Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Identification of drug-specific pathways based on gene expression data: application to drug induced lung injury†
Ioannis N.
Melas
a,
Theodore
Sakellaropoulos
b,
Francesco
Iorio
c,
Leonidas G.
Alexopoulos
bd,
Wei-Yin
Loh
e,
Douglas A.
Lauffenburger
f,
Julio
Saez-Rodriguez
*c and
Jane P. F.
Bai
*a
aOffice of Clinical Pharmacology, Office of Translational Science, Center for Drug Evaluation and Research, US Food and Drug Administration, Silver Spring, MD, USA. E-mail: jane.bai@fda.hhs.gov
bSchool of Mechanical Engineering, National Technical University of Athens, Athens, Greece
cEuropean Molecular Biology Laboratory, European Bioinformatics Institute (EMBL-EBI), Hinxton, Cambridgeshire, UK. E-mail: saezrodriguez@ebi.ac.uk
dProtATonce Ltd., Athens, Greece
eDepartment of Statistics, University of Wisconsin, Madison, WI, USA
fSchool of Biological Engineering, Massachusetts Institute of Technology, Cambridge, MA, USA
First published on 1st May 2015
Abstract
Identification of signaling pathways that are functional in a specific biological context is a major challenge in systems biology, and could be instrumental to the study of complex diseases and various aspects of drug discovery. Recent approaches have attempted to combine gene expression data with prior knowledge of protein connectivity in the form of a PPI network, and employ computational methods to identify subsets of the protein–protein-interaction (PPI) network that are functional, based on the data at hand. However, the use of undirected networks limits the mechanistic insight that can be drawn, since it does not allow for following mechanistically signal transduction from one node to the next. To address this important issue, we used a directed, signaling network as a scaffold to represent protein connectivity, and implemented an Integer Linear Programming (ILP) formulation to model the rules of signal transduction from one node to the next in the network. We then optimized the structure of the network to best fit the gene expression data at hand. We illustrated the utility of ILP modeling with a case study of drug induced lung injury. We identified the modes of action of 200 lung toxic drugs based on their gene expression profiles and, subsequently, merged the drug specific pathways to construct a signaling network that captured the mechanisms underlying Drug Induced Lung Disease (DILD). We further demonstrated the predictive power and biological relevance of the DILD network by applying it to identify drugs with relevant pharmacological mechanisms for treating lung injury.
Insight, innovation, integrationIn this manuscript we introduce a novel approach for the identification of signaling pathways that are functional in a specific biological context, by leveraging gene expression data and prior knowledge of protein connectivity. In more detail, we introduce a linear programming formulation to model signal transduction from one node to the next in a Prior Knowledge Network (PKN), and by minimizing the mismatch between model predictions and experimental data, we are able to identify subsets of the PKN that are most probably functional in the specific biological context. More specifically, we address the problem of identifying the modes of action of drugs that have been reported to induce respiratory side effects, based on their gene expression profiles, and subsequently, merge the drug specific pathways together to construct a signaling network that captures the signaling mechanisms underlying Drug Induced Lung Disease (DILD). Moreover, to demonstrate the predictive power and biological relevance of the DILD network, we use it to suggest potential drug repositioning for treating lung injury. |
1 Introduction
The identification and understanding of modes of drug action is at the core of pharmacology-based pharmaceutical R&D. For the many drugs that target signal transduction processes, this requires an understanding of the mode of action at the signaling level and in the specific tissue where the drug is to be used, along with other tissues that may be subject to off-target effects. Understanding this could have an enormous impact in many aspects of drug development and public health.1 Ideally, one would have dedicated (phospho)proteomic and chemoproteomic experiments,2 where the binding targets of the drug of interest are identified, and the amount and post-translational modifications of many proteins are measured upon perturbation with the drug. However, phospho- and chemo-proteomic data are still relatively hard to generate and hence very few datasets exist. In contrast, such data upon perturbation exist abundantly at the gene expression level,3 and they are an invaluable resource for comparative studies of drugs and cell lines, enabling the use of computational modeling for predicting drug efficacy or identifying potential drugs for repositioning.4 Thus, the development of novel approaches that leverage gene expression datasets to identify the modes of drug action is an important question in computational drug discovery.Most computational methodologies for identifying modes of drug action based on gene expression data use one of the following two workflows: (i) first, differentially expressed genes are identified upon perturbation with the interrogated drugs, and subsequently, enrichment analysis is performed to identify biological processes, signaling pathways, or other gene sets that are highly enriched in the differentially expressed genes and thus, are likely to be deregulated by the interrogated drugs. The gene sets could be either GO terms or genes that are deregulated upon perturbation with known, very specific stimulants.5 Because enrichment based strategies ignore the complex gene interactions that may drive cellular response, hybrid methods have also been proposed that take into account information from pathway maps to improve their prediction.6 (ii) Other approaches are primarily based on the incorporation of prior knowledge of signaling networks or transcription regulation in addition to the gene expression data.7–9 For example, in the work by Ziemek et al.,8 the Selventa knowledge-base was used that includes causal, condition specific relationships between signaling proteins and gene expressions, and a Bayesian inference approach was used to identify subsets of this knowledge base that are most probably active in the specific biological context. Ziemek et al. were able to identify the key regulators that govern gene expression, but they could only capture limited mechanistic aspects of the intermediates in signal transduction, i.e. how signal propagates from one protein to the next before translation into the gene expression level via the transcription factors (TFs). In another work by Chen et al.,10 a PPI network was used to represent protein connectivity, and an enrichment analysis method was implemented to infer the activity of TFs and signaling proteins based on the observed gene expression signatures. In similar fashion, Huang et al.9 used a PPI network to represent protein connectivity, and implemented a Prize Collecting Steiner Tree (PCST) algorithm to identify minimum subtrees of the PPI network that connect differentially expressed genes or proteins, discovering the backbone networks that are most probably functional in the specific biological context. In more detail, the PCST algorithm addresses the problem of connecting into a Steiner arborescence tree as much of the differentially expressed genes (or proteins) as possible, while minimizing the number of edges in the tree. The PCST does not impose the requirement that all differentially expressed genes/proteins are included in the solution, but identifies a subset of those whose connectivity is also strongly supported by the network, thus offering robust predictions even when noisy data are used. Also, the PCST can be formulated as an Integer Linear Programming (ILP) problem, which can be solved efficiently allowing the interrogation of genome-wide networks.
The use of PPI networks in general offers clear advantages over strictly data driven methods. Firstly, these methods combine gene expression data-sets with the wealth of published high throughput interaction data, making model predictions more biologically relevant. Secondly, the identification of network topologies implicated in drug response is easier to interpret, as it offers mechanistic insight into the mode of drug action. Finally, the use of networks allows the generation of predictions for signaling molecules that are not directly measured, for example nearest neighbours of the measured genes/proteins. Nevertheless, the use of PPI networks has its own shortcomings. PPIs are undirected; thus, the direction of signal flow from one protein to the next is not easily identifiable. While the original formulation of PCST considered undirected networks, extensions have been proposed11 to include directionality in the networks and to generalize from a single tree that connects together all differentially expressed genes (or proteins), to forests, thereby permitting different, unconnected neighborhoods of the PPI network to be functional at the same time. As more complexity is incorporated in the formulation, a global solution becomes intractable, forcing the use of heuristic methods in the optimization risking a suboptimal solution. Moreover, even these PCST extensions cannot incorporate signed data and interactions (positive vs. negative effects), while these effects are in fact key to defining the mechanisms underlying signal transduction.
In this paper, a methodology is introduced for the identification of the mode of drug action, based on gene expression data and prior knowledge of protein connectivity in the form of a large (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 956 proteins), directed signaling network. At the heart of our method is an Integer Linear Programming (ILP) formulation based on the one by Melas et al.,12 modified at key points to address the complexity of large-scale signaling networks. The methodology combines gene expression data with a Prior Knowledge Network (PKN) based on signed and directed causal interactions, such as those that can be curated from the literature, and it identifies subsets of the PKN that appear to be functional based on the data at hand. We addressed the modeling of signal transduction using rules that define signal propagation from one node to the next in the network, and incorporated the necessary intervention strategies to modify the network structure to best fit the experimental data at hand. Our method resembles the work by Tuncbag et al.11 with regards to the identification of minimum subsets of the PKN that fit the experimental data. However, by crafting the rules of signal transduction into our custom ILP formulation, we were able to additionally capture the valuable information contained in the sign of the interactions as well as to distinguish between positive and negative changes in the data (up- and down-regulations).
956 proteins), directed signaling network. At the heart of our method is an Integer Linear Programming (ILP) formulation based on the one by Melas et al.,12 modified at key points to address the complexity of large-scale signaling networks. The methodology combines gene expression data with a Prior Knowledge Network (PKN) based on signed and directed causal interactions, such as those that can be curated from the literature, and it identifies subsets of the PKN that appear to be functional based on the data at hand. We addressed the modeling of signal transduction using rules that define signal propagation from one node to the next in the network, and incorporated the necessary intervention strategies to modify the network structure to best fit the experimental data at hand. Our method resembles the work by Tuncbag et al.11 with regards to the identification of minimum subsets of the PKN that fit the experimental data. However, by crafting the rules of signal transduction into our custom ILP formulation, we were able to additionally capture the valuable information contained in the sign of the interactions as well as to distinguish between positive and negative changes in the data (up- and down-regulations).
To illustrate the value of our approach, the identification of the modes of action of drugs that are known to induce lung injury is addressed. Drug induced lung injury is a major safety concern and more than 800 drugs are listed as potential inducers13 of lung injury including asthma, fibrosis, or interstitial pneumonia. Thus, understanding the molecular mechanisms underlying Drug Induced Lung Disease (DILD) may have an impact in drug development and in public health. In this work, the modes of action of 200 drugs that are known to induce respiratory problems were identified, in terms of signaling pathways that start at the drug targets, go through the signaling level, and terminate at the genomic level with the regulation of genes that were differentially expressed upon perturbation with the toxic drugs. Subsequently, the drug specific pathways were merged together into a signaling network (i.e. DILD network) that captures the signaling mechanisms underlying DILD. Moreover, to demonstrate the predictive power of model predictions, our findings are used to suggest drugs with relevant pharmacological mechanisms for repositioning to treat DILD.
2 Results
2.1 Workflow
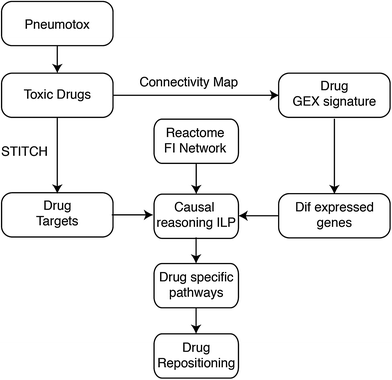
We propose a method to identify the mode of drug action based on gene expression data and prior knowledge of drug targets, protein connectivity and transcription regulation. The workflow of the proposed method is shown in Fig. 1. First, pharmacological targets were identified from the STITCH database,14 and differentially expressed genes upon perturbation with the interrogated drugs were identified from the Connectivity Map.3 Subsequently, an algorithm based on the Integer Linear Programming (ILP) formulation published in Melas et al.12 was used to identify functional interactions that model signal transduction from the drug targets to the differentially expressed genes. The identified pathways were functional subsets of a large signaling network, and originate at the drug targets, span across the signaling level, go through the affected transcription factors and terminate at the genomic level with the regulation of the differentially expressed genes (see also Fig. 2).The Pneumotox database13 was used to extract the drugs that were reported to cause lung injury. Pharmacological targets were extracted from STITCH and their gene expression profiles from the Connectivity Map, resulting in a list of 200 lung-toxic drugs with known drug–target interactions and gene expression profiles. Then, the Reactome Functional Interaction network15 was used to connect drug targets, transcription factors and gene expressions as illustrated in Fig. 1. Subsequently, the proposed ILP formulation identified a functional subset of the Reactome network, connecting drug targets and genes that were differentially expressed upon perturbation with the lung-toxic compounds. In particular, the ILP constructed a signaling pathway per toxic compound. At the end, the drug specific pathways were pooled together into a signal transduction network that captured the molecular mechanisms underlying DILD that we call the DILD network.
Finally, to demonstrate the biological relevance of the DILD network, it was leveraged to identify potential drugs for repositioning that could be useful to reverse DILD's phenotype. To this end, all remaining drugs in cMAP that were not in our DILD list were considered, and their targets were extracted from STITCH. If drug targets of these presumed non-toxic drugs overlapped with the DILD network, then their drug specific pathways were computed using the ILP algorithm, and the drugs were ranked based on how much their pathways disrupted the DILD network. The presumed non-toxic drugs that significantly disrupted the DILD network were considered candidates for repositioning.
2.2 Extraction of lung-toxic drugs, their known targets and identification of differentially expressed genes
| DNA inhibitor | 27 (62) | Cyclooxygenase 1,2 inhibitor | 18 (31) |
| Sodium channel alpha subunit blocker | 15 (47) | Serotonin 2a (5-HT2a) receptor antagonist | 11 (24) |
| GABA-A receptor; anion channel positive allosteric modulator | 11 (15) | Norepinephrine transporter inhibitor | 10 (19) |
| Serotonin transporter inhibitor | 9 (38) | Glucocorticoid receptor agonist | 9 (50) |
| Beta-1 adrenergic receptor antagonist | 9 (15) | Mu opioid receptor agonist | 8 (8) |
| Bacterial penicillin-binding protein inhibitor | 8 (36) | Angiotensin-converting enzyme inhibitor | 8 (8) |
| Bacterial 70S ribosome inhibitor | 7 (33) | Tubulin inhibitor | 7 (12) |
| Peroxisome proliferator-activated receptor gamma agonist | 7 (9) | D2-like dopamine receptor antagonist | 7 (17) |
| Beta-2 adrenergic receptor antagonist | 7 (26) | Progesterone receptor agonist | 5 (12) |
| Voltage-gated L-type calcium channel blocker | 5 (15) | Type-1 angiotensin II receptor antagonist | 5 (8) |
| RNA inhibitor | 5 (6) | Arachidonate 5-lipoxygenase inhibitor | 4 (4) |
| Thymidylate synthase inhibitor | 4 (4) | Serotonin 2c (5-HT2c) receptor antagonist | 4 (12) |
| Serotonin 1d (5-HT1d) receptor agonist | 4 (4) | Norepinephrine transporter releasing agent | 4 (18) |
| HMG-CoA reductase inhibitor | 4 (10) | FK506-binding protein 1A inhibitor | 4 (4) |
| Dopamine transporter inhibitor | 4 (17) | Dihydrofolate reductase inhibitor | 4 (7) |
| Androgen receptor agonist | 3 (8) | Adrenergic receptor alpha-2 agonist | 3 (7) |
| Vitamin K epoxide reductase complex subunit 1 isoform 1 inhibitor | 3 (4) | Ferriprotoporphyrin IX inhibitor | 3 (6) |
| Cytochrome P450 51 inhibitor | 3 (9) | Bacterial dihydropteroate synthase inhibitor | 3 (16) |
| Androgen receptor antagonist | 3 (8) |
As a positive control observation, DNA inhibitors are at the top of the table. This is expected since DNA inhibitors are often used as anti-neoplastic agents and are inherently toxic. Cyclooxygenase (COX) inhibitors are also at the top of the table. This is in good accordance with the literature where it has been reported that a range of COX inhibitors and other non steroid anti-inflammatory drugs (NSAIDs) (frequently used as analgesics) may cause respiratory problems.16 Beta-1 adrenergic receptor antagonists are also suspected of inducing respiratory distress, since beta adrenergic receptors are found to be desensitized in lung injury.17 Finally, tubulin inhibitors may contribute to lung injury via inducing oxidative stress.18 For the rest of the pharmacological effects there is no clear mechanism that could elucidate the etiology underlying Drug Induced Lung Disease.
The differentially expressed genes were used as a readout of the cellular response upon perturbation with the interrogated drugs, and they were used as the endpoints of the identified modes of drug action.
The lists of over-expressed and under-expressed genes are included in the ESI.† Subsequently, Gene Ontology (GO) enrichment analysis was performed to identify biological processes that are potentially linked to the differential gene expressions. Enriched terms with corrected p-value less than 0.05 are shown in Tables S1 and S2 (ESI†). Terms with corrected p-value less than 0.001 are shown in bold. As shown in Table S1 (ESI†), the over-expressed genes are mostly enriched in pro-apoptotic processes. This is expected, as a big part of the lung toxic compounds are chemotherapeutics or generally anti-neoplastic agents known to be toxic. Moreover, terms related to blood vessel development are present in Table S1 (ESI†). This is in good accordance with the literature, where vascular development has been reported in acute lung injury (ALI).19 The VEGF gene in particular is over-expressed in 12% of the lung toxic drugs. On the other hand, the GO terms corresponding to the under-expressed genes are mostly related to cell cycle, nuclear division, mitosis etc. From the physiological perspective, disruption of these processes by toxic compounds could result in lung injury.
2.3 Identification of modes of drug action and construction of DILD network
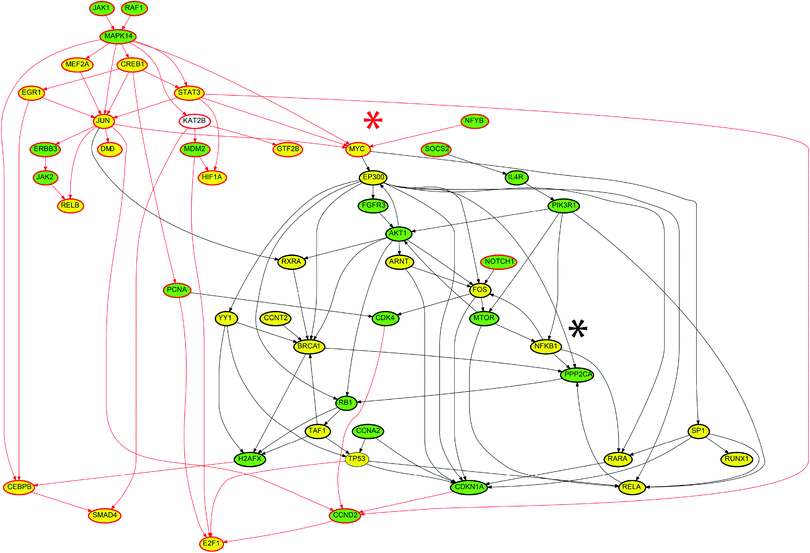
The ILP algorithm was employed to identify the modes of action of the 200 lung-toxic drugs, based on their gene expression signatures from the Connectivity Map and prior knowledge of protein connectivity, drug targets and transcription regulation. The proposed ILP algorithm identified for every drug its mode of action in terms of a signaling pathway starting at the drug targets (as these were extracted from STITCH), spanning across the signaling level, going through the layer of transcription factors and terminating at the gene expression level with the regulation of the differentially expressed genes. The ILP algorithm identified the minimum subset of the Prior Knowledge Network (PKN), that achieved the desired targets → gene expression connectivity. In this context, the drug targets correspond to the interface of the drugs with the cell's signaling machinery, and the differentially expressed genes represent the cellular responses upon perturbation with the interrogated drugs. Thus, the identified signaling pathways constitute cue–signal–response models,20 capturing cells responses to the toxic drugs. At the end, all drug specific signaling pathways were pooled together to obtain a signaling network that could best capture the molecular mechanisms underlying drug induced lung injury i.e. DILD network. All interactions in the DILD network were modeled using the same mathematical formalism presented in Section 4.1, regardless of the specific layer they belong to (e.g. drug–target interactions, protein–protein interactions, or TF–gene interactions).As shown in Fig. 3, only 22 targets were conserved in the solution, out of the 296 known targets for imatinib. The ILP, in an attempt to minimize the size of the network, conserved only the nodes that were required to propagate the signal from the drug targets to the differentially expressed genes. In this specific case, the observed gene expressions could be explained by using only the 22 targets, thus the remaining drug targets were removed.
The transcription factors to be conserved in the imatinib specific network were chosen in a similar way. The ILP conserved only the transcription factors that are required to propagate the signal to the differentially expressed genes. For example, NFKB1 was conserved with a positive sign (black asterisk in Fig. 3) because there are 2 genes downstream of NFKB1, which appeared to be over-expressed upon perturbation with imatinib (CFLAR and PIK3CD). Since NFKB1 is not one of the targets of imatinib, the ILP also conserved PIK3R1 (known to interact with imatinib) to activate NFKB1 via the PIK3R1 → NFKB1 interaction. Moreover, the NFKB1 → FOS interaction was conserved to activate FOS, inducing the expression of ETV5 and TNRC6B genes. NFKB1 also activates RARA. RARA serves to express the NCOA2 gene, that according to the Connectivity Map, is over-expressed upon perturbation with imatinib. FOS also interacts with MTOR and from there activates RELA facilitating the expression of PPARA and MMP14 genes. Even though the reaction FOS → MTOR is not necessary to activate MTOR, since MTOR is one of imatinib targets, this reaction was present in the PKN. This is due to the fact that our formulation minimized the number of nodes included, not the number of interactions, and it retained all edges that could not be disproven based on the experimental data.
Similar logic was applied to the down-regulated nodes in the imatinib specific network. For example the transcription factor MYC was conserved with a negative sign (red asterisk in Fig. 3), because 12 genes downstream of MYC appeared to be under-expressed upon perturbation with imatinib. MYC is not one of imatinib targets, thus signal has to originate from another target upstream of MYC, such as MAPK14 (P38 protein). Down-regulation of MAPK14 also lead to the down-regulation of STAT3, CREB1, MEF2A and JUN, all of which are transcription factors and have downstream genes that are downregulated upon perturbation with imatinib. There may be some interactions that were redundant, for example the downregulation of MAPK14 from JAK1 and RAF1. However, because both proteins were down-regulated by imatinib and there is an interaction between them in the PKN, the ILP could not disprove the presence of that reaction, it was thus conserved in the solution. The rest of the nodes and interactions were justified in a similar way.
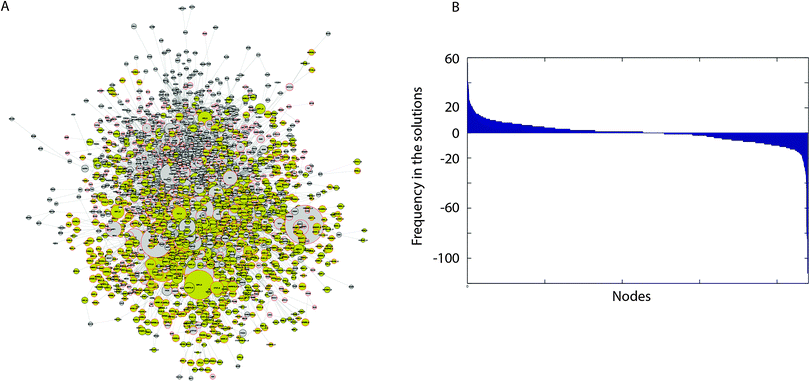 | ||
| Fig. 4 DILD network and analytics. (A) DILD network. It includes a total of 2197 nodes and 6480 reactions. Yellow nodes represent differentially expressed genes and grey nodes represent signaling proteins including drug targets and transcription factors. The size of the nodes corresponds to the number of solutions where this node is active. Thus, most significant nodes are plotted bigger than the rest. (B) An analytic showing the significance of the included nodes. The nodes of the network correspond to different coordinates of the x-axis and the y-axis corresponds to the number of drug specific pathways where each node is either up- or down-regulated. Consistently up-regulated nodes are placed on the left of the figure, while down-regulated nodes are placed on the right. There are a number of nodes that are consistently up- or down-regulated, and the signaling processes related to these nodes may play a key role in drug induced lung injury. Results were compared against predictions from randomized gene expression data, drug–target interactions and PKN connectivity (Section 2.5 and ESI†). | ||
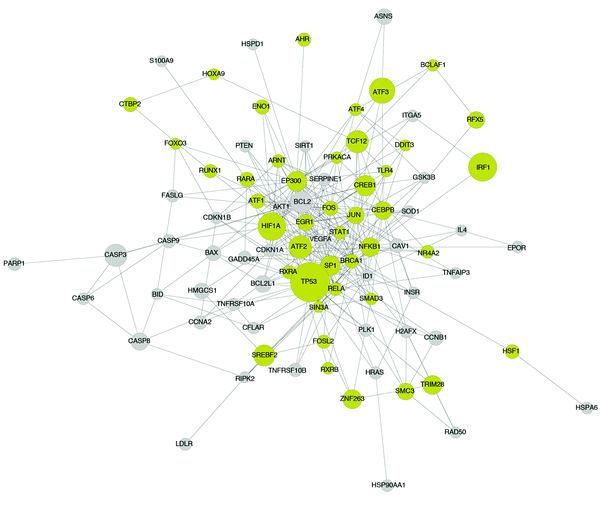
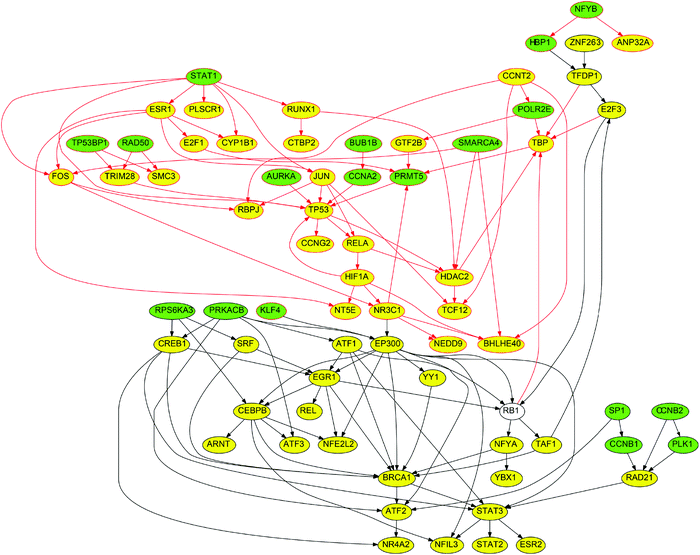
As shown in the consistently up-regulated network module of Fig. 5, a number of proteins related to DNA damage, apoptotic signaling, stress response and inflammation are present. For example TP53, CASP3, BCL2, BAX, CASP6, BCL2L1, CASP8, CASP9, BID, PARP1, CFLAR, GADD45A, FASLG, DDIT3, NFKB1, ATF2, ATF4, TNFRSF10A, TNFRSF10B, TNFAIP3, RIPK2, HSPD1, HSP90AA1, HSF1, HSPA6, IFNG, HIF1A and PTEN. Moreover, proteins with a broad role in signaling including JUN, CREB and FOS are present. The above findings are expected and are in good accordance with the Gene Ontology enrichment analysis applied on the differentially expressed genes, as discussed above, where the list of over-expressed genes was highly enriched in biological processes related to cell death and apoptosis. Here, the proposed ILP algorithm leveraged the differential gene expressions and prior knowledge of protein connectivity and transcription regulation to identify the signaling pathways underlying the observed gene expression signatures. Since the gene expression data revealed a strong correlation with biological processes related to cell death and apoptosis, the signaling pathways that yield this response are DNA damage and apoptotic signaling pathways as shown in Fig. 5. Pro-apoptotic and response to DNA damage pathways are also known to be implicated in various forms of lung injury.22 The agreement of the signaling pathways of Fig. 5 with the biological processes related to the differentially expressed genes (Table S2, ESI†), also validates that the breaching of the signaling and gene expression levels via the layer of transcription factors is accurate. Apart from the DNA damage and apoptosis pathways, proteins related to calcium signaling are present, such as FASLG, FOS, IL4 and JUN, which is in agreement with the literature where hypercalcemic activity has been observed in lung injury.23
Finally, in the network module of Fig. 5, some unrelated proteins appear, such as the EP300 protein. EP300 is a transcription factor and it affected the expression of 108 differentially expressed genes in the DILD network. Moreover, it took part in signaling and interacts with 170 proteins in the DILD network. Thus, it appeared to have a central role in the signaling pathways upon perturbation with the lung-toxic drugs. Since the ILP algorithm was agnostic to the biological function of the included proteins, and only uses the experimental data to identify subsets of the PKN that are functional in the specific biological context, proteins under-reported in the literature were expected to appear. These may constitute novel findings or they may be an artifact of the PKN structure, or the prior knowledge of transcription regulation. For example, the 108 differentially expressed genes connected to EP300 may also be expressed by another TF that is not included in the PKN, thus the ILP is forced to use the EP300 protein to fit these gene expressions, even though this is not the true mechanism. These advantages and pitfalls exist in all unbiased approaches including the proposed ILP formulation. In this context, EP300 is known to play a role in the WNT/β-catenin pathway which is found to induce IL1B expression and be implicated in interstitial pulmonary fibrosis, one of the lung injury phenotypes.24 Moreover, we performed GO enrichment analysis on the target genes of EP300, and found over-representation of programmed cell death and other apoptotic processes. In particular 42 genes related to apoptosis were regulated by EP300, which implies its potential role in pro-apoptotic response and consequently drug induced lung injury.
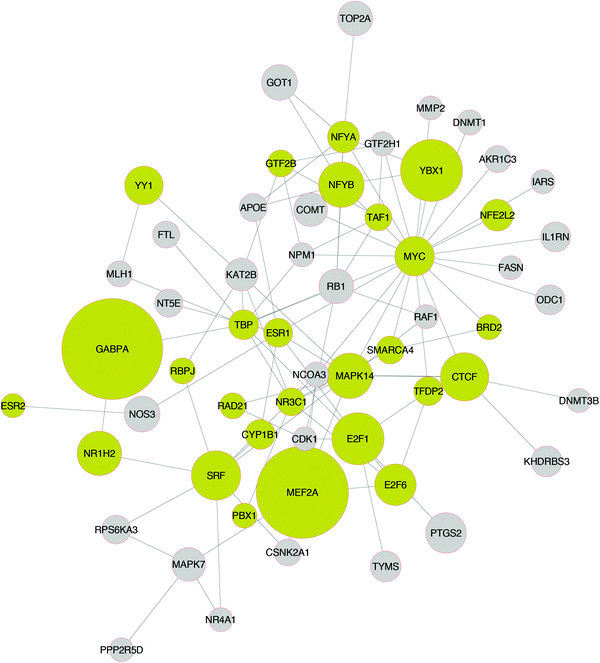
In Fig. 6, the network module of the consistently down-regulated proteins is shown. A number of proteins related to pro-growth and pro-survival pathways are present, such as MYC, E2F1, E2F6, CDK1, RAF1, SRF, RPS6A3, and MAPK7. This is in good accordance with the Gene Ontology enrichment analysis performed on the differentially expressed genes, as discussed above, where the list of under-expressed genes was highly enriched in biosynthetic and metabolic processes, and also processes affecting the cell cycle. Here, the signaling pathways underlying these biological processes are shown, as these were computed by the ILP algorithm based on the gene expression data. The under-expression of pro-growth, pro-survival and cell cycle pathways in lung injury has also been reported in literature.25 Apart from the major pro-growth pathways, TOP2A (DNA topoisomerase 2A) is also consistently down-regulated. This is expected as a large number of the lung toxic drugs are DNA inhibitors and target TOP2A. Finally, a number of proteins related to female hormone signaling are present in the network (ESR1, ESR2). This is in good accordance with the literature where estradiol and other estrogen receptor agonists are found to ameliorate the symptoms of, and protect, against lung injury.26
A number of unrelated proteins are also present in Fig. 6, such as MEF2A and GABPA. MEF2A mediates cellular functions mostly in the skeletal and cardiac muscle development. However, it is also found to play a diverse role in controlling cell growth survival and apoptosis via the MAPK14 (P38) signaling pathway.27 In good accordance with the literature, in Fig. 6, MEF2A was activated by MAPK14. Moreover, MEF2A regulated the expression of 28 genes, and also participated in signaling by interacting with 19 other proteins. GABPA was also found to play a significant role in the DILD network, regulating the expression of 40 genes and interacting with 8 proteins.
2.4 Identification of candidate drugs for treating DILD
Here we attempt to demonstrate the predictive power of the proposed ILP algorithm and the biological relevance of model predictions, by leveraging the DILD network in Fig. 4 to identify potential drugs for repositioning to treat DILD. To this end we focused in the non-toxic drugs of the Connectivity Map (a total of 1109 drugs). First, their targets were extracted from STITCH and their gene expressions from cMAP. Then, drug specific signaling pathways were computed for all drugs whose targets overlapped with the DILD network. Finally, the drugs were ranked according to how much their pathways disrupted the DILD network. A drug was considered to disrupt the DILD network if its signaling pathway upregulated proteins that were down-regulated in the network or down-regulated proteins that were upregulated in the network. The complete ranked list of overlapping drugs is included in the ESI.† The top 40, most highly ranked drugs are included in Table 2 together with their indication and relevant information supporting their usability for treating DILD (where that is available).| Score | Drug name | Indication and relevance to treating DILD |
|---|---|---|
| 119 | Ciclosporin | Anti-inflammatory. For treatment of transplant (kidney, liver, and heart) rejection, rheumatoid arthritis, severe psoriasis. Shown to be effective treatment for interstitial lung disease of unknown etiology28 |
| 115 | Tretinoin | Immunosuppressor. For the induction of remission in patients with acute promyelocytic leukemia (APL), French–American–British (FAB) classification M3 (including the M3 variant); for the topical treatment of acne vulgaris, flat warts and other skin conditions (psoriasis, ichthyosis congenita, ichthyosis vulgaris, lamellar ichthyosis, keratosis palmaris et plantaris, epidermolytic hyperkeratosis, senile comedones, senile keratosis, keratosis follicularis (Darier's disease), and basal cell carcinomas); for palliative therapy to improve fine wrinkling, mottled hyperpigmentation, roughness associated with photodamage. |
| 108 | Quercetin | Flavonol. Has anti-inflammatory properties. Used to prevent the progression of obstructive pulmonary diseases.29 |
| 92 | Resveratrol | Experimental, being investigated for the treatment of Herpes labialis infections (cold sores). Has anti-inflammatory and antioxidant effects. Has been shown to alleviate COPD lung injury in rats30 |
| 91 | Paracetamol | For temporary relief of fever, minor aches, and pains. Demonstrates weak anti-inflammatory action. Has been shown to be potentially induce asthma in long term use31 |
| 88 | Genistein | Flavonoid. Has anti-inflammatory action. Currently being studied in clinical trials as a treatment for prostate cancer. Reverses severe pulmonary hypertension and prevents right heart failure in rats32 |
| 78 | Diethylstilbestrol | Estrogen. For the treatment of hypertension, angina, and cluster headache prophylaxis. |
| 78 | Copper sulfate | NA |
| 76 | Fulvestrant | For the treatment of hormone receptor positive metastatic breast cancer in postmenopausal women with disease progression following anti-estrogen therapy. |
| 73 | Wortmannin | Used in research. Has been shown to reduce immediate-type allergic response and late phase pulmonary inflammation induced by allergen challenge in the ovalbumin-sensitised Brown Norway rat33 |
| 70 | ly-294002 | Potent inhibitor of phosphoinositide 3-kinases (PI3Ks). Has been shown to reduce allergic airway inflammation in rats.34 |
| 69 | Melatonin | Used orally for jet lag, insomnia, shift-work disorder, circadian rhythm disorders in the blind, and benzodiazepine and nicotine withdrawal. Evidence indicates that melatonin is likely effective for treating circadian rhythm sleep disorders in blind children and adults. May be effective for treating sleep-wake cycle disturbances in children and adolescents with mental retardation, autism, and other central nervous system disorders. It may also improve secondary insomnia associated with various sleep-wake cycle disturbances. Demonstrates anti-inflammatory activity in the CNS. Reduces lung oxidative stress in patients with chronic obstructive pulmonary disease35 |
| 68 | Celastrol | Plant extract. Potent antioxidant and anti-inflammatory drug. |
| 65 | Cyclic adenosine monophosphate | Experimental, targets: potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2, cAMP-dependent protein kinase type I-alpha regulatory subunit, cAMP-dependent protein kinase type II-beta regulatory subunit, adenylate cyclase, cyclic nucleotide-gated potassium channel mll3241, cAMP-activated global transcriptional regulator CRP. Decreases pulmonary edema in experimental acid-induced lung injury36 |
| 63 | sb-202190 | Experimental, target: P38MAPK. |
| 63 | Dopamine | For the correction of hemodynamic imbalances present in the shock syndrome due to myocardial infarction, trauma, endotoxic septicemia, open-heart surgery, renal failure, and chronic cardiac decompensation as in congestive failure. Has immunomodulatory action. Has been shown inhibit pulmonary edema through the VEGF–VEGFR2 axis in a murine model of acute lung injury37 |
| 62 | Dinoprostone | Prostaglandin E2. Up-regulation of PGE2 expression protects against the development of fibrosis after lung injury.38 |
| 62 | Acetylsalicylic acid | Aspirin. For use in the temporary relief of various forms of pain, inflammation associated with various conditions (including rheumatoid arthritis, juvenile rheumatoid arthritis, systemic lupus erythematosus, osteoarthritis, and ankylosing spondylitis), and is also used to reduce the risk of death and/or nonfatal myocardial infarction in patients with a previous infarction or unstable angina pectoris. Was found to improve outcome in animal models of acute lung injury39 |
| 61 | sb-203580 | Experimental, target: P38MAPK. |
| 61 | Rottlerin | Experimental, conductance potassium channel (BKCa++) opener. May cause pulmonary edema in vivo40 |
| 60 | Sulfinpyrazone | For the treatment of gout and gouty arthritis. |
| 60 | Staurosporine | Experimental, targets: tyrosine-protein kinase Lck, serine/threonine-protein kinase pim-1, tyrosine-protein kinase ITK/TSK, tyrosine-protein kinase SYK, MAP kinase-activated protein kinase 2, glycogen synthase kinase-3 beta, tyrosine-protein kinase CSK, cyclin-dependent kinase 2, phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, 3-phosphoinositide-dependent protein kinase 1, protein kinase C theta type, protein kinase C theta type. |
| 59 | Nocodazole | Experimental, target: hematopoietic prostaglandin D synthase. |
| 59 | Chrysin | Plant extract. Suppresses inflammation. Attenuates allergic airway inflammation in mice.41 |
| 58 | Pirinixic acid | Experimental, under investigation for prevention of severe cardiac dysfunction, cardiomyopathy and heart failure as a result of lipid accumulation within cardiac myocytes. |
| 58 | Lidocaine | A local anesthetic and cardiac depressant used as an antiarrhythmia agent. Demonstrates anti-inflammatory action. Attenuates acute lung injury induced by a combination of phospholipase A2 and trypsin42 |
| 58 | Ketoconazole | For the treatment of the following systemic fungal infections: candidiasis, chronic mucocutaneous candidiasis, oral thrush, candiduria, blastomycosis, coccidioidomycosis, histoplasmosis, chromomycosis, and paracoccidioidomycosis. Has been tested for early treatment of acute lung injury and acute respiratory distress syndrome in a randomized controlled trial, but was ineffective.43 |
| 58 | Kanamycin | For treatment of infections where one or more of the following are the known or suspected pathogens: E. coli, Proteus species (both indole-positive and indole-negative), E. aerogenes, K. pneumoniae, S. marcescens, and Acinetobacter species. |
| 58 | Arachidonic acid | Experimental, targets: fatty acid-binding protein, prostaglandin G/H synthase 1. |
| 57 | Thioridazine | For the treatment of schizophrenia and generalized anxiety disorder. |
| 57 | Nortriptyline | For the treatment of depression, chronic pain, irritable bowel syndrome, sleep disorders, diabetic neuropathy, agitation and insomnia, and migraine prophylaxis. |
| 57 | Cycloheximide | Experimental, inhibitor of protein biosynthesis in eukaryotic organisms |
| 56 | Furosemide | For the treatment of edema associated with congestive heart failure, cirrhosis of the liver, and renal disease, including the nephrotic syndrome. Also for the treatment of hypertension alone or in combination with other antihypertensive agents. |
| 56 | Clioquinol | Withdrawn. Used as a topical antifungal treatment. |
| 55 | Zaprinast | Unsuccessful clinical drug candidate that was a precursor to the chemically related PDE5 inhibitors, such as sildenafil, which successfully reached the market. |
| 55 | Thiamazole | For the treatment of hyperthyroidism, goiter, Graves disease and psoriasis. Has anti-inflammatory action. |
| 55 | Ouabain | For the treatment of atrial fibrillation and flutter and heart failure. |
| 55 | Indometacin | For moderate to severe rheumatoid arthritis including acute flares of chronic disease, ankylosing spondylitis, osteoarthritis, acute painful shoulder (bursitis and/or tendinitis) and acute gouty arthritis. Has been shown to attenuate lung injury in surfactant-deficient rabbits44 |
| 55 | Chenodeoxycholic acid | For patients with radiolucent stones in well-opacifying gallbladders, in whom selective surgery would be undertaken except for the presence of increased surgical risk due to systemic disease or age. Chenodiol will not dissolve calcified (radiopaque) or radiolucent bile pigment stones. |
| 54 | Kaempferol | Experimental, target: UDP-glucuronosyltransferase 3A1. Has anti-inflammatory action. Has preventive and curative effects in TH2-driven allergic airway disease45 |
The drugs at the top of the list are in good accordance with our previous predictions, are expected to strongly disrupt the DILD network calculated above, and have also been shown to a great extent to ameliorate the symptoms of lung injury. For example, ciclosporin is an immunosuppressant drug widely used in organ transplantation. It reduces the activity of immune system by interfering with the activity of T cells. It is also effective in rheumatoid arthritis and severe psoriasis, 2 auto-immune disorders with strong inflammatory component. Moreover, it has been shown to be an effective therapy for interstitial lung disease of unknown aetiology.28 Its signaling pathway is shown in Fig. 7. We observed proteins that were strongly upregulated in the DILD network, and implicated in apoptotic and inflammatory processes, were downregulated by ciclosporin and vice versa. For example, the proteins TP53, TRIM28, RELA, HIF1A, FOS and JUN that were consistently up-regulated upon exposure to the lung toxic drugs, they were down-regulated upon perturbation with ciclosporin. Moreover, the proteins RPS6KA3 and SRF, related to cell cycle and consistently down-regulated upon perturbation with the toxic compounds, were up-regulated upon perturbation with ciclosporin. In total, ciclosporin upregulated 13 proteins that were down-regulated in the DILD network: CCNB2, ESR2, NFE2L2, NFYA, RAD21, RB1, REL, RPS6KA3, SRF, TAF1, TFDP1, YBX1, YY1, and down-regulated 20 proteins that were up-regulated in the DILD network: AURKA, BHLHE40, BUB1B, CCNA2, CCNG2, CTBP2, FOS, HBP1, HIF1A, JUN, POLR2E, RAD50, RELA, RUNX1, SMC3, STAT1, TCF12, TP53, TP53BP1, and TRIM28. These results suggested that cyclosporine could have a potential disease modifying action.
Apart from ciclosporin, the flavonols quercetin and genistein, ranked third and sixth in the list, have strong anti-inflammatory action and been shown to be beneficial in treating pulmonary disease. The protective effect of flavonoids on lung injury has been reported.46 Resveratrol (ranked 4th) is another plant extract that has been shown to alleviate COPD injury in rats.30 Tretinoin, ranked second in the list, is also an immunosuppressant, and was able to ameliorate the symptoms of oxygen induced lung injury in the newborn rat.47 However, it has also been reported in Medsfacts.com to have caused traumatic lung injury in at least one patient out of 957 reports of any other side effects of tretinoin. Of the 933 physicians that expressed their opinions on the report, 295 were highly suspicious of tretinoin as the cause of the incident. Whether treatment effect or toxicity dominates could be attributed to differences in dosing regimens and duration of use.
In addition to the immunosuppressants and other anti-inflammatory drugs, estrogen diethylstilbestrol is also present (ranked 7th). Even though diethylstilbestrol has not been shown to treat DILD, it upregulated ESR1 and ESR2, that according to our predictions were consistently down-regulated in DILD (see Fig. 6). Moreover, estradiol and other estrogen receptor agonists are found to ameliorate the symptoms and protect against lung injury,48 Implying diethylstilbestrol could be a novel finding of this analysis.49 Similarly, dinoprostone (prostaglandin E2), ranked 16th in the list, significantly disrupted the DILD network, which is in good accordance with finding of prostaglandin-endoperoxide synthase 2 (also known as COX-2) being consistently downregulated in lung injury. In addition, dinoprostone has been found to protect against lung fibrosis.38,50 Similar observations could be made for other drugs in the list.
Next, we wanted to explore how our approach relates to data-driven signature matching methods. Towards this end, we applied the standard gene expression matching algorithm available on the cMAP website (http://https://www.broadinstitute.org/cmap/) that scores drugs based on their “connection” (i.e. a measure of consistency based on a non parametric statistical method) with a user defined signature. As a query signature we used the top 500 over- and under-expressed genes upon perturbation with the lung toxic drugs. Thus, the cMAP algorithm identified (i) drugs that were predicted to exert effects similar to those of lung toxic drugs and (ii) drugs whose transcriptional response was anti-correlated with that of the toxic drugs, therefore were predicted as potential candidates for repositioning to ameliorate the lung injury phenotype. The candidate drugs, as identified from the disease network, that also match the cMAP predictions are shown in bold.
We observed that 15 of the 40 drugs in Table 2 (in bold fonts), were also identified by gene expression matching to anti-correlate with the lung toxic drugs and thus constituted candidates for repositioning. Gene expression matching does not consider mechanistic information on the mode of drug action, and only identifies drugs that reverse the gene expression signature related to lung toxicity. However, as seen in Table 2, not all drugs that disrupted the DILD network also reversed gene expression, including ciclosporin, the top candidate extracted from the DILD network.
2.5 Investigation of the statistical significance of the ILP predictions
To establish the statistical significance of the disease network and model predictions, the following analysis was performed. First, GUIDE – a classification and regression trees algorithm – was applied to calculate correlations between drug targets and differentially expressed genes.51,52 The drug targets from STITCH were used as predictors and the differentially expressed genes were used as a response variable. GUIDE constructed regression trees that correlate the drug targets to the expressed genes. Subsequently, these correlations were compared with the ILP predictions.In brief, of the 4478 drug targets in total present in the PKN, the GUIDE algorithm identified 78 to be related to differential gene expressions, with 71 being present in the optimized network. The ILP algorithm conserved in the solution 1056 drug targets (of the original 4478). If GUIDE and ILP are orthogonal, the significance of the overlap can be calculated using the hypergeometric cdf and equals to 2.0630 × 10−37 (highly significant), see the ESI† for more details.
Furthermore, the performance of the ILP algorithm for different values of model parameters was examined. The ILP formulation incorporates two user defined parameters α and β that determine the weight of the measurement-prediction mismatch (parameter α) and the solution size (parameter β) in the objective function. To demonstrate the effect of these parameters on the ILP performance we repeated the pathway construction procedure for 12 different α/β ratios, while monitoring the solution size, the goodness of fit to the data and the predicted signaling activities for the consistently up- and down-regulated nodes. In brief, we observe that almost all of the consistently up-/down-regulated nodes demonstrate the same trend for all ratio values, demonstrating the robustness and statistical significance of model predictions. Detailed results are shown in the ESI.†
In addition, we randomized the gene expression data, drug targets and PKN connectivity, repeated the pathway construction procedure, and compared our findings with the protein activities calculated based on the original PKN and data. Overall, when comparing the protein activities predicted from the original data with those predicted across all these randomized setups, we observed significant divergences from expected values.
3 Conclusions
In this work, an approach is presented based on an ILP formulation that combined available gene expression data with prior knowledge of protein connectivity and transcription regulation, in the form of a Prior Knowledge Network (PKN), to identify subsets of the network that appeared to be functional based on the data. As a case study to demonstrate the utility of this approach, the identification of mode of action of drugs that are known to induce lung injury was addressed. A signaling network was constructed spanning across both the signaling and gene expression levels, through a layer of transcription factors, that elucidates the signaling mechanisms underlying drug induced lung injury (DILD). Manual inspection of the DILD network revealed that pathways related to DNA damage, inflammation and apoptosis were consistently upregulated, while pathways related to cell cycle were down-regulated. This is in good accordance with the GO enrichment analysis performed on the differentially expressed genes upon perturbation with the toxic compounds, which uncovered biological processes related to cell death highly enriched in the over-expressed genes, and processes related to cell cycle highly enriched in the under-expressed genes. This supports that our method successfully bridges the signaling and genomic levels through the layer of transcription factors. Pathways related to DNA damage, inflammation and apoptosis have also been reported in the literature to be implicated in lung injury,22 while pathways related to cell cycle have been reported to be under-expressed.25 Note that Reactome, like any other pathway resource, is biased in favour of the highly studied proteins. For example TP53 was connected to 153 signaling proteins and regulated the transcription of 281 genes. Thus, the ILP will conserve it in the solution to branch out to as many of the differentially expressed genes as possible while incorporating the smallest number of nodes. This implies that other signaling proteins, possibly important in lung injury, may be missed because the same connectivity was accomplished via hubs in the network. Network rewiring (i.e. randomization preserving node degrees) studies can be used to identify such artifacts.Moreover, to establish the statistical significance of the ILP predictions we: (i) implemented an independent classification and regression trees analysis using the GUIDE algorithm and evaluated its overlap with our findings using the ILP; (ii) repeated the pathway construction procedure for a wide range of user defined parameters, to evaluate the robustness of our analysis; and (iii) randomized the gene expression data, drug–target interactions and PKN connectivity, repeated the pathway construction procedure and compared our findings with the protein activities calculated based on the original PKN and data. Importantly, the network randomization strategy that we used is actually a rewiring (i.e. it preserves the degrees of the nodes, which indicates the level of characterisation of the corresponding protein). Therefore, a highly studied protein, which has a very high degree, will conserve its high degree in the null model.59 In brief, the predictions using GUIDE significantly overlapped with the ILP predictions, supporting the relevance of the drug specific signaling pathways; the sensitivity analysis established the robustness of our findings for a wide range of user defined parameters; and the randomization studies uncovered the individual contribution of the gene expression data, drug–target interactions and PKN connectivity to the compound specific pathways and protein activities.
Finally, to demonstrate its usability, the DILD network was leveraged to identify suitable drugs that could be repositioned to treat lung injury. To this end, drugs whose targets overlap with the DILD network were considered, and their signaling pathways were constructed using the ILP algorithm. The drugs were ranked according to how much their pathways disrupted the DILD network, which was presumed to indicate a potential disease modifying action. We observed that most drugs at the top of the list were good candidates for treating DILD. They have strong anti-inflammatory action and many of them have also been shown to ameliorate the symptoms and/or protect against lung injury. Nevertheless, given that the cMAP gene expression data we used as a starting point to our analysis are not measured in the lung, and also the drug targets in STITCH have been extracted from in vitro binding experiments, the analysis presented herein serves an exploratory purpose and subsequent, targeted, experiments should be performed to prove the relevance of the DILD network or candidate drugs for treating lung injury.
A key feature of our proposed method for reconstructing signaling networks based on gene expression data, and fundamental for its predictive power, is working with directed, signed signaling reactions, rather than undirected and unsigned PPIs, along the ability of our ILP algorithm to efficiently handle this information. When working with PPI networks, the lack of directionality and sign of the interactions makes it difficult to interpret the results. In most cases connectivity metrics are employed such as node centrality, betweenness, communicability, etc. to evaluate the significance of every node in the network. However, these metrics fail to capture the mechanistic component of signal flow. In this work, we crafted the very rules that define signal transduction into an ILP, and also allowed the algorithm to arbitrarily remove nodes from the network to best fit the experimental data at hand. The ILP formulation not only offered the required flexibility but also provided a global solution.
Overall, we have presented a novel pathway construction algorithm for identifying functional/deregulated signaling pathways based on gene expression data and prior knowledge of protein connectivity and transcription regulation. We demonstrated its usefulness by addressing the challenging problem of identifying the modes of action of drugs known to induce lung injury, and validated the model predictions by suggesting potential drugs for repositioning to treat DILD.
4 Methods
4.1 ILP formulation – basic definitions and core formulation
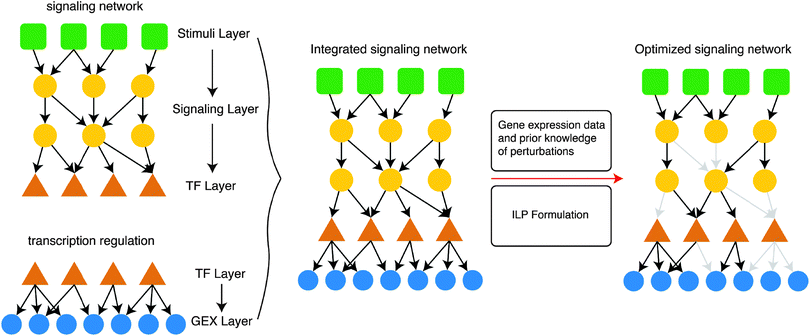
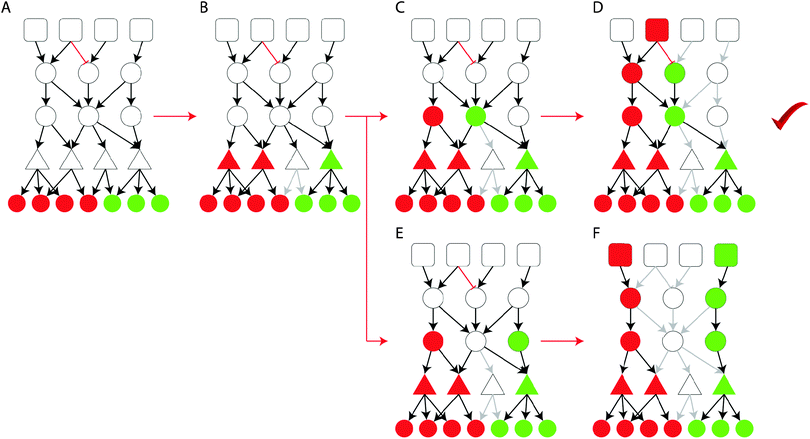
The proposed ILP formulation is based on the formulation by Melas et al.,12 modified at key points to address the computational complexity of very large (tens of thousands of nodes) signaling networks, and attempts to combine gene expression data upon perturbation with the interrogated drugs with prior knowledge of protein connectivity and transcription regulation, and identify the interactions that appear to be functional based on the data at hand. The resulting/optimized network originates at the drug targets, spans across the signaling level, goes through the layer of transcription factors and terminates at the gene expression level at the deregulated genes. Of all the subsets of the Prior Knowledge Network that achieve the desired targets → genes connectivity, the ILP algorithm selects the one numbering the fewest nodes. See Fig. 8 for an illustration of the pathway construction procedure on a toy model.Assuming a signaling network G defined as a set of reactions i = 1,…,nr and a set of species (i.e. nodes) j = 1,…,ns. Each reaction i is an ordered pair of species of the form Si → Ti, where Si, Ti ∈ {1,…,ns} are the source and target species respectively. Moreover, the sign of i is denoted with σi ∈ {−1,1}, distinguishing between activations (σi = 1) and inhibitions (σi = −1). We also define a set of experiments k = 1,…,ne. Where in each experiment a set of species are perturbed Ij,k ∈ {−1,0,1} and a set of species are measured mj,k ∈ {−1,0,1}. Variables xj,k ∈ {−1,0,1} are introduced to denote the predicted activation state of species j in experiment k.
We introduce variables u+i,k ∈ {0,1} and u−i,k ∈ {0,1}; i = 1,…,nr; k = 1,…,ne to denote the activity of reaction i in experiment k. If u+ = 1 then reaction i is active and can potentially up-regulate its target node; else if u− = 1 then reaction i is active and can potentially down-regulate its target node. A reaction i: Si → Ti is active and may up-regulate Ti (u+i,k = 1), if xj,k = 1 and σi = 1 or xj,k = −1 and σi = −1; j = Si. On the other hand, a reaction i: Si → Ti is active and may down-regulate Ti (u−i,k = 1), if xj,k = 1 and σi = −1 or xj,k = −1 and σi = 1; j = Si.
Moreover, we introduce variables x+j,k ∈ {0,1} and x−j,k ∈ {0,1} to denote the potential of node j being up (or down) regulated. node j may be up-regulated (x+j,k = 1) if ∃i: u+i,k = 1 or Ij,k = 1. On the other hand a node may be down-regulated (x−j,k = 1) if ∃i: u−i,k = 1 or Ij,k = −1. The activation state that node j will ultimately assume (xj,k) is the sum of x+ and x−. Thus, if x+j,k = 1 and x−j,k = 0, then xj,k = 1, else if x+j,k = 0 and x−j,k = 1, then xj,k = −1, else if x+j,k = 1 and x−j,k = 1, then xj,k = 1, else xj,k = 0.
Aim of the formulation is to identify the minimum subset of G that minimizes the mismatch between measurements and model predictions, thus:
| min∑α|xj,k − mj,k| + ∑βx+j,k + ∑βx−j,k | (1) |
The rules of signal transduction discussed above can be modelled as linear equality/inequality constraints as follows:
| u+i,k ≥ σixj,k; i ∈ {1,…,nr}; j = Si; k = 1,…,ne | (2a) |
| u−i,k ≥ −σixj,k; i ∈ {1,…,nr}; j = Si; k = 1,…,ne | (2b) |
| u+i,k ≤ 1 − u−i,k; i ∈ {1,…,nr}; k = 1,…,ne | (2c) |
| u+i,k ≤ σixj,k + u−i,k; i ∈ {1,…,nr}; j = Si; k = 1,…,ne | (2d) |
| u−i,k ≤ −σixj,k + u+i,k; i ∈ {1,…,nr}; j = Si; k = 1,…,ne | (2e) |
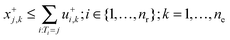 | (2f) |
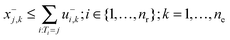 | (2g) |
| xj,k = x+j,k − x−j,k + Ij,k; j ∈ {1,…,ns}; k = 1,…,ne | (2h) |
The objective function in 1, together with the constraints in 2 formulate an Integer Linear Program (ILP) solved using IBM ILOG CPLEX. The main difference with the ILP formulation published in ref. 12 lies in the eqn (2a–e) for calculating the reaction activities u−i,k and u+i,k. In the work by Melas et al., auxiliary variables d1i,k, d2i,k, d3i,k, and d4i,k were used to calculate u−i,k and u+i,k as a function of xj,k. Here a different, more efficient formulation was developed that did not require the auxiliary variables, resulting in a smaller overall number of variables and constraints. A side-effect of this representation is that the minimization of the network size is not enforced by minimizing the number of edges though the yi variables, as it was performed in the formulation by Melas et al., but enforced through the minimization of active nodes using the x+j,k and x−j,k variables. This is a result of decoupling the u−i,k and u+i,k variables from yi.
4.2 ILP formulation – removal of feedback loops from the signaling network
Next, the removal of feedback loops from the signaling network is addressed. Positive feedback loops break the inference of pathway activities in our framework, as they allow for signal flow to be generated without an external perturbation. For example, for node j to be active (xj,k = 1), it either has to be directly perturbed Ij,k = 1, or be activated by an upstream reaction i, such that j = Ti and u+i,k = 1. However, if n nodes form a positive cycle (a cycle where all reactions are positive), then one node will be able to activate the next all the way around the cycle, without the need for an external perturbation (or an incoming interaction transitively connected to a perturbation). In the formulation by Melas et al.,12 positive feedback cycles had been removed manually before the optimization procedure. However, when very large signaling networks are interrogated, manual curation is not feasible. To address the removal of feedback cycles in an automated way, we introduce variables dj,k ≥ 0 to represent the distance of node j from a perturbed node in experiment k. If node j is not connected to a perturbed node, then dj,k = 0, else dj,k > 0. For node j to be active, dj,k > 0 has to hold true. If dj,k = 0, then xj,k = 0. The distance of node j has to be greater than all of its upstream nodes at least by one (to enforce that the distance grows the further away from the input nodes we move), unless, the upstream reactions are not active (i.e. u+i,k = u−i,k = 0). Finally, the distance of any given node cannot be greater than the total number of reactions in the signaling network. The above may be formulated using linear constraints in the following manner:| x+j,k ≤ dj |
| x−j,k ≤ dj |
| dTi ≥ dSi + 1 − M + u+i,k·M |
| dTi ≥ dSi + 1 − M + u−i,k·M |
| dj ≤ M | (3) |
The above constraints prohibit the ILP algorithm from conserving a positive feedback loop in the solution and all the included reactions to be active, unless there is an input node in the loop. Assuming a loop like that is conserved, then the distance dj,k would increase indefinitely in the loop, making the ILP infeasible since dj,k is bound by M. Where M is a sufficiently big number.
4.3 Construction of Prior Knowledge Network (PKN)
The Prior Knowledge Network (PKN) is used to represent prior knowledge of protein connectivity and transcription regulation and serves as a scaffold for the ILP algorithm presented above. It was constructed by merging the Functional Interaction Network (FIN) by Reactome15 and information on transcription regulation in the form of set of transcription factor regulons (i.e. sets of targeted genes) assembled from public available resources, such as ChEA, Transfac, and Jaspar.10,53,54 All connections from TFs to their target genes are modeled with a positive sign (i.e. TF up-regulation leads to target gene over-expression and TF down-regulation leads to target gene under-expression). Interactions that model gene suppression or other types of negative connections have been omitted from the PKN. Before using the FIN other networks were considered including the Human Signaling Network,55 Signalink,56 and the network by Kirouac et al.57 The FIN was chosen because it offered the best coverage of the transcription factors for which there is an available regulon, while being the sparsest of all, facilitating the performance of the ILP algorithm.The FIN consists of 209![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 988 functional interactions between 10
988 functional interactions between 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 956 proteins. The regulons implement 16
956 proteins. The regulons implement 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 043 interactions between 153 transcription factors and 12
043 interactions between 153 transcription factors and 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 376 target genes. Before merging, undirected and unsigned interactions were removed, as well as interactions that were predicted computationally without experimental validation. We also removed interactions between proteins that appear not to be expressed (or take part in the signaling processes) in the lung tissue. To this end the lung specific PPI network published by Guan et al.58 was leveraged. In the work by Guan et al., the authors compiled tissue specific PPI networks. Here we account for tissue specificity by including in the PKN only the interactions whose both interacting proteins are present in the lung specific PPI network. The resulting PKN spans across the protein and genetic levels going through a layer of transcription factors and includes a total of 64
376 target genes. Before merging, undirected and unsigned interactions were removed, as well as interactions that were predicted computationally without experimental validation. We also removed interactions between proteins that appear not to be expressed (or take part in the signaling processes) in the lung tissue. To this end the lung specific PPI network published by Guan et al.58 was leveraged. In the work by Guan et al., the authors compiled tissue specific PPI networks. Here we account for tissue specificity by including in the PKN only the interactions whose both interacting proteins are present in the lung specific PPI network. The resulting PKN spans across the protein and genetic levels going through a layer of transcription factors and includes a total of 64![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 801 reactions between 2585 signaling proteins and 12
801 reactions between 2585 signaling proteins and 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 376 genes. All interactions in the PKN are modelled using the same mathematical formalism presented in Section 4.1, regardless of the specific layer they belong to (e.g. drug–target interactions, protein–protein interactions, or TF–gene interactions).
376 genes. All interactions in the PKN are modelled using the same mathematical formalism presented in Section 4.1, regardless of the specific layer they belong to (e.g. drug–target interactions, protein–protein interactions, or TF–gene interactions).
4.4 Gene expression matching using the cMAP online query tool
The query tool on cMAP web site (http://https://www.broadinstitute.org/cmap/) was used to identify drugs that anti-correlate with the lung toxic gene expression signature and thus, constitute candidates for repositioning. The top 500 over- and under-expressed genes upon perturbation with the toxic drugs were used to construct a query, that we uploaded in the cMAP web service. The cMAP algorithm scored every drug in cMAP based on its correlation with that search query. Drugs with positive score may potentially induce lung injury, while drugs with negative score reverse the lung toxic gene expression signature. See also ref. 3.Disclaimer
The views expressed are those of the authors and do not necessarily represent the position of, nor imply endorsement from, the US Food and Drug Administration or the US government.Code availability
The ILP code is available in the ESI.†Acknowledgements
The core methodology of this paper on the integration of gene expression data into signaling pathways, by leveraging prior knowledge or protein connectivity and transcription regulation via regular optimization formulations, was conceptualized and motivated by INM, DAL, LGA and JSR. The ILP formulation was developed by INM, TS and JSR. The prior knowledge of transcription regulation in the form of TF regulons and the rewired versions of the original network, used for null model simulations, were provided by FI. All aspects of the DILD application were conceptualised, developed and implemented by INM and JPFB. The GUIDE algorithm was developed and implemented by WYL. Computational infrastructure was provided by EMBL-EBI. All authors edited the manuscript. We gratefully acknowledge IBM for providing the IBM ILOG CPLEX, free of charge. INM was supported as an ORISE fellow through 2013 Medical Countermeasures grant to JPFB. DAL was partially supported by ARO W911NF-09-D-0001 Institute for Collaborative Biotechnologies.References
- S. Zhao and R. Iyengar, Annu. Rev. Pharmacol. Toxicol., 2012, 52, 505–521 CrossRef CAS PubMed.
- J. Li, U. Rix, B. Fang, Y. Bai, A. Edwards, J. Colinge, K. L. Bennett, J. Gao, L. Song, S. Eschrich, G. Superti-Furga, J. Koomen and E. B. Haura, Nat. Chem. Biol., 2010, 6, 291–299 CrossRef CAS PubMed.
- J. Lamb, E. D. Crawford, D. Peck, J. W. Modell, I. C. Blat, M. J. Wrobel, J. Lerner, J.-P. Brunet, A. Subramanian, K. N. Ross, M. Reich, H. Hieronymus, G. Wei, S. A. Armstrong, S. J. Haggarty, P. A. Clemons, R. Wei, S. A. Carr, E. S. Lander and T. R. Golub, Science, 2006, 313, 1929–1935 CrossRef CAS PubMed.
- F. Iorio, T. Rittman, H. Ge, M. Menden and J. Saez-Rodriguez, Drug Discovery Today, 2013, 18, 350–357 CrossRef CAS PubMed.
- J. R. Parikh, B. Klinger, Y. Xia, J. A. Marto and N. Bluthgen, Nucleic Acids Res., 2010, 38, W109–W117 CrossRef CAS PubMed.
- A. L. Tarca, S. Draghici, P. Khatri, S. S. Hassan, P. Mittal, J.-S. Kim, C. J. Kim, J. P. Kusanovic and R. Romero, Bioinformatics, 2009, 25, 75–82 CrossRef CAS PubMed.
- S. Jaeger, J. Min, F. Nigsch, M. Camargo, J. Hutz, A. Cornett, S. Cleaver, A. Buckler and J. L. Jenkins, J. Biomol. Screening, 2014, 19, 791–802 CrossRef PubMed.
- K. Zarringhalam, A. Enayetallah, A. Gutteridge, B. Sidders and D. Ziemek, Bioinformatics, 2013, 29, 3167–3173 CrossRef CAS PubMed.
- S.-s. C. Huang and E. Fraenkel, Sci. Signaling, 2009, 2, ra40 Search PubMed.
- E. Y. Chen, H. Xu, S. Gordonov, M. P. Lim, M. H. Perkins and A. Ma'ayan, Bioinformatics, 2012, 28, 105–111 CrossRef CAS PubMed.
- N. Tuncbag, A. Braunstein, A. Pagnani, S.-S. C. Huang, J. Chayes, C. Borgs, R. Zecchina and E. Fraenkel, J. Comput. Biol., 2013, 20, 124–136 CrossRef CAS PubMed.
- I. N. Melas, R. Samaga, L. G. Alexopoulos and S. Klamt, PLoS Comput. Biol., 2013, 9, e1003204 CAS.
- P. Camus, A. Fanton, P. Bonniaud, C. Camus and P. Foucher, Respiration, 2004, 71, 301–326 CrossRef PubMed.
- M. Kuhn, D. Szklarczyk, S. Pletscher-Frankild, T. H. Blicher, C. von Mering, L. J. Jensen and P. Bork, Nucleic Acids Res., 2014, 42, D401–D407 CrossRef CAS PubMed.
- G. Wu, X. Feng and L. Stein, Genome Biol., 2010, 11, R53 CrossRef PubMed.
- M. Schwaiblmair, W. Behr, T. Haeckel, B. Markl, W. Foerg and T. Berghaus, Open Respir. Med. J., 2012, 6, 63–74 CrossRef PubMed.
- S. M. Kabir, S. Mukherjee, V. Rajaratnam, M. G. Smith and S. K. Das, J. Biochem. Mol. Toxicol., 2009, 23, 59–70 CrossRef CAS PubMed.
- E. Kratzer, Y. Tian, N. Sarich, T. Wu, A. Meliton, A. Leff and A. A. Birukova, Am. J. Respir. Cell Mol. Biol., 2012, 47, 688–697 CrossRef CAS PubMed.
- A. R. L. Medford and A. B. Millar, Thorax, 2006, 61, 621–626 CrossRef CAS PubMed.
- K. A. Janes, J. R. Kelly, S. Gaudet, J. G. Albeck, P. K. Sorger and D. A. Lauffenburger, J. Comput. Biol., 2004, 11, 544–561 CrossRef CAS PubMed.
- M. M. Peerzada, T. P. Spiro and H. A. Daw, Clin. Adv. Hematol. Oncol., 2011, 9, 824–836 Search PubMed.
- T. R. Martin, M. Nakamura and G. Matute-Bello, Crit. Care Med., 2003, 31, S184–S188 CrossRef PubMed.
- E. J. Seeley, P. Rosenberg and M. A. Matthay, J. Clin. Invest., 2013, 123, 1015–1018 CrossRef CAS PubMed.
- M. Kolb, P. J. Margetts, D. C. Anthony, F. Pitossi and J. Gauldie, J. Clin. Invest., 2001, 107, 1529–1536 CrossRef CAS PubMed.
- M. A. O'Reilly, Am. J. Physiol.: Lung Cell. Mol. Physiol., 2001, 281, L291–L305 Search PubMed.
- S. P. McGee, H. Zhang, W. Karmaus and T. Sabo-Attwood, Int. J. Mol. Epidemiol. Genet., 2014, 5, 71–86 Search PubMed.
- M. Zhao, L. New, V. V. Kravchenko, Y. Kato, H. Gram, F. di Padova, E. N. Olson, R. J. Ulevitch and J. Han, Mol. Cell. Biol., 1999, 19, 21–30 CrossRef CAS PubMed.
- J. A. Moolman, P. G. Bardin, D. J. Rossouw and J. R. Joubert, Thorax, 1991, 46, 592–595 CrossRef CAS PubMed.
- S. Ganesan, A. Faris, A. Comstock, S. Chattoraj, A. Chattoraj, J. Burgess, J. Curtis, F. Martinez, S. Zick, M. Hershenson and U. Sajjan, Respir. Res., 2010, 11, 131 CrossRef PubMed.
- M. Zhou, J.-L. He, S.-Q. Yu, R.-F. Zhu, J. Lu, F.-Y. Ding and G.-L. Xu, Yaoxue Xuebao, 2008, 43, 128–132 CAS.
- S. Shaheen, J. Sterne, C. Songhurst and P. Burney, Thorax, 2000, 55, 266–270 CrossRef CAS PubMed.
- H. Matori, S. Umar, R. D. Nadadur, S. Sharma, R. Partow-Navid, M. Afkhami, M. Amjedi and M. Eghbali, Hypertension, 2012, 60, 425–430 CrossRef CAS PubMed.
- B. Tigani, J. P. Hannon, L. Mazzoni and J. R. Fozard, Eur. J. Pharmacol., 2001, 433, 217–223 CrossRef CAS PubMed.
- W. Duan, A. M. K. Aguinaldo Datiles, B. P. Leung, C. J. Vlahos and W. S. F. Wong, Int. Immunopharmacol., 2005, 5, 495–502 CrossRef CAS PubMed.
- A. G. de Matos Cavalcante, P. F. C. de Bruin, V. M. S. de Bruin, D. M. Nunes, E. D. B. Pereira, M. M. Cavalcante and G. M. Andrade, J. Pineal Res., 2012, 53, 238–244 CrossRef PubMed.
- D. F. McAuley, J. A. Frank, X. Fang and M. A. Matthay, Crit. Care Med., 2004, 32, 1470–1476 CrossRef CAS PubMed.
- P. K. Vohra, L. H. Hoeppner, G. Sagar, S. K. Dutta, S. Misra, R. D. Hubmayr and D. Mukhopadhyay, Am. J. Physiol.: Lung Cell. Mol. Physiol., 2012, 302, L185–L192 CrossRef CAS PubMed.
- R. J. Hodges, R. G. Jenkins, C. P. D. Wheeler-Jones, D. M. Copeman, S. E. Bottoms, G. J. Bellingan, C. B. Nanthakumar, G. J. Laurent, S. L. Hart, M. L. Foster and R. J. McAnulty, Am. J. Pathol., 2004, 165, 1663–1676 CrossRef CAS PubMed.
- P. R. Tuinman, M. C. Muller, G. Jongsma, M. A. Hegeman and N. P. Juffermans, Shock, 2013, 40, 334–338 CrossRef CAS PubMed.
- J. R. Klinger, J. D. Murray, B. Casserly, D. F. Alvarez, J. A. King, S. S. An, G. Choudhary, A. N. Owusu-Sarfo, R. Warburton and E. O. Harrington, J. Appl. Physiol., 2007, 103, 2084–2094 CrossRef CAS PubMed.
- Q. Du, X. Gu, J. Cai, M. Huang and M. Su, Mol. Med. Rep., 2012, 6, 100–104 CAS.
- Y. Kiyonari, K. Nishina, K. Mikawa, N. Maekawa and H. Obara, Crit. Care Med., 2000, 28, 484–489 CrossRef CAS PubMed.
- The ARDS Network Authors for the ARDS Network, J. Am. Med. Assoc., 2000, 283, 1995–2002 CrossRef.
- R. Burger, D. Fung and A. C. Bryan, J. Appl. Physiol., 1990, 69, 2067–2071 CAS.
- K. C. P. Medeiros, L. Faustino, E. Borduchi, R. J. B. Nascimento, T. M. S. Silva, E. Gomes, M. R. Piuvezam and M. Russo, Int. Immunopharmacol., 2009, 9, 1540–1548 CrossRef CAS PubMed.
- J. Wang, H.-W. Xu, B.-S. Li, J. Zhang and J. Cheng, Asian Pac. J. Cancer Prev., 2012, 13, 6441–6446 CrossRef PubMed.
- E. A. Ozer, A. Kumral, E. Ozer, N. Duman, O. Yilmaz, S. Ozkal and H. Ozkan, Pediatr. Pulmonol., 2005, 39, 35–40 CrossRef PubMed.
- M. K. Glassberg, R. Choi, V. Manzoli, S. Shahzeidi, P. Rauschkolb, R. Voswinckel, M. Aliniazee, X. Xia and S. J. Elliot, Endocrinology, 2014, 155, 441–448 CrossRef CAS PubMed.
- H.-P. Yu, Y.-C. Hsieh, T. Suzuki, T. Shimizu, M. A. Choudhry, M. G. Schwacha and I. H. Chaudry, Am. J. Physiol.: Lung Cell. Mol. Physiol., 2006, 290, L1004–L1009 CrossRef CAS PubMed.
- V. Ivanova, O. B. Garbuzenko, K. R. Reuhl, D. C. Reimer, V. P. Pozharov and T. Minko, Eur. J. Pharm. Biopharm., 2013, 84, 335–344 CrossRef CAS PubMed.
- W.-Y. Loh, Stat. Sin., 2002, 361–386 Search PubMed.
- J. Hur, A. Y. Guo, W. Y. Loh, E. L. Feldman and J. P. F. Bai, CPT: Pharmacometrics Syst. Pharmacol., 2014, 3, e114 CrossRef CAS PubMed.
- V. Matys, E. Fricke, R. Geffers, E. Gossling, M. Haubrock, R. Hehl, K. Hornischer, D. Karas, A. E. Kel, O. V. Kel-Margoulis, D.-U. Kloos, S. Land, B. Lewicki-Potapov, H. Michael, R. Munch, I. Reuter, S. Rotert, H. Saxel, M. Scheer, S. Thiele and E. Wingender, Nucleic Acids Res., 2003, 31, 374–378 CrossRef CAS PubMed.
- G. D. Stormo, Bioinformatics, 2000, 16, 16–23 CrossRef CAS PubMed.
- N. Zaman, L. Li, M. L. Jaramillo, Z. Sun, C. Tibiche, M. Banville, C. Collins, M. Trifiro, M. Paliouras, A. Nantel, M. O'Connor-McCourt and E. Wang, Cell Rep., 2013, 5, 216–223 CrossRef CAS PubMed.
- D. Fazekas, M. Koltai, D. Turei, D. Modos, M. Palfy, Z. Dul, L. Zsakai, M. Szalay-Beko, K. Lenti, I. J. Farkas, T. Vellai, P. Csermely and T. Korcsmaros, BMC Syst. Biol., 2013, 7, 7 CrossRef PubMed.
- D. Kirouac, J. Saez-Rodriguez, J. Swantek, J. Burke, D. Lauffenburger and P. Sorger, BMC Syst. Biol., 2012, 6, 29 CrossRef PubMed.
- Y. Guan, D. Gorenshteyn, M. Burmeister, A. K. Wong, J. C. Schimenti, M. A. Handel, C. J. Bult, M. A. Hibbs and O. G. Troyanskaya, PLoS Comput. Biol., 2012, 8, e1002694 CAS.
- A. Gobbi, F. Iorio, K. J. Dawson, D. C. Wedge, D. Tamborero, L. B. Alexandrov, N. Lopez-Bigas, M. J. Garnett, G. Jurman and J. Saez-Rodriguez, Bioinformatics, 2014, 30, i617–i623 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c4ib00294f |
| This journal is © The Royal Society of Chemistry 2015 |