RNA-seq based transcriptomic analysis of single bacterial cells†
Abstract
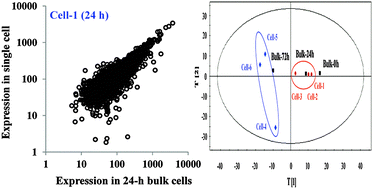
Gene-expression heterogeneity among individual cells determines the fate of a bacterial population. Here we report the first bacterial single-cell RNA sequencing (RNA-seq), BaSiC RNA-seq, a method integrating RNA isolation, cDNA synthesis and amplification, and RNA-seq analysis of the whole transcriptome of single cyanobacterium Synechocystis sp. PCC 6803 cells which typically contain approximately 5–7 femtogram total RNA per cell. We applied the method to 3 Synechocystis single cells at 24 h and 3 single cells at 72 h after nitrogen-starvation stress treatment, as well as their bulk-cell controls under the same conditions, to determine the heterogeneity upon environmental stress. With 82–98% and 31–48% of all putative Synechocystis genes identified in single cells of 24 and 72 h, respectively, the results demonstrated that the method could achieve good identification of the transcripts in single bacterial cells. In addition, the preliminary results from nitrogen-starved cells also showed a possible increasing gene-expression heterogeneity from 24 h to 72 h after nitrogen starvation stress. Moreover, preliminary analysis of single-cell transcriptomic datasets revealed that genes from the “Mobile elements” functional category have the most significant increase of gene-expression heterogeneity upon stress, which was further confirmed by single-cell RT-qPCR analysis of gene expression in 24 randomly selected cells.

 Please wait while we load your content...
Please wait while we load your content...