen of the crystallized homo-tetramer gave low nanomolar binders, which bind on the dimer–dimer interface.142
Little direct research on type I synthase KRs has been accomplished. In one study, chicken liver fatty acid synthase was digested with proteases and the 3-ketoacyl reductase isolated.143 This isolated enzyme was active on N-acetyl-S-acetoacetyl cysteamine but inactive on acetoacetyl-CoA, whereas the full synthase is active on both substrate mimics. The mammalian KR has received some attention as a potential anti-cancer target, and compounds have been identified as selective inhibitors.144
The structures of KR currently available do not explain the binding of ACP and how this influences cooperativity. To better understand this phenomenon, and its differences and similarities with ER, it is necessary to obtain structural data on the transient KR–ACP complexes.
FAS dehydration: dehydratases
After the initial reduction of ketone to alcohol by KR, the β-hydroxyacyl alcohol is dehydrated to an enoyl moiety by DH enzymes via the elimination of water (Fig. 6a). A conserved active site histidine catalyzes this reaction.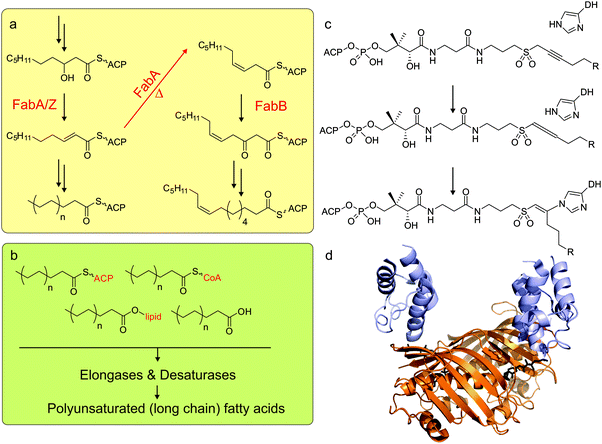 | ||
| Fig. 6 Dehydratases. (a) The bacterium E. coli has only one way to produce unsaturated fatty acids: FabA can not only reduce 3-hydroxydecanoyl-ACP, but also isomerize the double bond from trans-2-decenoyl-ACP to cis-3-decenoyl-ACP. Subsequent chain elongation by action of the other fatty acid synthase enzymes leads to the fatty acid C16:1 and C18:1. (b) Other organisms (including cyanobacteria, plants, algae) have elongases and desaturases (green panel) that can extend and desaturate the saturated C16:0 or C18:0 fatty acids, as either acyl-ACP, acyl-CoA, acyl-lipid or free fatty acids. (c) Mechanistic crosslinking between ACP and DH using a sulfonyl alkynyl pantetheinamide probe. (d) The X-ray crystal structure of mechanistically crosslinked E. coli AcpP with FabA (PDB: 4KEH). | ||
The resolved DH in the mammalian type I FAS is positioned above the ‘waist’ region of the FAS, with one pseudodimer observed per chain and two pseudodimers per complex.49 These pseudodimers are two non-identical but similar subdomains that form a single active site together.146 Several type II FAS DH structures are available, such as the homodimeric E. coli FabA (PDB: 1MKB)147 and the hexameric (trimer of homodimers) P. aeruginosa FabZ (PDB: 1U1Z).148 Both type I and type II FAS DH complexes are made up of monomers demonstrating a double-hotdog topology in which α-helixes are wrapped by β-sheets.
Membrane plasticity is controlled in part by the ratio of saturated and unsaturated fatty acids. To this end, careful control of the synthesis of unsaturated fatty acids is critical for life. Organisms employing type I FAS systems synthesize fully saturated fatty acids, then achieve unsaturation using oxygen dependent desaturases (Fig. 6b). Some bacteria, cyanobacteria, plants, and algae have desaturases as well. In contrast, many anaerobes and bacteria with type II FAS systems must use different approaches to achieve unsaturation.
In E. coli, the DH FabA offers a route to unsaturation. Both FabA and FabZ functionally eliminate the alcohol to the trans double bond, but FabA can further isomerize this to a cis 3-alkene which is not acted on by the downstream ER (Fig. 6a).149 In E. coli, FabA and the KSI FabB are co-expressed under the same operon and both are necessary for unsaturated fatty acid production. Gram positive bacteria, such as S. pneumoniae and P. falciparum lack the gene and must use a discrete isomerase to isomerize the double bond and produce unsaturated fatty acids.149B. subtilis was also found to have an independent desaturase.150 The C. acetobutylicium FabZ can replace E. coli FabZ but cannot replace FabA, and it remains unknown how C. acetobutylicium produces unsaturated fatty acids since this organism lacks a FabA.
Comparing specificity has revealed that FabZ is more promiscuous than FabA, the latter being more active on acyl-ACPs of intermediate length.151 This was demonstrated during the reconstitution of a FAS in vitro, which revealed activity relationships with the FabZ concentration consistent with promiscuity.120 Additionally, the ratio of FabB to FabA correlated to the degree of unsaturation observed.152 Taken together, these studies suggest that the positions of the unsaturations produced are controlled by substrate specificity in the FabA, while FabZ simply dehydrates what is available. However, while manipulating the FabB to FabA ratio may prove useful in an engineering scenario, in E. coli protein level feedback inhibition is necessary since FabA and FabB are co-expressed under the same operon.
Engineering attempts to manipulate unsaturation have succeeded in identifying functional replacements for the FabA gene in E. coli.13 Substitutions of the β-strands have suggested that the isomerase activity is achieved by the shape of the active site tunnel, and disruptions to the tunnel eliminated this activity.153 Later studies have found that FabZ can be inhibited by either occupying this active site tunnel's entrance or blocking the tunnel itself, both with the net effect of preventing access to the active site.154 Work on the P. falciparum FabZ has suggested that regulation of this DH is achieved by manipulating the oligomeric state of the FabZ, which may offer another mechanism to target with antibiotics.155
Similar to ERs, DHs offer an interesting drug target due to differences between type II and type I systems. To this end, activity assays using crotonoyl-CoA have been developed and P. falciparum FabZ156 and H. pylori FabZ157 have been screened against multiple inhibitors. Significant effort towards engineering mechanism-based inhibitors has led to the development of the crosslinking probe used to study the ACP FabA interaction discussed previously in the ACP section (Fig. 6c).48,158,159 A sulfonyl 3-alkynyl pantetheinamide generates a reactive allene that forms a trapped covalent bond with the active site histidine of the dehydratase (Fig. 6c).
The crosslinking probe allowed us to see that structurally, the DH offers charge-complementary electrostatic interactions to anchor helix II of the ACP and a positive patch to pry helix III away and allow substrate translocation via the chain-flipping mechanism (PDB: 4KEH, Fig. 2 and 6d).48 Interestingly, the interacting face is part of one DH monomer while the active site histidine is provided by the other, with the chain translocating to the interface between the monomers. This may offer insight into the requirement of pseudodimers in type I systems, since a simple dimer is prohibited by special requirements and two pseudodimers may offer reactive and interactive elements albeit with only one active site per chain.
Previous work on DHs included studying regulation, substrate specificity, and mechanism. With tools engineered from this data, including crotonyl-CoA assays and mechanism-based crosslinkers, we may be on the verge of engineering DHs in earnest. New structural data is becoming available offering us our first real looks at how these machines function with their partners, and the promise of manipulating unsaturation highlights the potential of producing specific chemical feedstock targets.
Reduction of enoyl intermediates: enoyl-ACP reductases
After DH activity, the resulting alkene is further reduced by an NAD(P)H dependent enoyl-ACP reductase (ER). Norris and Bloch160–162 first discovered ER during their initial studies of the FAS. Further work and characterization163,164 demonstrated functionality only on crotonyl-ACP and not crotonyl-CoA or crotonyl-pantetheine. However, it was later shown that this enzyme also works on crotonyl-CoA.165 Two different ERs were identified in E. coli, one binding NADPH and one NADH, with different specificity and substrate promiscuity characteristics.164 Thirty years later it was found that only one ER is responsible for both saturated and unsaturated fatty acid biosynthesis in E. coli.20Around the same time, the ER was also isolated from the chloroplastic type II FAS from the oilseed plant B. napus,166,167 further characterized, and its homo-tetrameric structure (Fig. 7) with Lys and Tyr active-site residues determined.168 The ER resembles a hydroxysteroid dehydrogenase and is part of the large short-chain dehydrogenase reductase (SDR) family, although it uses a different Tyr and Lys-containing motif than the classical SDR members.169 Similar to the E. coli enzyme, ER from B. napus exhibits some substrate promiscuity, with a Km of 1 μM for crotonyl-ACP and 178 μM for crotonyl-CoA. The ER gene was found by identifying the target of antibacterial diazaborines, and renamed from EnvM to FabI. In addition to diazaborines, the enzyme is inhibited to a lesser extent by palmitoyl-CoA and -ACP, and one single point-mutation (Ser241Phe) results in the loss of NADPH-while retaining NADH-dependent activity.165
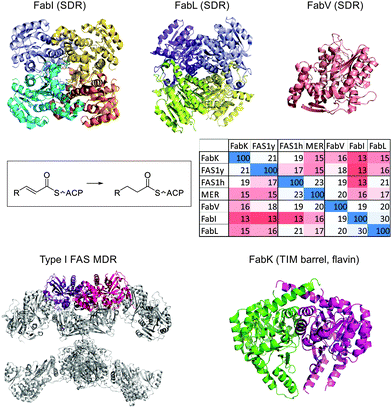 | ||
| Fig. 7 Structures of enoyl-ACP reductases. Top: FabI from E. coli (PDB: 2FHS), FabL from B. subtilis (PDB: 3OIC) and FabV from X. oryzae (PDB: 3S8M) are all members of the SDR superfamily, utilizing NAD(P)H. Whereas FabI and FabL are tetramers, FabV is a monomer. Clustal Omega was used to generate a sequence identity matrix, including FAS1y (the yeast FAS ER), FAS1h (the human FAS ER) and MER (the human mitochondrial ER). Bottom: The ER dimer in type I FAS from pig (2VZ8) is shown in pink/purple, and FabK from S. pneumoniae (2Z6I). | ||
Although many structures are available, only a few structural studies focus on the substrates of ER and not the NAD(P)H co-factor or inhibitors. InhA, the ER from the type II FAS of M. tuberculosis has been co-crystallized with a C16 fatty acid moiety170 and the E. coli FabI tetramer has been co-crystallized with two ACPs.50 In the latter work, only two ACPs are observed, whereas the tetramer has four active sites. Comparing the orientation of ACP in relation to ER with other ACP-partner protein structures (see Fig. 8) draws question to the significance of the structure.
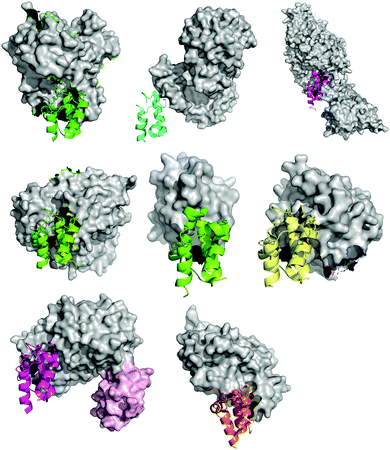 | ||
| Fig. 8 Comparison of all eight ACP-partner protein X-ray crystal structures deposited in the PDB, from the perspective of the ACP, showing the similar binding orientation of the ACP, but the different binding motifs of the partner proteins. From top to bottom and left to right: PDB: 1F80, E. coli PPTase AcpS with E. coli holo-AcpP, PDB: 2FHS, E. coli FabI with E. coli AcpP, PDB: 2XZ0, Ricinus communis stearoyl desaturase with R. communis ACP bearing a phosphoserine, PDB: 3EJB, B. subtilis P450BioI with E. coli AcpP bearing tetradecanoic acid, PDB: 3NY7, STAS domain of E. coli YchM with E. coli AcpP bearing a terminal acid propionic acid thioester, PDB: 4ETW, E. coli BioH with E. coli pimeloyl-AcpP, PDB: 4KEH, E. coli FabA mechanistically crosslinked with E. coli AcpP bearing a non-hydrolyzable pantetheinamide crosslinker (in light pink surface the second ACP) and PDB: 2CG5, Homo sapiens AASDHPPT with human FAS apo-ACP, the latter excised from the human type I FAS. | ||
ERs have received much attention in the field of antimicrobial research,171,172 especially with the discovery that the M. tuberculosis ER, InhA, is a target of isoniazid.173 FabI of the malarial causative apicomplexan P. falciparum has also been targeted.174 However, after the discovery that P. falciparum requires only de novo fatty acid biosynthesis in late growth stages, it is heavily under debate whether FAS is still a viable antimicrobial target.175
Triclosan is the most widely-used ER inhibitor, and was added to products such as hand-sanitizer and toothpaste even prior to understanding its target. Four papers in 1998 and 1999 signaled the start of triclosan-ER related research.176–179 Interestingly, triclosan also inhibits the human fatty acid synthase in vitro, albeit with IC50 values many orders of magnitude larger than those observed in E. coli or P. falciparum.180 While some bacteria are severely inhibited by triclosan, several other species have been identified that tolerate it. Either triclosan resistance originates from a mutation in FabI181 or, more commonly, several species harbor multiple ERs or different types of ERs.182B. subtilis has two ERs: one highly homologous to E. coli FabI and an alternate named FabL.23 Other bacteria, like S. pneumoniae, use the flavin dependent TIM-barrel reductase FabK.22 Interestingly, the yeast type I FAS also contains an ER with homology to flavin-binding FabK.37,183 Other bacteria, such as Vibrio cholerae, use FabV, another short-chain dehydrogenase/reductase (SDR)-fold ER but monomeric and considerably larger (Fig. 7).28 The mammalian type I FAS harbors a different MDR subfamily ER, dissimilar to the SDR-fold (FabI, FabL, and FabV), the TIM-barrel fold (FabK), or the mitochondrial MDR subfamily found in eukaryotes (Fig. 7).49 Thus, nature has found many different solutions to do the same reaction.
Very little metabolic engineering has been completed with ERs, but some insight in its effect on fatty acid levels has been gleaned from overproduction or knockout studies. While toxic, E. coli FabI could be overexpressed by supplementation with low levels of triclosan.184 Two theories about FabI toxicity involve a reduction in lipid A building blocks or enhanced competition with FabA for substrate, leading to a deficiency in unsaturated fatty acids.182 Recently, overexpression of FabI in E. coli did not result in growth retardation or any effect on fatty acid accumulation.145 In plants, increased ER expression was observed during TAG deposition.185 The ratio of plant FAS enzymes was observed to change significantly during growth, suggesting a life-cycle dependence or regulation.186
FAS termination: acyltransferases and thioesterases
After chain elongation, fatty acid biosynthesis is terminated either by offloading fatty acids from ACP by acyl-ACP TEs, releasing free fatty acids, or by direct transesterification onto a lipid by acyltransferases (AT) (Fig. 9a). In general, prokaryotes utilize ATs whereas eukaryotes make use of dedicated TEs.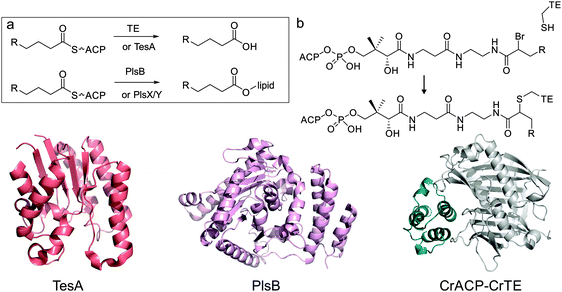 | ||
| Fig. 9 Fatty acid biosynthesis termination by thioesterases or acyltransferases. Top: (a) in plants, green algae and some bacteria, dedicated hotdog-fold acyl-ACP thioesterases (e.g. CrTE, Chlamydomonas reinhardtii Fat1) are responsible for hydrolyzing fatty acids, with a certain chain-length off the ACP (e.g. CrACP, C. reinhardtii ACP). TesA is a multifunctional enzyme that can hydrolyze fatty acids off the ACP, but this is presumably not its primary function in E. coli. In bacteria, dedicated acyl-transferases, like PlsB, trans-esterify lipid headgroups with a fatty acid, directly from the ACP. Alternatively, E. coli uses PlsX to synthesize phospho-fatty acids, which are loaded onto lipids using PlsY. (b) Mechanistic crosslinking of ACP to TE using α-bromo acid pantetheinamide probe (R = C3H7 or C13H27). Bottom: structures of TesA (PDB: 1IVN), PlsB (PDB: 1K30) and a docked structure of chloroplastic C. reinhardtii ACP (model) with C. reinhardtii TE FAT1 (model). | ||
E. coli however does harbor two α/β hydrolase TEs, TesA (Fig. 9) and TesB,187 but these have much higher activity with acyl-CoAs over acyl-ACPs and are not believed to be directly involved in fatty acid or lipid biosynthesis. TesA is a general purpose thioesterase/lipase/hydrolase that is localized in the periplasm. Overexpression of TesA in the cytosol increases the pool of free fatty acids in E. coli.188–190E. coli uses two different methods to transesterify acyl chains from acyl-ACP to lipids, involving PlsB, PlsY and PlsX, further discussed in the “Other Enzymes” section of this review.
Dedicated acyl-ACP TEs were discovered in 1991 in oil seed plants that show an unusual shorter fatty acid pattern in storage lipids.191,192 Dedicated short/medium chain TEs, FatBs,185,188,190 are responsible for the accumulation of these fatty acids. Later, it was found that plants in addition to FatB harbor FatA,193 a more general TE showing some specificity for oleic acid. These enzymes have now also been annotated in some anaerobic bacteria, mosses and algae.31 Although other eukaryotic species (e.g. fungi and mammals) also use dedicated TEs embedded in their type I fatty acid megasynthases, these TEs have a different fold (α/β hydrolase)194 than their plant/algal counterparts, which have a typical hotdog-fold.30,195 In plants these hotdog-fold TEs solely can determine the chain length of fatty acids, and there has been considerable interest in metabolic engineering and mutagenizing these enzymes.
When plant TEs are overexpressed in E. coli, fatty acid biosynthesis is altered according to the specificity of the TE.189,196–199 For example, overexpressing the C12:0-ACP selective Umbellularia californica TE “UcFatB1” in E. coli lead to a 500 fold increase in C12:0 fatty acid production.196 Mutagenesis on TEs has met mixed success and seems unpredictable, possibly because of limited structural information on these enzymes.200,201 In one case of success, it was shown that mutagenesis of UcFatB1 can change its specificity to C14:0-ACP.202 Metabolic engineering of fatty acid biosynthesis in plants203 has been demonstrated by the expression of FatB192 and FatA204 in B. napus seeds. Introduction of UcFatB1 in B. napus seeds resulted in lauric acid levels of ∼60% of the total fatty acids. Such a remarkable change is the exception; for example introduction of C8:0-ACP selective Cuphea hookeriana TE “ChTE” into rapeseed gives only 12% octanoic acid whereas the source species contains 50%.205 In general, production of short-chain fatty acids in plants has been marginally successful. When a FatB TE was overexpressed in an acyl-ACP synthetase (AasS, see “Other Enzymes”) knock-out strain of A. thaliana, a 2-fold increase in octanoic acid production was observed, suggesting that AasS not only works on exogenous acids but also plastidically produced acids.206
In contrast to plants, green algae only have one general purpose hotdog-fold TE (Fat1), and brown/red algae and diatoms seem to lack any.32,66 The green microalgae C. reinhardtii has recently been engineered with FatBs from different seed oil plants, with very limited success.32,66 Only the strain overexpressing its native Fat1 TE showed a modest increase in total fatty acids, due to the mismatch in protein–protein interactions between foreign TEs and ACP. This was observed by mechanistic crosslinking (Fig. 9b), and highlights the importance of specific interaction. Engineering of cyanobacteria (blue green algae), with plant TEs resulted in the predicted increases in short/medium chain fatty acids,207 although engineering of an algal TE into cyanobacteria did not result in this effect.208 Cyanobacteria are an attractive species since they are photosynthetic, can fix nitrogen, are genetically amenable and secrete large portions of produced fatty acids.209
Metabolic engineering of fatty acid synthases
In the past decade, we have seen large strides in the metabolic engineering of fatty acid synthases in bacteria. However, the dream of designer strains producing specific fats and feedstock remains distant. Here, we will discuss select hallmarks of metabolic engineering of fatty acid synthases in various organisms, focusing on E. coli.Important requisites for fatty acid biosynthesis are holo-ACP, malonyl-ACP and malonyl-CoA. It is assumed that the ACP itself is not rate limiting because it is present in high concentration, overexpression does not change the fatty acid profile, and because apo-ACP is toxic.61 Overexpression of AcpS (the FAS PPTase responsible for transforming apo-ACP into holo-ACP) stops growth due to strong inhibition of glycerol-3-phosphate acyltransferase,211 but can be partially alleviated by AcpH coexpression.
MCAT and KSIII are responsible for chain-initiation and thus formation of malonyl-ACP. Malonyl-CoA is produced by the ACCase complex. Overexpression of these proteins resulted in modest (1.2–1.6 fold) increases in total lipids, with a 5-fold increase in palmitic acid production when all three enzymes were overexpressed.212 Similarly, engineering ACCase subunits accA, accB, accC, MCAT and a TE from Streptococcus pyogenes into E. coli resulted in a 2.4 fold increase in fatty acid production.105 Interestingly, overexpressing the ACCase by itself did not change fatty acid production,213 presumably due to inhibition by acyl-ACPs. A combination of deletion of β-oxidation (fatty acid degradation) and overexpression of an ACCase and a plant TE (at a low copy number) did however result in a 5-fold increase in fatty acid production.213 Overexpression of MCATs from various species only resulted in slight increases in fatty acid production,214 whereas deletion of MCATs was lethal.136,215
Studies of E. coli strains overexpressing its own FAS enzymes found that overexpression of KR (FabG) and DH (FabZ) produced the largest amount of fatty acids. Additional overexpression of the ER (FabI) did not improve yield, and overexpression of the KSIII (FabH) actually reduced the amount of fatty acids, confirming the inhibitory effect of high concentrations of FabI and FabH on FAS in E. coli.216 All strains containing FabZ show a significant increase in cis-C16:1 and cis-C18:1. Overexpression of KSII (FabF) gave a 1.3-fold increase in fatty acid production at 37 °C and a 2.7-fold increase at 20 °C, in which in the latter case the amount of unsaturated fatty acids increased by 7-fold.216 Combination of FabF with other FAS genes resulted in enhanced fatty acid production at lower temperatures, except in the case of FabI, which appeared to be inhibitory.
Reconstituting the FAS of E. coli120 and a cyanobacterium18in vitro allowed for a comparison of these bacterial synthases. Surprisingly, where FabI and FabZ are the rate limiting enzymes in E. coli, in Synechococcus sp. PCC 7002, FabH is solely rate limiting.18 Cyanobacteria have desaturases at their disposal, whereas E. coli needs to regulate fatty acid unsaturation via the ratio of FabA/FabZ and FabB.
Elimination of fatty acid β-oxidation as a means to improve the accumulation of fatty acids is a popular approach. For example, knocking out FadD (fatty acid CoA-ligase), while overexpressing a plant TE, the endogenous TE TesA and ACCase, resulted in high fatty acid titers in E. coli.189 A combination of fadE (acyl-CoA dehydrogenase) deletion and overexpression of TesA resulted in a strain producing 1.2 g L−1 fatty acids,188 whereas deletion of FadD, combined with the overexpression of dedicated seed plant TEs resulted in strains producing up to 4.5 g L−1 fatty acids.213,217
Free fatty acid secretion is sought after, since it would facilitate the production of biofuel. Looking specifically at extracellular fatty acid production, a FadD knockout did not result in an increase, whereas a FadL (outer membrane fatty acid transporter) knockout, in combination with TesA overexpression resulted in a large increase in extracellular fatty acid production (from 5.5 mg L−1 to 4500 mg L−1).218 Interestingly, identification and overexpression of several E. coli fatty acid transporters did not result in a superior strain.219
Besides initiation, transport, and degradation, some core enzymes of the E. coli FAS have been overexpressed with the goal of enhancing yield. FabA, FabZ, and FabG were overexpressed in a hallmark study in which the fatty acid synthase from E. coli was reconstituted in vitro,217 allowing for detailed studies of rate-limiting steps in the synthase. Overexpression of FabZ alone also resulted in a higher fatty acid production, and a combination of fadD knockout, sucC (glycolysis) knockout, and FabZ overexpression gave a strain producing 5.7 g L−1 C14–C16 fatty acids.220 Overexpression of FabG leads to a 2–3 fold increase in fatty acid production,145 whereas overexpression of FabI showed no effect, FabH was semi-lethal, FabF was lethal and FabB in combination with FabA showed modest increases in fatty acid production. Lastly, TEs which are responsible for off-loading the fatty acid from the ACP, have been engineered and overexpressed in many studies, utilizing TEs from various plant, algal or endogenous sources, resulting in shifting the fatty acid profile towards the specificity of the TE (except in algal engineering with foreign TEs and algal TE expression in cyanobacteria).220
Besides fatty acids themselves, there is an increasing interest in coupling fatty acid biosynthesis to other metabolic products. For example, fatty alcohols can be made by overexpressing acyl-CoA synthase and acyl-CoA reductase.188,221 The discovery of an acyl-ACP reductase facilitates this process further.222 Methylketones, fatty acid methylesters, and alkanes can also be derived from fatty acids. Methylketones are made from fatty acids via β-oxidation. Fatty acids are initially converted into β-ketoacyl-CoAs, prior to hydrolysis by a TE (producing β-keto-fatty acids) and decarboxylation yielding the final methylketones.223 Fatty acid methylesters can be made directly in a living organism by expressing a wax-ester synthase224 or an S-adenosylmethionine-dependent methyltransferase.225 Alkanes or alkenes can be derived from fatty acids using multiple pathways. These include, the combination of acyl-ACP reductase and aldehyde-decarbonylase,222 or a P450-enzyme,226 or head-to-head condensation of two fatty acids.227 Recently, the identification of a wax-ester/diacylglyceride acyltransferase (WS/DGAT, AtfA) in Acinetobacter baylii228,229 and subsequent expression in E. coli, resulted in the production of triacylglycerides (TAGs) in bacteria.230 Overexpression of AtfA, FadD, and deletion of diacylglycerol kinase, resulted in a strain with high TAG titers.112
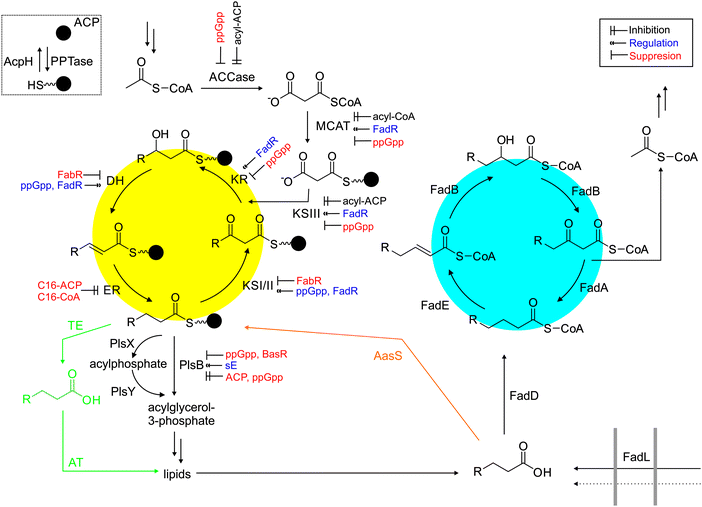
Regulation of fatty acid biosynthesis is currently a crucial bottleneck in metabolic engineering (Fig. 10).112 For example, long chain acyl-ACPs seems to regulate the flux through fatty acid synthases. Cultures starved of glycerol (limiting phospholipid biosynthesis) show a decreased rate of acyl-ACP biosynthesis. Fatty acid biosynthesis is also downregulated when PlsB (sn-glycerol-3-phosphate acyltransferase) is inhibited by the alarmone ppGpp.122 When TesA is overexpressed, acyl-CoAs and acyl-ACPs are hydrolyzed, depleting long-chain ACPs, increasing the rate of fatty acid biosynthesis. Acyl-ACPs directly inhibit the ACCase, KSIII and ER. In vitro, ER or ER/DH are the rate limiting enzymes in the fatty acid synthase, but how transcriptional or translational regulation influence fatty acid synthases is still an open question. FabR and FadR are regulators of fatty acid biosynthesis and fatty acid degradation but do not control all enzymes involved. Interestingly, coexpression of FadR and TesA resulted in a large increase of fatty acid production.231 A FadR-derived regulator, FadE deletion and TesA overexpression also resulted in robust fatty acid production (5 g L−1) in minimal media.232 FadM is another regulator only recently discovered,233 and more details of the complex regulation of fatty acid biosynthesis in E. coli continue to be elucidated.234–237
In cyanobacteria, bacterial and plant TEs have been overexpressed with success,207,238 but in general there has been relatively little development in metabolic engineering of cyanobacterial fatty acid synthases.239 In one promising work, acyl-ACP synthetase was knocked out and a plant acyl-ACP TE overexpressed, leading to accumulation of free fatty acids.238 The same strategy was later applied to Synechocystis sp. PCC 6803240 and Synechococcus elongatus PCC 7942.241 Based on this work, six generations of Synechocystis sp. PCC 6803 were constructed with a variety of genes knocked out or overexpressed (either native or from foreign hosts).209 The in vitro reconstitution of Synechococcus sp. PCC 7002 FAS opens up avenues to study the cyanobacterial FAS in more detail.18 The increased interest in biofuels during the last five years has created an explosion of papers on improving lipid content of photosynthetic organisms,242 primarily by engineering acyl-ACP thioesterases. In addition, the beneficial effect of omega fatty acids has led to many studies of overexpression of fatty acid modifying enzymes, like elongases and desaturases (see “Other Enzymes” section). We recently engineered various plant acyl-ACP thioesterases in the green microalgae C. reinhardtii, with limited success.66 Only overexpression of the native acyl-ACP thioesterase, Fat1, increased fatty acid production. When Fat1 was expressed in cyanobacteria, very little effect was observed.208 In diatoms work has focused on TEs,243 desaturases, and elongases.244 In plants, several efforts have led to strains with improved and manipulated oil content,203,241 again focused on acyl-ACP TEs,203 desaturases,245 and elongases.246
Engineering of type I synthases (e.g. fungal and mammalian) is in its infancy. Yeast FAS contains subunits FAS1 (β-subunit, AT-ER-DH and malonyl/palmitoyl-transferase) and FAS2 (α-subunit, ACP-KR-KS-PPTase, see Fig. 1), forming a heterododecameric enzyme complex of 2.6 MDa. Creating chimeric FAS by mixing FAS1 and FAS2 from S. cerevisiae and H. polymorpha, produced interesting alterations to the fatty acid profiles, but rational design is still elusive. Swapping PPTase domains did not show any difference, whereas swapping KS domains gave marked changes in fatty acid chain length.127 Recently, human type I FAS was inserted into a yeast FAS knockout strain, bearing different TE domains, and co-expressing different PPTases.247 Although the yields were modest, (∼50–100 mg L−1) short-chain fatty acids showed a 64-fold increase compared to the wild type.
Other enzymes and other functions
The fatty acid synthase is surrounded by dozens of other enzymes that either input metabolites or remove fatty acids for further modification. ACCases are responsible for the production of malonyl-CoA (Fig. 10), one of the essential starting materials of fatty acid biosynthesis. In E. coli, four proteins form the ACCase complex: biotin carboxylase, biotin carboxylic carrier protein, CoA-carboxylase and carboxyl transferase. In this reaction, biotin is carboxylated and the carboxyl group transferred to acetyl-CoA, forming malonyl-CoA. Acetyl-CoA stems from pyruvate, which is a product of glycolysis. The ACCase is inhibited by acyl-ACPs, presumably preventing the accumulation of fatty acids.249 Overexpression of the ACCase in combination with TesA resulted in significant increases in fatty acid production.250Some organisms are able to convert holo-ACP to acyl-ACP by shunting exogenous fatty acids directly into fatty acid or lipid metabolism (and not catabolism). This process is carried out by a dedicated acyl-ACP synthetase (AasS, Fig. 3 and 10), which catalyzes the activation of a free fatty acid to its AMP-ester and subsequent thioester formation with the free thiol of the PPant arm of holo-ACP. E. coli harbors a unique bifunctional membrane-bound protein251 which shows this activity in vitro, but is unable to utilize supplemented fatty acids in vivo. In contrast, V. harveyi has a soluble AasS, which can use exogenously supplied fatty acids with a preference for medium chain fatty acids.252,253 AasS has recently been found in cyanobacteria254 and plants.206
In many bacteria, fatty acids get transferred directly from acyl-ACP to acyl-glycerol-3-phosphate. On the other hand, fatty acids from algae and plants get hydrolyzed off acyl-ACP by a dedicated TE (Fig. 10). There are, at least, two ways for bacteria to transfer acyl chains from ACP to glycerophosphate. PlsX catalyzes the formation of acyl-phosphate, which is the substrate for glycerophosphate-acyltransferase PlsY. Alternatively, PlsB can directly transfer the fatty acid from ACP to glycerol-3-phosphate. The product acyl-glycerol-3-phosphate (also called mono-acylglyceride) is the substrate for enzymes that catalyze the formation of di-acylglycerides, and subsequently membrane lipids.
Lipoic acid is produced by mitochondrial type II fatty acid synthases in eukaryotes and relies on type II FAS in bacteria, starting with LipB catalyzed acyl-transfer from octanoyl-ACP to the apo-octanoyl-domain, and followed by the LipA catalyzed insertion of two sulfur atoms.255 FASs are also directly involved in the biosynthesis of biotin, elucidated in 2010. BioC is responsible for producing malonyl-CoA methyl ester, which is condensed with malonyl-ACP by KSIII. This product gets taken through a complete cycle of fatty acid biosynthesis, forming pimeloyl-ACP methyl ester, the substrate for BioH. This then forms pimeloyl-ACP, the substrate for the subsequent biotin-biosynthetic enzymes.256
Fatty acids are often found in natural products, from which calcium dependent antibiotic and daptomycin are possibly the most famous examples.257 There is also a suite of natural products that are made from mature fatty acids, including falcarinol alkynes, cicutoxin, panaxytriol, wyerone, amides of fatty acids like oleamide, ricinoleic acid, vernolic acid, lipstatin, prostaglandins, thromboxanes, leukotrienes, urushiol, jasmonic acid, ginkgolic acids, coniine, and various modified fatty acids with epoxides, methyl side chains, cyclopropanes, and even unsaturated cyclopropanes embedded in the chain. Also, many polyketide synthases require hexanoyl-CoA starter units, which are most likely derived from FAS. Fatty acids are also often found attached to proteins. This post-translational modification plays important roles in regulation of protein trafficking, signaling and behavior.258 Most common is myristoylation, the irreversible attachment of myrystic acid to an N-terminal glycine residue via an amide bond. Palmitoylation is reversible and encompasses the thioesterification of cysteine thiols.
Fatty acids are degraded by a dedicated pathway that closely resembles the reverse biosynthesis of fatty acids, but with the critical difference that CoA, and not ACP, carries the acyl cargo (Fig. 10). FadL is a transporter that can transport exogenous acids into a cell while FadD is an acyl-CoA synthetase that loads these (and other cellular) free acids onto CoA. FadE catalyzes the oxidation of acyl-CoA to enoyl-CoA. FadB hydrates enoyl-CoA to 3-hydroxyacyl-CoA and oxidizes it to 3-ketoacyl-CoA, which is the substrate for the ketothiolase FadA, forming acetyl-CoA and acyl-CoA. FadH, a 2,4-dienoyl-CoA reductase, is necessary for the processing of unsaturated fatty acids and FadM is a recently identified TE. Fatty acid degradation is stringently regulated by the acyl-CoA controlled regulator FadR, the cyclic adenosine monophosphate receptor protein–cAMP complex, (p)ppGpp, and sigma-factor RpoS.
Conclusion and outlook
Fatty acid synthases are fascinating biosynthetic machines that offer many opportunities to study biology, biochemistry, and chemistry of biological processes. In contrast to the early discovery of fatty acid degradation (β-oxidation) in 1904 by Knoop, the elucidation of fatty acid biosynthesis259 started much later with the publication of five papers by Barker and Stadtman in 1949.260–264 They exploited the then newly available 14C isotope, labeling acetate and feeding this to cell-free extracts of Clostridium kluyverii. This was followed by a large body of work by Vagelos and co-workers in the 1960s, using enzymology.265 In the 1970s, Wakil266 and co-workers showed that the eukaryotic synthase is a multidomain megasynthase, while Bloch and co-workers described fatty acid metabolism and regulation.267 Building on these milestones, the labs of Cronan, Rock and Ohlrogge have dug deeper into various aspects of fatty acid biosynthesis in bacteria and plants.It is fascinating to note that, although studied for 75 years and present in all biochemistry textbooks, we are only now uncovering the detailed features of the complex machinery of fatty acid biosynthesis. Regulation by many factors is being discovered, and we expect that many more regulatory elements will be elucidated in the coming years. Protein–protein interactions between ACP and partner proteins seem to govern processivity, and with access to new chemical biology tools268 we hope to see many more structures and predictions of these protein–protein interactions. Phylogeny of the individual FAS enzymes might also be a useful tool to anticipate differences and similarities between species' FAS. With enhanced insight into the biochemistry of this important biosynthetic process, metabolic engineering for designer oils or other FAS-derived products should become possible. Modern spectroscopic269 and ACP modifying techniques appear to be the 14C of our time, offering us the exciting opportunity to explore and engineer fatty acid biosynthesis in a new manner. Indeed, considering global energy demands and the increasing thirst for renewable liquid fuels, this opportunity may be crucial to our future.
Acknowledgements
We thank K. Finzel and N. Schoepp for critical advice. This work was supported by California Energy Commission CILMSF 500-10-039, DOE DE-EE0003373 and NIH R01GM095970.References
- R. W. Howarth, A. Ingraffea and T. Engelder, Nature, 2011, 477, 271–275 CrossRef CAS PubMed.
- S. Mayfield, Genome, 2013, 56, 551–555 CrossRef PubMed.
- A. D. McCarthy and D. G. Hardie, Trends Biochem. Sci., 1984, 9, 60–63 CrossRef CAS.
- H. Wada, D. Shintani and J. Ohlrogge, Proc. Natl. Acad. Sci. U. S. A., 1997, 94, 1591–1596 CrossRef CAS.
- J. Lombard, P. López-García and D. Moreira, Archaea, 2012, 2012 Search PubMed.
- J. Lombard, P. López-García and D. Moreira, Mol. Biol. Evol., 2012, 29, 3261–3265 CrossRef CAS PubMed.
- D. Chan and H. Vogel, Biochem. J., 2010, 430, 1–19 CrossRef CAS PubMed.
- D. M. Byers and H. Gong, Biochem. Cell Biol., 2007, 85, 649–662 CrossRef CAS PubMed.
- J. Crosby and M. P. Crump, Nat. Prod. Rep., 2012, 29, 1111–1137 RSC.
- J. Thomas, D. J. Rigden and J. E. Cronan, Biochemistry, 2007, 46, 129–136 CrossRef CAS PubMed.
- J. Beld, E. C. Sonnenschein, C. R. Vickery, J. P. Noel and M. D. Burkart, Nat. Prod. Rep., 2014, 31, 61–108 RSC.
- R. J. Heath and C. O. Rock, J. Biol. Chem., 1996, 271, 27795–27801 CrossRef CAS PubMed.
- H. Wang and J. E. Cronan, J. Biol. Chem., 2004, 279, 34489–34495 CrossRef CAS PubMed.
- C. Jiang, S. Y. Kim and D.-Y. Suh, Mol. Phylogenet. Evol., 2008, 49, 691–701 CrossRef CAS PubMed.
- C. Oefner, H. Schulz, A. D'Arcy and G. E. Dale, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2006, 62, 613–618 CrossRef PubMed.
- J. E. Cronan Jr and G. L. Waldrop, Prog. Lipid Res., 2002, 41, 407–435 CrossRef.
- C.-Y. Lai and J. E. Cronan, J. Bacteriol., 2004, 186, 1869–1878 CrossRef CAS PubMed.
- J. Kuo and C. Khosla, Metab. Eng., 2014, 53–59 CrossRef CAS PubMed.
- J.-T. Tsay, W. Oh, T. Larson, S. Jackowski and C. Rock, J. Biol. Chem., 1992, 267, 6807–6814 CAS.
- R. J. Heath and C. O. Rock, J. Biol. Chem., 1995, 270, 26538–26542 CrossRef CAS PubMed.
- K. Magnuson, M. R. Carey and J. Cronan, J. Bacteriol., 1995, 177, 3593–3595 CAS.
- H. Marrakchi, W. DeWolf Jr, C. Quinn, J. West, B. Polizzi, C. So, D. Holmes, S. Reed, R. Heath and D. Payne, Biochem. J., 2003, 370, 1055–1062 CrossRef CAS PubMed.
- R. J. Heath, N. Su, C. K. Murphy and C. O. Rock, J. Biol. Chem., 2000, 275, 40128–40133 CrossRef CAS PubMed.
- E. M. Fozo and R. G. Quivey, J. Bacteriol., 2004, 186, 4152–4158 CrossRef CAS PubMed.
- H. Bi, H. Wang and J. E. Cronan, Chem. Biol., 2013, 20, 1157–1167 CrossRef CAS PubMed.
- J. B. Ohlrogge and J. G. Jaworski, Annu. Rev. Plant Biol., 1997, 48, 109–136 CrossRef CAS PubMed.
- A. Jerga and C. O. Rock, J. Biol. Chem., 2009, 284, 15364–15368 CrossRef CAS PubMed.
- R. P. Massengo-Tiassé and J. E. Cronan, J. Biol. Chem., 2008, 283, 1308–1316 CrossRef PubMed.
- J. J. Salas and J. B. Ohlrogge, Arch. Biochem. Biophys., 2002, 403, 25–34 CrossRef CAS PubMed.
- D. C. Cantu, Y. Chen and P. J. Reilly, Protein Sci., 2010, 19, 1281–1295 CrossRef CAS PubMed.
- F. Jing, D. C. Cantu, J. Tvaruzkova, J. P. Chipman, B. J. Nikolau, M. D. Yandeau-Nelson and P. J. Reilly, BMC Biochem., 2011, 12, 1–16 CrossRef PubMed.
- J. Beld, J. L. Blatti, C. Behnke, M. Mendez and M. D. Burkart, J. Appl. Phycol., 2014, 26, 1619–1629 CrossRef CAS PubMed.
- E. Ploskoń, C. J. Arthur, S. E. Evans, C. Williams, J. Crosby, T. J. Simpson and M. P. Crump, J. Biol. Chem., 2008, 283, 518–528 CrossRef PubMed.
- G. Bunkoczi, S. Pasta, A. Joshi, X. Wu, K. L. Kavanagh, S. Smith and U. Oppermann, Chem. Biol., 2007, 14, 1243–1253 CrossRef CAS PubMed.
- T. Maier, M. Leibundgut and N. Ban, Science, 2008, 321, 1315–1322 CrossRef CAS PubMed.
- S. Jenni, M. Leibundgut, D. Boehringer, C. Frick, B. Mikolásek and N. Ban, Science, 2007, 316, 254–261 CrossRef CAS PubMed.
- S. Jenni, M. Leibundgut, T. Maier and N. Ban, Science, 2006, 311, 1263–1267 CrossRef CAS PubMed.
- C. Anselmi, M. Grininger, P. Gipson and J. D. Faraldo-Gómez, J. Am. Chem. Soc., 2010, 132, 12357–12364 CrossRef CAS PubMed.
- P. Johansson, B. Mulinacci, C. Koestler, R. Vollrath, D. Oesterhelt and M. Grininger, Structure, 2009, 17, 1063–1074 CrossRef CAS PubMed.
- P. Gipson, D. J. Mills, R. Wouts, M. Grininger, J. Vonck and W. Kühlbrandt, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 9164–9169 CrossRef CAS PubMed.
- M. Grininger, Curr. Opin. Struct. Biol., 2014, 25, 49–56 CrossRef CAS PubMed.
- P. Lu, C. Vogel, R. Wang, X. Yao and E. M. Marcotte, Nat. Biotechnol., 2007, 25, 117–124 CrossRef CAS PubMed.
- C. O. Rock and J. E. Cronan Jr, Methods Enzymol., 1981, 71, 341–351 CAS.
- J. E. Cronan, Biochem. J., 2014, 460, 157–163 CrossRef CAS PubMed.
- A. Roujeinikova, W. J. Simon, J. Gilroy, D. W. Rice, J. B. Rafferty and A. R. Slabas, J. Mol. Biol., 2007, 365, 135–145 CrossRef CAS PubMed.
- A. S. Flaman, J. M. Chen, S. C. Van Iderstine and D. M. Byers, J. Biol. Chem., 2001, 276, 35934–35939 CrossRef CAS PubMed.
- G. A. Zornetzer, B. G. Fox and J. L. Markley, Biochemistry, 2006, 45, 5217–5227 CrossRef CAS PubMed.
- C. Nguyen, R. W. Haushalter, D. J. Lee, P. R. Markwick, J. Bruegger, G. Caldara-Festin, K. Finzel, D. R. Jackson, F. Ishikawa, B. O'Dowd, J. A. McCammon, S. J. Opella, S.-C. Tsai and M. D. Burkart, Nature, 2013, 505, 427–431 CrossRef PubMed.
- T. Maier, S. Jenni and N. Ban, Science, 2006, 311, 1258–1262 CrossRef CAS PubMed.
- S. Rafi, P. Novichenok, S. Kolappan, X. Zhang, C. F. Stratton, R. Rawat, C. Kisker, C. Simmerling and P. J. Tonge, J. Biol. Chem., 2006, 281, 39285–39293 CrossRef CAS PubMed.
- E. Płoskoń, C. J. Arthur, A. L. Kanari, P. Wattana-amorn, C. Williams, J. Crosby, T. J. Simpson, C. L. Willis and M. P. Crump, Chem. Biol., 2010, 17, 776–785 CrossRef PubMed.
- D. I. Chan, B. C. Chu, C. K. Lau, H. N. Hunter, D. M. Byers and H. J. Vogel, J. Biol. Chem., 2010, 285, 30558–30566 CrossRef CAS PubMed.
- T. Ritsema, A. Gehring, A. Stuitje, K. Van der Drift, I. Dandal, R. Lambalot, C. Walsh, J. Thomas-Oates, B. Lugtenberg and H. Spaink, Mol. Gen. Genet., 1998, 257, 641–648 CrossRef CAS PubMed.
- M. R. Mofid, R. Finking and M. A. Marahiel, J. Biol. Chem., 2002, 277, 17023–17031 CrossRef CAS PubMed.
- A. S. Worthington, G. H. Hur and M. D. Burkart, Mol. BioSyst., 2011, 7, 365–370 RSC.
- J. Yin, P. D. Straight, S. M. McLoughlin, Z. Zhou, A. J. Lin, D. E. Golan, N. L. Kelleher, R. Kolter and C. T. Walsh, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 15815–15820 CrossRef CAS PubMed.
- J. Yin, A. J. Lin, D. E. Golan and C. T. Walsh, Nat. Protoc., 2006, 1, 280–285 CrossRef CAS PubMed.
- A. W. Barb, J. R. Cort, J. Seetharaman, S. Lew, H. W. Lee, T. Acton, R. Xiao, M. A. Kennedy, L. Tong and G. T. Montelione, Protein Sci., 2011, 20, 396–405 CrossRef CAS PubMed.
- A. Battesti and E. Bouveret, J. Bacteriol., 2009, 191, 616–624 CrossRef CAS PubMed.
- A. Battesti and E. Bouveret, Mol. Microbiol., 2006, 62, 1048–1063 CrossRef CAS PubMed.
- D. H. Keating, M. R. Carey and J. E. Cronan, J. Biol. Chem., 1995, 270, 22229–22235 CrossRef CAS PubMed.
- N. R. De Lay and J. E. Cronan, J. Bacteriol., 2006, 188, 287–296 CrossRef CAS PubMed.
- N. R. De Lay and J. E. Cronan, J. Biol. Chem., 2007, 282, 20319–20328 CrossRef CAS PubMed.
- M. C. Oswood, Y. Kim, J. B. Ohlrogge and J. H. Prestegard, Proteins, 1997, 27, 131–143 CrossRef CAS.
- E. Murugan, R. Kong, H. Sun, F. Rao and Z.-X. Liang, Protein Expression Purif., 2010, 71, 132–138 CrossRef CAS PubMed.
- J. L. Blatti, J. Beld, C. A. Behnke, M. Mendez, S. P. Mayfield and M. D. Burkart, PLoS One, 2012, 7, e42949 CAS.
- N. M. Kosa, R. W. Haushalter, A. R. Smith and M. D. Burkart, Nat. Methods, 2012, 981–984 CrossRef CAS PubMed.
- N. M. Kosa, K. M. Pham and M. D. Burkart, Chem. Sci., 2014, 5, 1179–1186 RSC.
- C. Andre, R. P. Haslam and J. Shanklin, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 10107–10112 CrossRef CAS PubMed.
- J. Thomas and J. E. Cronan, J. Biol. Chem., 2005, 280, 34675–34683 CrossRef CAS PubMed.
- V. Joshi and S. J. Wakil, Arch. Biochem. Biophys., 1971, 143, 493–505 CrossRef CAS PubMed.
- F. E. Ruch and P. R. Vagelos, J. Biol. Chem., 1973, 248, 8086–8094 CAS.
- S. R. Stapleton and J. G. Jaworski, Biochim. Biophys. Acta, 1984, 794, 240–248 CrossRef CAS.
- S. Stapleton and J. Jaworski, Fed. Proc., 1982, 41, 1193 Search PubMed.
- I. Caughey and R. G. O. Kekwick, Eur. J. Biochem., 1982, 123, 553–561 CrossRef CAS PubMed.
- D. J. Guerra and J. B. Ohlrogge, Arch. Biochem. Biophys., 1986, 246, 274–285 CrossRef CAS PubMed.
- F. M. Brück, R. Schuch and F. Spener, J. Plant Physiol., 1994, 143, 550–555 CrossRef.
- J. Simon and A. Slabas, FEBS Lett., 1998, 435, 204–206 CrossRef CAS PubMed.
- Y. Liu, Y. Zhang, X. Cao and S. Xue, Acta Crystallogr., Sect. F: Struct. Biol. Cryst. Commun., 2013, 69, 1256–1259 CrossRef CAS PubMed.
- S. K. Hong, K. H. Kim, J. K. Park, K.-W. Jeong, Y. Kim and E. E. Kim, FEBS Lett., 2010, 584, 1240–1244 CrossRef CAS PubMed.
- L. Zhang, A. K. Joshi and S. Smith, J. Biol. Chem., 2003, 278, 40067–40074 CrossRef CAS PubMed.
- J. Molnos, R. Gardiner, G. E. Dale and R. Lange, Anal. Biochem., 2003, 319, 171–176 CrossRef CAS PubMed.
- M. Sun, G. Zhu, Z. Qin, C. Wu, M. Lv, S. Liao, N. Qi, M. Xie and J. Cai, Mol. Biochem. Parasitol., 2012, 184, 20–28 CrossRef CAS PubMed.
- S. T. Prigge, X. He, L. Gerena, N. C. Waters and K. A. Reynolds, Biochemistry, 2003, 42, 1160–1169 CrossRef CAS PubMed.
- M. Sreshty, A. Surolia, G. N. Sastry and U. S. Murty, Mol. Inf., 2012, 31, 281–299 CrossRef CAS.
- L. Serre, E. C. Verbree, Z. Dauter, A. R. Stuitje and Z. S. Derewenda, J. Biol. Chem., 1995, 270, 12961–12964 CrossRef CAS PubMed.
- L. Zhang, W. Liu, J. Xiao, T. Hu, J. Chen, K. Chen, H. Jiang and X. Shen, Protein Sci., 2007, 16, 1184–1192 CrossRef CAS PubMed.
- A. T. Keatinge-Clay, A. A. Shelat, D. F. Savage, S.-C. Tsai, L. J. Miercke, J. D. O'Connell III, C. Khosla and R. M. Stroud, Structure, 2003, 11, 147–154 CrossRef CAS PubMed.
- C. J. Arthur, C. Williams, K. Pottage, E. Płoskon, S. C. Findlow, S. G. Burston, T. J. Simpson, M. P. Crump and J. Crosby, ACS Chem. Biol., 2009, 4, 625–636 CrossRef CAS PubMed.
- J. Dreier, Q. Li and C. Khosla, Biochemistry, 2001, 40, 12407–12411 CrossRef CAS PubMed.
- A. E. Szafranska, T. S. Hitchman, R. J. Cox, J. Crosby and T. J. Simpson, Biochemistry, 2002, 41, 1421–1427 CrossRef CAS PubMed.
- L. Kremer, K. M. Nampoothiri, S. Lesjean, L. G. Dover, S. Graham, J. Betts, P. J. Brennan, D. E. Minnikin, C. Locht and G. S. Besra, J. Biol. Chem., 2001, 276, 27967–27974 CrossRef CAS PubMed.
- Y.-S. Huang, J. Ge, H.-M. Zhang, J.-Q. Lei, X.-L. Zhang and H.-H. Wang, Protein Expression Purif., 2006, 45, 393–399 CrossRef CAS PubMed.
- Z. Li, Y. Huang, J. Ge, H. Fan, X. Zhou, S. Li, M. Bartlam, H. Wang and Z. Rao, J. Mol. Biol., 2007, 371, 1075–1083 CrossRef CAS PubMed.
- H. Ghadbane, A. K. Brown, L. Kremer, G. S. Besra and K. Futterer, Acta Crystallogr., Sect. F: Struct. Biol. Cryst. Commun., 2007, 63, 831–835 CrossRef CAS PubMed.
- S. Natarajan, J.-K. Kim, T.-K. Jung, T. T. N. Doan, M.-K. Hong, S. Kim, V. P. Tan, S. J. Ahn, S. H. Lee and Y. Han, Mol. Cells, 2012, 33, 19–25 CrossRef CAS PubMed.
- A.-L. Matharu, R. J. Cox, J. Crosby, K. J. Byrom and T. J. Simpson, Chem. Biol., 1998, 5, 699–711 CrossRef CAS PubMed.
- C. J. Arthur, A. Szafranska, S. E. Evans, S. C. Findlow, S. G. Burston, P. Owen, I. Clark-Lewis, T. J. Simpson, J. Crosby and M. P. Crump, Biochemistry, 2005, 44, 15414–15421 CrossRef CAS PubMed.
- A. Misra, S. K. Sharma, N. Surolia and A. Surolia, Chem. Biol., 2007, 14, 775–783 CrossRef CAS PubMed.
- P. Zhou, G. Florova and K. A. Reynolds, Chem. Biol., 1999, 6, 577–584 CrossRef CAS PubMed.
- G. Florova, G. Kazanina and K. A. Reynolds, Biochemistry, 2002, 41, 10462–10471 CrossRef CAS PubMed.
- I. I. Verwoert, K. H. van der Linden, H. J. J. Nijkamp and A. R. Stuitje, Plant Mol. Biol., 1994, 26, 189–202 CrossRef CAS PubMed.
- R. Cheng, Y. Ge, B. Yang, X. Zhong, X. Lin and Z. Huang, World J. Microbiol. Biotechnol., 2013, 29, 959–967 CrossRef CAS PubMed.
- J. Tian, M. Zheng, G. Yang, L. Zheng, J. Chen and B. Yang, Gene, 2013, 530, 33–38 CrossRef CAS PubMed.
- E. Jeon, S. Lee, J.-I. Won, S. O. Han, J. Kim and J. Lee, Enzyme Microb. Technol., 2011, 49, 44–51 CrossRef CAS PubMed.
- D. Schneidman-Duhovny, Y. Inbar, R. Nussinov and H. J. Wolfson, Nucleic Acids Res., 2005, 33, W363–W367 CrossRef CAS PubMed.
- A. Tovchigrechko and I. A. Vakser, Nucleic Acids Res., 2006, 34, W310–W314 CrossRef CAS PubMed.
- S. R. Comeau, D. W. Gatchell, S. Vajda and C. J. Camacho, Bioinformatics, 2004, 20, 45–50 CrossRef CAS PubMed.
- K.-H. Choi, R. J. Heath and C. O. Rock, J. Bacteriol., 2000, 182, 365–370 CrossRef CAS PubMed.
- K. Taguchi, Y. Aoyagi, H. Matsusaki and T. Fukui, FEMS Microbiol. Lett., 1999, 176, 183–190 CrossRef CAS PubMed.
- C. T. Nomura, K. Taguchi, S. Taguchi and Y. Doi, Appl. Environ. Microbiol., 2004, 70, 999–1007 CrossRef CAS PubMed.
- H. J. Janßen and A. Steinbüchel, Appl. Microbiol. Biotechnol., 2014, 98, 1913–1924 CrossRef PubMed.
- Y. Feng and J. E. Cronan, J. Biol. Chem., 2009, 284, 29526–29535 CrossRef CAS PubMed.
- J. Garwin, A. Klages and J. Cronan, J. Biol. Chem., 1980, 255, 3263–3265 CAS.
- S. Subrahmanyam and J. E. Cronan, J. Bacteriol., 1998, 180, 4596–4602 CAS.
- W. Zha, S. B. Rubin-Pitel, Z. Shao and H. Zhao, Metab. Eng., 2009, 11, 192–198 CrossRef CAS PubMed.
- R. M. Morgan-Kiss and J. E. Cronan, Arch. Microbiol., 2008, 190, 427–437 CrossRef CAS PubMed.
- L. Zhu, J. Cheng, B. Luo, S. Feng, J. Lin, S. Wang, J. E. Cronan and H. Wang, BMC Microbiol., 2009, 9, 119 CrossRef PubMed.
- J. P. Torella, T. J. Ford, S. N. Kim, A. M. Chen, J. C. Way and P. A. Silver, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 11290–11295 CrossRef CAS PubMed.
- X. Yu, T. Liu, F. Zhu and C. Khosla, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 18643–18648 CrossRef CAS PubMed.
- R. J. Heath and C. O. Rock, J. Biol. Chem., 1996, 271, 10996–11000 CrossRef CAS PubMed.
- R. J. Heath and C. O. Rock, J. Biol. Chem., 1996, 271, 1833–1836 CrossRef CAS PubMed.
- Q. Wei, J. Li, L. Zhang, P. Wu, Y. Chen, M. Li, H. Jiang and G. Wu, J. Plant Physiol., 2012, 169, 816–824 CrossRef CAS PubMed.
- A. S. Worthington, H. Rivera Jr, M. D. Alexander and M. D. Burkart, ACS Chem. Biol., 2006, 1, 687–691 CrossRef CAS PubMed.
- K. Dehesh, H. Tai, P. Edwards, J. Byrne and J. G. Jaworski, Plant Physiol., 2001, 125, 1103–1114 CrossRef CAS PubMed.
- I. I. Verwoert, K. H. van der Linden, M. C. Walsh, H. J. J. Nijkamp and A. R. Stuitje, Plant Mol. Biol., 1995, 27, 875–886 CrossRef CAS PubMed.
- J. Sangwallek, Y. Kaneko, M. Sugiyama, H. Ono, T. Bamba, E. Fukusaki and S. Harashima, Arch. Microbiol., 2013, 195, 843–852 CrossRef CAS PubMed.
- J. G. Olsen, A. Kadziola, P. von Wettstein-Knowles, M. Siggaard-Andersen and S. Larsen, Structure, 2001, 9, 233–243 CrossRef CAS PubMed.
- W. Huang, J. Jia, P. Edwards, K. Dehesh, G. Schneider and Y. Lindqvist, EMBO J., 1998, 17, 1183–1191 CrossRef CAS PubMed.
- C. Davies, R. J. Heath, S. W. White and C. O. Rock, Structure, 2000, 8, 185–195 CrossRef CAS PubMed.
- X. Qiu, C. A. Janson, A. K. Konstantinidis, S. Nwagwu, C. Silverman, W. W. Smith, S. Khandekar, J. Lonsdale and S. S. Abdel-Meguid, J. Biol. Chem., 1999, 274, 36465–36471 CrossRef CAS PubMed.
- Y. Chen, E. E. Kelly, R. P. Masluk, C. L. Nelson, D. C. Cantu and P. J. Reilly, Protein Sci., 2011, 20, 1659–1667 CrossRef CAS PubMed.
- Y.-M. Zhang, M. S. Rao, R. J. Heath, A. C. Price, A. J. Olson, C. O. Rock and S. W. White, J. Biol. Chem., 2001, 276, 8231–8238 CrossRef CAS PubMed.
- R. E. Toomey and S. J. Wakil, Biochim. Biophys. Acta, 1966, 116, 189–197 CrossRef CAS.
- Y.-H. Sun, Q. Cheng, W.-X. Tian and X.-D. Wu, J. Biochem. Biophys. Methods, 2008, 70, 850–856 CrossRef CAS PubMed.
- Y. Zhang and J. E. Cronan, J. Bacteriol., 1998, 180, 3295–3303 CAS.
- H. Wang and J. E. Cronan, Biochemistry, 2004, 43, 11782–11789 CrossRef CAS PubMed.
- Q. Ren, N. Sierro, B. Witholt and B. Kessler, J. Bacteriol., 2000, 182, 2978–2981 CrossRef CAS PubMed.
- M. Fisher, J. Kroon, W. Martindale, A. R. Stuitje, A. R. Slabas and J. B. Rafferty, Structure, 2000, 8, 339–347 CrossRef CAS PubMed.
- A. C. Price, Y.-M. Zhang, C. O. Rock and S. W. White, Biochemistry, 2001, 40, 12772–12781 CrossRef CAS PubMed.
- A. C. Price, Y.-M. Zhang, C. O. Rock and S. W. White, Structure, 2004, 12, 417–428 CrossRef CAS PubMed.
- C. D. Cukier, A. G. Hope, A. A. Elamin, L. Moynie, R. Schnell, S. Schach, H. Kneuper, M. Singh, J. H. Naismith and Y. Lindqvist, ACS Chem. Biol., 2013, 8, 2518–2527 CrossRef CAS PubMed.
- H. Wong, J. Mattick and S. Wakil, J. Biol. Chem., 1983, 258, 15305–15311 CAS.
- M. J. Vázquez, W. Leavens, R. Liu, B. Rodríguez, M. Read, S. Richards, D. Winegar and J. M. Domínguez, FEBS J., 2008, 275, 1556–1567 CrossRef PubMed.
- E. Y. Jeon, S. H. Lee and Y. J. Yoon, J. Microbiol. Biotechnol., 2012, 22, 990–999 CrossRef CAS PubMed.
- S. Pasta, A. Witkowski, A. K. Joshi and S. Smith, Chem. Biol., 2007, 14, 1377–1385 CrossRef CAS PubMed.
- M. Leesong, B. S. Henderson, J. R. Gillig, J. M. Schwab and J. L. Smith, Structure, 1996, 4, 253–264 CrossRef CAS PubMed.
- M. S. Kimber, F. Martin, Y. Lu, S. Houston, M. Vedadi, A. Dharamsi, K. M. Fiebig, M. Schmid and C. O. Rock, J. Biol. Chem., 2004, 279, 52593–52602 CrossRef CAS PubMed.
- H. Marrakchi, Y. Zhang and C. Rock, Biochem. Soc. Trans., 2002, 30, 1050–1055 CrossRef CAS PubMed.
- P. S. Aguilar, J. E. Cronan and D. De Mendoza, J. Bacteriol., 1998, 180, 2194–2200 CAS.
- G. M. Helmkamp, R. Rando, D. Brock and K. Bloch, J. Biol. Chem., 1968, 243, 3229–3231 CAS.
- X. Xiao, X. Yu and C. Khosla, Biochemistry, 2013, 52, 8304–8312 CrossRef CAS PubMed.
- Y.-J. Lu, S. W. White and C. O. Rock, J. Biol. Chem., 2005, 280, 30342–30348 CrossRef CAS PubMed.
- L. Zhang, W. Liu, T. Hu, L. Du, C. Luo, K. Chen, X. Shen and H. Jiang, J. Biol. Chem., 2008, 283, 5370–5379 CrossRef CAS PubMed.
- P. L. Swarnamukhi, S. K. Sharma, P. Bajaj, N. Surolia, A. Surolia and K. Suguna, FEBS Lett., 2006, 580, 2653–2660 CrossRef CAS PubMed.
- S. K. Sharma, M. Kapoor, T. Ramya, S. Kumar, G. Kumar, R. Modak, S. Sharma, N. Surolia and A. Surolia, J. Biol. Chem., 2003, 278, 45661–45671 CrossRef CAS PubMed.
- W. Liu, C. Luo, C. Han, S. Peng, Y. Yang, J. Yue, X. Shen and H. Jiang, Biochem. Biophys. Res. Commun., 2005, 333, 1078–1086 CrossRef CAS PubMed.
- J. L. Meier, R. W. Haushalter and M. D. Burkart, Bioorg. Med. Chem. Lett., 2010, 20, 4936–4939 CrossRef CAS PubMed.
- F. Ishikawa, R. W. Haushalter, D. J. Lee, K. Finzel and M. D. Burkart, J. Am. Chem. Soc., 2013, 135, 8846–8849 CrossRef CAS PubMed.
- K. Bloch, P. Baronowsky, H. Goldfine, W. Lennarz, R. Light, A. Norris and G. Scheuerbrandt, Fed. Proc., 1961, 20, 921–927 CAS.
- A. T. Norris and K. Bloch, J. Biol. Chem., 1963, 238, PC3133–PC3134 CAS.
- W. Lennarz, R. Light and K. Bloch, Proc. Natl. Acad. Sci. U. S. A., 1962, 48, 840 CrossRef CAS.
- P. W. Majerus, A. Alberts and P. R. Vagelos, J. Biol. Chem., 1965, 240, 618–621 CAS.
- G. Weeks and S. J. Wakil, J. Biol. Chem., 1968, 243, 1180–1189 CAS.
- H. Bergler, S. Fuchsbichler, G. Högenauer and F. Turnowsky, Eur. J. Biochem., 1996, 242, 689–694 CAS.
- A. Slabas, C. Sidebottom, A. Hellyer, R. Kessell and M. Tombs, Biochim. Biophys. Acta, 1986, 877, 271–280 CrossRef CAS.
- P. S. Sheldon, R. G. Kekwick, C. G. Smith, C. Sidebottom and A. R. Slabas, Biochim. Biophys. Acta, 1992, 1120, 151–159 CrossRef CAS.
- J. B. Rafferty, J. W. Simon, C. Baldock, P. J. Artymiuk, P. J. Baker, A. R. Stuitje, A. R. Slabas and D. W. Rice, Structure, 1995, 3, 927–938 CrossRef CAS PubMed.
- M. Baker, Biochem. J., 1995, 309, 1029–1030 CrossRef CAS PubMed.
- D. A. Rozwarski, C. Vilchèze, M. Sugantino, R. Bittman and J. C. Sacchettini, J. Biol. Chem., 1999, 274, 15582–15589 CrossRef CAS PubMed.
- J. B. Parsons and C. O. Rock, Curr. Opin. Microbiol., 2011, 14, 544–549 CrossRef CAS PubMed.
- Y. Wang and S. Ma, ChemMedChem, 2013, 8, 1589–1608 CAS.
- A. Banerjee, E. Dubnau, A. Quemard, V. Balasubramanian, K. S. Um, T. Wilson, D. Collins, G. de Lisle and W. Jacobs, Science, 1994, 263, 227–230 CAS.
- N. Surolia and A. Surolia, Nat. Med., 2001, 7, 167–173 CrossRef CAS PubMed.
- C. Y. Botté, F. Dubar, G. I. McFadden, E. Maréchal and C. Biot, Chem. Rev., 2011, 112, 1269–1283 CrossRef PubMed.
- L. M. McMurry, M. Oethinger and S. B. Levy, Nature, 1998, 394, 531–532 CrossRef CAS PubMed.
- R. J. Heath, Y.-T. Yu, M. A. Shapiro, E. Olson and C. O. Rock, J. Biol. Chem., 1998, 273, 30316–30320 CrossRef CAS PubMed.
- C. W. Levy, A. Roujeinikova, S. Sedelnikova, P. J. Baker, A. R. Stuitje, A. R. Slabas, D. W. Rice and J. B. Rafferty, Nature, 1999, 398, 383–384 CrossRef CAS PubMed.
- W. H. Ward, G. A. Holdgate, S. Rowsell, E. G. McLean, R. A. Pauptit, E. Clayton, W. W. Nichols, J. G. Colls, C. A. Minshull and D. A. Jude, Biochemistry, 1999, 38, 12514–12525 CrossRef CAS PubMed.
- B. Liu, Y. Wang, K. L. Fillgrove and V. E. Anderson, Cancer Chemother. Pharmacol., 2002, 49, 187–193 CrossRef CAS PubMed.
- H. P. Schweizer, FEMS Microbiol. Lett., 2001, 202, 1–7 CrossRef CAS PubMed.
- R. P. Massengo-Tiassé and J. E. Cronan, Cell. Mol. Life Sci., 2009, 66, 1507–1517 CrossRef PubMed.
- I. B. Lomakin, Y. Xiong and T. A. Steitz, Cell, 2007, 129, 319–332 CrossRef PubMed.
- S. Goh and L. Good, BMC Biotechnol., 2008, 8, 61 CrossRef PubMed.
- T. Fawcett, W. J. Simon, R. Swinhoe, J. Shanklin, I. Nishida, W. W. Christie and A. R. Slabas, Plant Mol. Biol., 1994, 26, 155–163 CrossRef CAS PubMed.
- P. O'Hara, A. R. Slabas and T. Fawcett, Plant Physiol., 2002, 129, 310–320 CrossRef PubMed.
- A. K. Spencer, A. D. Greenspan and J. E. Cronan Jr, J. Biol. Chem., 1978, 253, 5922–5926 CAS.
- E. J. Steen, Y. Kang, G. Bokinsky, Z. Hu, A. Schirmer, A. McClure, S. B. Del Cardayre and J. D. Keasling, Nature, 2010, 463, 559–562 CrossRef CAS PubMed.
- X. Lu, H. Vora and C. Khosla, Metab. Eng., 2008, 10, 333–339 CrossRef CAS PubMed.
- P. Jiang and J. Cronan, J. Bacteriol., 1994, 176, 2814–2821 CAS.
- M. R. Pollard, L. Anderson, C. Fan, D. J. Hawkins and H. M. Davies, Arch. Biochem. Biophys., 1991, 284, 306–312 CrossRef CAS PubMed.
- T. A. Voelker, A. C. Worrell, L. Anderson, J. Bleibaum, C. Fan, D. J. Hawkins, S. E. Radke and H. M. Davies, Science, 1992, 257, 72–74 CAS.
- P. Dormann, T. A. Voelker and J. B. Ohlrogge, Arch. Biochem. Biophys., 1995, 316, 612–618 CrossRef CAS PubMed.
- C. W. Pemble, L. C. Johnson, S. J. Kridel and W. T. Lowther, Nat. Struct. Mol. Biol., 2007, 14, 704–709 CAS.
- S. C. Dillon and A. Bateman, BMC Bioinf., 2004, 5, 109 CrossRef PubMed.
- T. A. Voelker and H. M. Davies, J. Bacteriol., 1994, 176, 7320–7327 CAS.
- X. Zhang, M. Li, A. Agrawal and K. Y. San, Metab. Eng., 2011, 13, 713–722 CrossRef CAS PubMed.
- P. Handke, S. A. Lynch and R. T. Gill, Metab. Eng., 2011, 13, 28–37 CrossRef CAS PubMed.
- Y. Cao, J. Yang, M. Xian, X. Xu and W. Liu, Appl. Microbiol. Biotechnol., 2010, 87, 271–280 CrossRef CAS PubMed.
- K. M. Mayer and J. Shanklin, BMC Plant Biol., 2007, 7, 1 CrossRef PubMed.
- K. G. Srikanta Dani, K. S. Hatti, P. Ravikumar and A. Kush, Plant Biol., 2011, 13, 453–461 CrossRef PubMed.
- L. Yuan, T. A. Voelker and D. J. Hawkins, Proc. Natl. Acad. Sci. U. S. A., 1995, 92, 10639–10643 CrossRef CAS.
- J. J. Thelen and J. B. Ohlrogge, Metab. Eng., 2002, 4, 12–21 CrossRef CAS PubMed.
- D. J. Hawkins and J. C. Kridl, Plant J., 1998, 13, 743–752 CrossRef CAS PubMed.
- K. Dehesh, A. Jones, D. S. Knutzon and T. A. Voelker, Plant J., 1996, 9, 167–172 CAS.
- H. Tjellström, M. Strawsine, J. Silva, E. B. Cahoon and J. B. Ohlrogge, FEBS Lett., 2013, 587, 936–942 CrossRef PubMed.
- S. A. Kay, E. Lis, S. Golden, M. Melnick, D. M. Adin and J. W. Golden, US 20120184004, 2010 Search PubMed.
- A. M. Ruffing, J. Appl. Phycol., 2013, 25, 1495–1507 CrossRef CAS.
- X. Liu, J. Sheng and R. Curtiss III, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 6899–6904 CrossRef CAS PubMed.
- B. J. Dunn and C. Khosla, Interface, 2013, 10, 1–13 Search PubMed.
- C. Rock, S. Goelz and J. Cronan, J. Biol. Chem., 1981, 256, 736–742 CAS.
- S. Lee, E. Jeon, H. S. Yun and J. Lee, Biotechnol. Bioprocess Eng., 2011, 16, 706–713 CrossRef CAS.
- R. M. Lennen, D. J. Braden, R. M. West, J. A. Dumesic and B. F. Pfleger, Biotechnol. Bioeng., 2010, 106, 193–202 CrossRef CAS PubMed.
- X. Zhang, A. Agrawal and K. Y. San, Biotechnol. Prog., 2012, 28, 60–65 CrossRef PubMed.
- I. I. Verwoert, E. F. Verhagen, K. H. van der Linden, E. C. Verbree, H. J. J. Nijkamp and A. R. Stuitje, FEBS Lett., 1994, 348, 311–316 CrossRef CAS PubMed.
- S. Lee, S. Lee, Y. J. Yoon and J. Lee, Appl. Biochem. Biotechnol., 2013, 169, 462–476 CrossRef CAS PubMed.
- T. Liu, H. Vora and C. Khosla, Metab. Eng., 2010, 12, 378–386 CrossRef CAS PubMed.
- H. Liu, C. Yu, D. Feng, T. Cheng, X. Meng, W. Liu, H. Zou and M. Xian, Microb. Cell Fact., 2012, 11, 41–54 CrossRef CAS PubMed.
- R. M. Lennen, M. G. Politz, M. A. Kruziki and B. F. Pfleger, J. Bacteriol., 2013, 195, 135–144 CrossRef CAS PubMed.
- T. W. Tee, A. Chowdhury, C. D. Maranas and J. V. Shanks, Biotechnol. Bioeng., 2014, 111, 849–857 CrossRef CAS PubMed.
- S. Reiser and C. Somerville, J. Bacteriol., 1997, 179, 2969–2975 CAS.
- A. Schirmer, M. A. Rude, X. Li, E. Popova and S. B. Del Cardayre, Science, 2010, 329, 559–562 CrossRef CAS PubMed.
- E.-B. Goh, E. E. Baidoo, J. D. Keasling and H. R. Beller, Appl. Environ. Microbiol., 2012, 78, 70–80 CrossRef CAS PubMed.
- R. Kalscheuer, T. Stölting and A. Steinbüchel, Microbiology, 2006, 152, 2529–2536 CrossRef CAS PubMed.
- P. Nawabi, S. Bauer, N. Kyrpides and A. Lykidis, Appl. Environ. Microbiol., 2011, 77, 8052–8061 CrossRef CAS PubMed.
- M. A. Rude, T. S. Baron, S. Brubaker, M. Alibhai, S. B. Del Cardayre and A. Schirmer, Appl. Environ. Microbiol., 2011, 77, 1718–1727 CrossRef CAS PubMed.
- D. J. Sukovich, J. L. Seffernick, J. E. Richman, J. A. Gralnick and L. P. Wackett, Appl. Environ. Microbiol., 2010, 76, 3850–3862 CrossRef CAS PubMed.
- A. F. Alvarez, H. M. Alvarez, R. Kalscheuer, M. Wältermann and A. Steinbüchel, Microbiology, 2008, 154, 2327–2335 CrossRef CAS PubMed.
- T. Stöveken, R. Kalscheuer, U. Malkus, R. Reichelt and A. Steinbüchel, J. Bacteriol., 2005, 187, 1369–1376 CrossRef PubMed.
- F. Lin, Y. Chen, R. Levine, K. Lee, Y. Yuan and X. N. Lin, PLoS One, 2013, 8, e78595 CAS.
- J. Cronan, J. Bacteriol., 1997, 179, 1819–1823 CAS.
- F. Zhang, M. Ouellet, T. S. Batth, P. D. Adams, C. J. Petzold, A. Mukhopadhyay and J. D. Keasling, Metab. Eng., 2012, 14, 653–660 CrossRef CAS PubMed.
- Y. Feng and J. E. Cronan, J. Bacteriol., 2009, 191, 6320–6328 CrossRef CAS PubMed.
- Y. Feng and J. E. Cronan, J. Bacteriol., 2010, 192, 4289–4299 CrossRef CAS PubMed.
- Y. Feng and J. E. Cronan, Mol. Microbiol., 2011, 80, 195–218 CrossRef CAS PubMed.
- Y. Feng and J. E. Cronan, Mol. Microbiol., 2011, 81, 1020–1033 CrossRef CAS PubMed.
- Y. Feng and J. E. Cronan, PLoS One, 2012, 7, e46275 CAS.
- P. G. Roessler, Y. Chen, B. Liu and C. N. Dodge, US 20090298143, 2008 Search PubMed.
- N. Quintana, F. Van der Kooy, M. D. Van de Rhee, G. P. Voshol and R. Verpoorte, Appl. Microbiol. Biotechnol., 2011, 91, 471–490 CrossRef CAS PubMed.
- P. Hu, S. Borglin, N. A. Kamennaya, L. Chen, H. Park, L. Mahoney, A. Kijac, G. Shan, K. L. Chavarría and C. Zhang, Appl. Energy, 2013, 102, 850–859 CrossRef CAS.
- M. Rogalski and H. Carrer, Plant Biotechnol. J., 2011, 9, 554–564 CrossRef CAS PubMed.
- J. L. Blatti, J. Michaud and M. D. Burkart, Curr. Opin. Chem. Biol., 2013, 17, 496–505 CrossRef CAS PubMed.
- R. Radakovits, P. M. Eduafo and M. C. Posewitz, Metab. Eng., 2011, 13, 89–95 CrossRef CAS PubMed.
- M. L. Hamilton, R. P. Haslam, J. A. Napier and O. Sayanova, Metab. Eng., 2014, 22, 3–9 CrossRef CAS PubMed.
- H. T. Nguyen, G. Mishra, E. Whittle, M. S. Pidkowich, S. A. Bevan, A. O. Merlo, T. A. Walsh and J. Shanklin, Plant Physiol., 2010, 154, 1897–1904 CrossRef CAS PubMed.
- M. Venegas-Calerón, O. Sayanova and J. A. Napier, Prog. Lipid Res., 2010, 49, 108–119 CrossRef PubMed.
- C. Leber and N. A. Da Silva, Biotechnol. Bioeng., 2014, 111, 347–358 CrossRef CAS PubMed.
- J. Schellenberger, R. Que, R. M. Fleming, I. Thiele, J. D. Orth, A. M. Feist, D. C. Zielinski, A. Bordbar, N. E. Lewis and S. Rahmanian, Nat. Protoc., 2011, 6, 1290–1307 CrossRef CAS PubMed.
- M. S. Davis and J. E. Cronan, J. Bacteriol., 2001, 183, 1499–1503 CrossRef CAS PubMed.
- M. S. Davis, J. Solbiati and J. E. Cronan, J. Biol. Chem., 2000, 275, 28593–28598 CrossRef CAS PubMed.
- S. Jackowski, P. D. Jackson and C. O. Rock, J. Biol. Chem., 1994, 269, 2921–2928 CAS.
- Y. Jiang, R. M. Morgan-Kiss, J. W. Campbell, C. H. Chan and J. E. Cronan, Biochemistry, 2010, 49, 718–726 CrossRef CAS PubMed.
- Y. Jiang, C. H. Chan and J. E. Cronan, Biochemistry, 2006, 45, 10008–10019 CrossRef CAS PubMed.
- D. Kaczmarzyk and M. Fulda, Plant Physiol., 2010, 152, 1598–1610 CrossRef CAS PubMed.
- J. E. Cronan, X. Zhao and Y. Jiang, Adv. Microb. Physiol., 2005, 50, 103–146 CrossRef CAS PubMed.
- J. E. Cronan and S. Lin, Curr. Opin. Chem. Biol., 2011, 15, 407–413 CrossRef CAS PubMed.
- R. H. Baltz, V. Miao and S. K. Wrigley, Nat. Prod. Rep., 2005, 22, 717–741 RSC.
- R. N. Hannoush and J. Sun, Nat. Chem. Biol., 2010, 6, 498–506 CrossRef CAS PubMed.
- N. Kresge, R. D. Simoni and R. L. Hill, J. Biol. Chem., 2005, 280, e23 CAS.
- E. Stadtman and H. Barker, J. Biol. Chem., 1949, 180, 1085–1093 CAS.
- E. Stadtman and H. Barker, J. Biol. Chem., 1949, 180, 1095–1115 CAS.
- E. Stadtman and H. Barker, J. Biol. Chem., 1949, 180, 1117–1124 CAS.
- E. Stadtman and H. Barker, J. Biol. Chem., 1949, 181, 221–235 CAS.
- E. Stadtman and H. Barker, J. Biol. Chem., 1949, 180, 1169–1186 CAS.
- J. J. Volpe and P. R. Vagelos, Annu. Rev. Biochem., 1973, 42, 21–60 CrossRef CAS PubMed.
- S. J. Wakil, J. K. Stoops and V. C. Joshi, Annu. Rev. Biochem., 1983, 52, 537–579 CrossRef CAS PubMed.
- K. Bloch and D. Vance, Annu. Rev. Biochem., 1977, 46, 263–298 CrossRef CAS PubMed.
- J. Beld, K. Finzel and M. D. Burkart, Chem. Biol., 2014 DOI:10.1016/j.chembiol.2014.08.015.
- J. Beld, H. Cang and M. D. Burkart, Angew. Chem., Int. Ed., 2014 Search PubMed , in press.
| This journal is © The Royal Society of Chemistry 2015 |