Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Non-monotonic cellular responses to heterogeneity in talin protein expression-level†
Alexa
Kiss‡
,
Xiaowei
Gong‡
,
Jacob M.
Kowalewski
,
Hamdah
Shafqat-Abbasi
,
Staffan
Strömblad‡
* and
John G.
Lock‡
*
Center for Innovative Medicine, Department of Biosciences and Nutrition, Karolinska Institutet, Novum, Hälsov. 7-9, G-building floor 6, S-141 83 Huddinge, Sweden. E-mail: staffan.stromblad@ki.se; john.lock@ki.se; Tel: +46-8-52481122 Tel: +46-730-968-078
First published on 14th May 2015
Abstract
Talin is a key cell–matrix adhesion component with a central role in regulating adhesion complex maturation, and thereby various cellular properties including adhesion and migration. However, knockdown studies have produced inconsistent findings regarding the functional influence of talin in these processes. Such discrepancies may reflect non-monotonic responses to talin expression-level variation that are not detectable via canonical “binary” comparisons of aggregated control versus knockdown cell populations. Here, we deployed an “analogue” approach to map talin influence across a continuous expression-level spectrum, which we extended with sub-maximal RNAi-mediated talin depletion. Applying correlative imaging to link live cell and fixed immunofluorescence data on a single cell basis, we related per cell talin levels to per cell measures quantitatively defining an array of cellular properties. This revealed both linear and non-linear correspondences between talin expression and cellular properties, including non-monotonic influences over cell shape, adhesion complex-F-actin association and adhesion localization. Furthermore, we demonstrate talin level-dependent changes in networks of correlations among adhesion/migration properties, particularly in relation to cell migration speed. Importantly, these correlation networks were strongly affected by talin expression heterogeneity within the natural range, implying that this endogenous variation has a broad, quantitatively detectable influence. Overall, we present an accessible analogue method that reveals complex dependencies on talin expression-level, thereby establishing a framework for considering non-linear and non-monotonic effects of protein expression-level heterogeneity in cellular systems.
Insight, innovation, integrationInference of talin’s functional influence through “binary” comparison of aggregated control and knockdown cell populations has produced inconsistent conclusions. These may reflect undetected non-monotonicity in talin’s influence at different expression-levels. We therefore established an “analogue” sampling approach using correlative imaging, where per cell immunofluorescent talin-level measurement follows live single-cell imaging. Combining control and RNAi-treated cell data populated a broad talin expression-level continuum, with talin levels linked per cell to quantitative cellular properties – extracted via automated image analysis. This provided proof-of-principle for non-monotonic dependencies between talin expression and several inconsistently described properties (cell shape, adhesion localization, adhesion-F-actin association), while also revealing talin-level-dependent changes in inter-property relationships. This accessible, analogue method offers important advantages over canonical binary, knockdown-based approaches to protein function-inference. |
Introduction
Cell adhesion and migration play central roles in many physiological and disease-related processes.1 Integrin-mediated cell–matrix adhesion complexes (CMACs) and the F-actin cytoskeleton are core machineries in the cell migration system.2 Talin, in turn, is a fundamental regulator of adhesion complexes and their association with F-actin, being intimately involved in multiple stages of adhesion complex maturation. Specifically, talin promotes adhesion assembly through integrin activation,3–5 facilitates the mechanical linkage between integrins and F-actin to support adhesion reinforcement6–10 and contributes to adhesion disassembly.11,12 These molecular scale functions also modulate cellular scale features, such as cell morphology and motility. Yet despite talin's clearly pivotal role, data from numerous knockdown and knockout studies fail to establish a clear consensus regarding the functional influence of talin over properties arising at either adhesion complex or cellular scales.13–17 For example, some studies have indicated that talin depletion produces less mature adhesions with reduced F-actin association,13,15,16,18,19 while others have shown adhesions to be relatively unchanged, retaining strong F-actin association,14,20 or even becoming more mature.17 Similarly, the localization of adhesions within cells has been differentially affected by talin depletion, with adhesions becoming visibly more peripheral15,16 more central,13,21 or having a normal spatial distribution.17 At the cellular scale, many studies have shown that talin depletion leads to rounder, less protrusive, more compact cell shapes,13–15,19 while others indicate less compact, more protrusive cells.16,18 Understanding the source of such inconsistencies is potentially of high importance, especially given recent indications that each of these features (adhesion-F-actin association, adhesion localization, cell compactness) is causally related to variability in cell migration behaviors.22The inconsistencies exemplified above may reflect diversity in the experimental assays and cell lines used to study talin function. However, such an interpretation adds little to our understanding of this crucial molecular component. Alternatively, these differences could reflect variations in the strength of experimental perturbations (e.g. talin depletion) coupled with the insensitivity of analysis methods to complex (e.g. non-linear) dependencies on protein expression-level, including non-monotonic responses. Indeed, most knockdown analyses are binary, allowing pairwise comparison of aggregated data from control and talin-depleted cell populations. This aggregative analysis effectively reduces data to two values (e.g. control median versus knockdown median), providing a basis for the inference of exclusively linear, monotonic relationships between protein expression-level and functional readouts. At face value, such linear interpretations imply that responses to protein level fluctuation (increase or decrease) are equivalent regardless of the starting protein expression value (high or low). More conservatively, binary analyses certainly provide no evidence on which to ground more nuanced interpretations (e.g. non-linear expression-level dependencies). If non-linear, or more significantly, non-monotonic sensitivities to protein expression-level do exist, then reliance on binary analyses may result in an oversimplified view of relationships between protein expression and associated functional consequences.
To overcome the potential insensitivity of binary approaches, methods for the analogue (continuous, semi-continuous) measurement of protein expression-levels are needed. In particular, methods exploiting the information associated with natural protein expression heterogeneity have the capability to reveal how endogenous protein level regulation (as opposed to extreme RNAi-induced depletion) affects cellular processes.23,24 In a recent study of noise genetics, cell-to-cell heterogeneity in fluorescently tagged endogenous protein levels was correlated to phenotypic differences between individual migrating cells.25 Importantly, while the authors identified putative cell migration genes based on this natural variation, they also forecast the existence of non-monotonic relationships between protein expression-levels and cell migration speed. Given the noted discrepancies in published talin effects, defining any such non-monotonic dependencies may contribute to a more coherent understanding of how talin regulates specific cell characteristics, such as cell adhesion and migration.
Understanding talin's influence also requires consideration of how talin expression-levels affect the relationships between cellular properties, not just the values of the properties themselves. Geiger and colleagues addressed this issue, showing that talin knockdown caused the “breaking” of specific correlative relationships between properties.17 Though highly instructive, the binary nature of this analysis did not allow a more detailed assessment of talin expression-level dependencies. For example, are different inter-property correlations sensitive to distinct thresholds of talin expression, implying complex, context-sensitive dependencies? Notably, we and others have recently demonstrated related context-dependent plasticity in the causal relationships between cell migration system properties – over time and between perturbed conditions.22,26 Defining similar adaptability linked to heterogeneity in talin expression-level may expand our understanding of talin's regulatory influence.
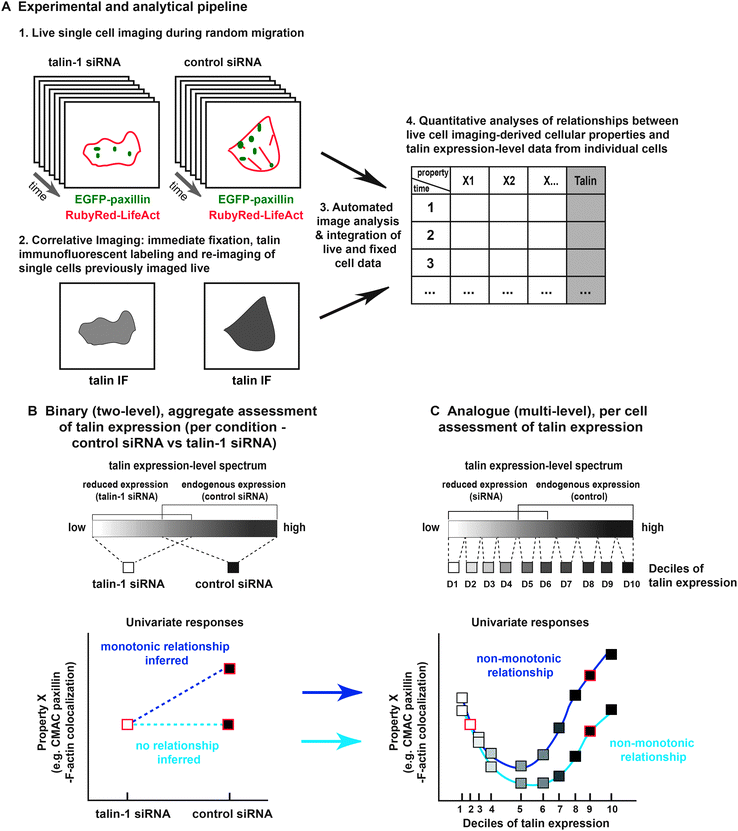
Here, we address the potential both for non-monotonicity in feature responses and for the restructuring of inter-feature correlative networks, as consequences of both natural and induced talin expression-level heterogeneity. To this end, we have established an analogue sampling method based on correlative imaging of single cells – first during live random migration – and again following fixation and fluorescent labeling of endogenous talin protein (Fig. 1). Importantly, this correlative imaging approach was performed in both control and RNAi-treated cells, producing a broad, continuous spectrum of talin expression, with talin levels linked on a per cell basis to quantitative cellular properties – extracted via automated image analysis.22 This integrated data provides the first evidence of non-monotonic dependencies upon talin expression-level. Indeed, while these findings may help to resolve inconsistencies in the talin field, they in fact represent a general proof-of-principle for the existence of non-monotonic protein expression-level dependencies. In addition, by mapping correlative inter-feature networks, we show that correlative relationships between a broader range of cell migration and adhesion properties are sensitive to talin level variation. Collectively, this study advances our understanding of talin's influence on cellular properties and their inter-relations, while also emphasizing more generally the importance of applying analogue approaches to the study of protein function.
Results
Correlative imaging, talin expression-level measurement and quantitative data extraction
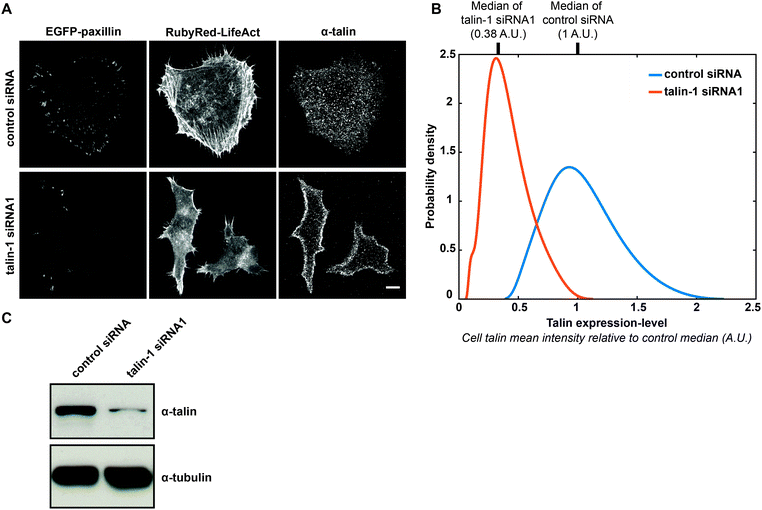
In this study, we used H1299 (human non-small cell lung carcinoma) cells stably expressing EGFP-paxillin (CMAC marker) and RubyRed-LifeAct (F-actin marker). In these cells, termed H1299 P/L cells, we detected no influence of exogenous proteins on the expression of a number of commonly studied cell–matrix adhesion components (Fig. S1, ESI†). H1299 P/L cells were treated for 48 h with either a non-targeting control siRNA or siRNA against talin-1, then plated onto fibronectin and imaged for 6 h (as depicted in Fig. 1A). Following live cell imaging, cells were immediately fixed and immunofluorescently labeled for talin expression. Cell fields captured during live imaging were then re-imaged to assess talin levels on a per cell basis (as described in Fig. 1A, 2A and Fig. S2, ESI†). Importantly, the end-point measurement of talin expression is expected to effectively indicate talin levels during live imaging based on the finding that protein levels in single H1299 cells are stable (relative to population distributions) over long periods, ranging between 0.8 and 2.5 cell cycle lengths.27 Also note that immunofluorescence conditions were optimized to retain maximal talin, including the cytoplasmic pool (Fig. 2A and Fig. S2A, ESI†), as opposed to using standard conditions that extract the cytoplasmic pool (thereby limiting the inference of expression-levels) to emphasize CMAC-associated talin labeling (Fig. S2A, ESI†). Importantly, comparison of immunofluorescence signal levels under non-extractive conditions in control siRNA or talin-1 siRNA1-treated cells, in the presence or absence of anti-talin primary antibody, confirmed the specificity of fluorescence intensities as a measure of talin expression-level (Fig. S2B and C, ESI†). Furthermore, immunoblotting showed that talin knockdown had no significant impact on the expression of a range of adhesion complex components (Fig. S3, ESI†). Given these controls, automated image analysis of live cell data was performed to segment and track individual cells and their CMAC cohorts (Fig. S4, Movies S1 and S2, ESI†). This enabled the quantitative extraction of properties defining morphology, localization, intensity and dynamics over macromolecular and cellular scales (Fig. 1A and Table S1, ESI†), as applied previously.22 Mean talin fluorescence intensity values were also extracted from the same individual cells. Ultimately, this correlative single cell imaging-based analysis, integrating live and fixed data, facilitates the analogue mapping of relationships between talin expression-level and quantitative cellular properties.Binary aggregative analysis of talin knockdown and talin-depletion effects
Before continuing with our analogue approach, we performed analyses mimicking canonical binary strategies, thereby establishing a direct point of comparison between the inferences accessible via binary or analogue methods. To this end, we first evaluated the degree of RNAi-mediated talin depletion in aggregated cell populations by immunoblotting (Fig. S5A, ESI†), comparing cells treated with control siRNA, or two distinct oligonucleotides against talin-1. Since H1299 cells express both talin-1 and talin-2,28 we applied a pan-anti-talin antibody (8d4) to detect both isoforms.13 The sharp decrease in signal upon talin-1 knockdown implies that in this cell line talin-1 is the dominant isoform and that there is little or no compensation by talin-2 expression. Both anti-talin oligonucleotides reduced talin expression to a similar extent. Importantly, equivalent results were observed when assessing talin depletion based on aggregated populations of single cell immunofluorescence imaging data (Fig. S5B, ESI†). It is noteworthy that the boxplots in Fig. S5B (ESI†) indicate overlap in talin expression values between control siRNA and talin-1 siRNA1 conditions. This important result is even more clearly exemplified by the overlapping talin expression-level probability distribution functions fitted to control and talin-1 siRNA1 conditions in Fig. 2B. Unsurprisingly, it is not possible to infer the existence of this overlap from the immunoblot comparing these conditions (Fig. 2C). Such an overlap was not observed between the control and talin-1 siRNA2 conditions (Fig. S5B, ESI†). Therefore, given the importance to our analogue analysis of generating a continuous talin expression spectrum, the majority of analyses hereafter (excluding Fig. S5, ESI†) focus on the control and talin-1 siRNA1 conditions only.Continuing to assess the single cell imaging-derived data in aggregated form, we found that talin-depletion significantly affected a range of cell adhesion and cell morphology properties, exemplified in Fig. S5C–H (ESI†). Specifically, in cells treated with either of the two talin-1 siRNAs, talin depletion significantly reduced: CMAC paxillin content (Fig. S5C, ESI,† indicative of CMAC maturity); the rate of CMAC area change (Fig. S5E, ESI,† indicative of CMAC stability), and; intra-CMAC paxillin-F-actin colocalization (Fig. S5G, ESI,† indicative of CMAC-F-actin association,29). Conversely, talin depletion increased: the rate of CMAC paxillin intensity change (Fig. S5D, ESI,† indicative of CMAC-associated paxillin net kinetics), and CMAC distance from cell border (Fig. S5F, ESI,† indicative of CMAC localization within cells). The effects of talin depletion on cell compactness (Fig. S5H, ESI,† high values indicate increasing cell protrusivity or non-roundness) were weak and inconsistent between talin-1 siRNA oligonucleotides 1 and 2. Excluding effects on cell compactness, however, talin depletion-induced changes were highly consistent between the two talin-1 siRNAs. Similar reproducibility in talin depletion effects was evident when comparing aggregate results for control and talin-1 siRNA1 conditions over 7 independent experimental repeats (Fig. S6, ESI†). Again, only cell compactness failed to show consistent effects, reflecting the weak aggregate effect of talin depletion on this feature. Importantly, most effects of talin depletion quantified in Fig. S5 (ESI†) are directly observable in representative images of cells from the control siRNA and talin-1 siRNA1 conditions (Fig. S7, ESI†). Overall, it is noteworthy that the features affected by talin depletion in this study overlap significantly with those observable in previous studies.13–17
Analogue analysis delineates both monotonic and non-monotonic correspondences to talin expression-level variation
The continuous talin expression-level spectrum achieved by combining data from the partially overlapping control siRNA and talin-1 siRNA1 conditions (as shown in Fig. 2B) provides the basis for our analogue approach. This overlap is the product of natural heterogeneity within these populations, which is undetected or unutilized using aggregative, binary methods. Leveraging of this heterogeneity provides the opportunity to differentiate linear, non-linear and even non-monotonic responses to talin expression variation.30 To this end, in Fig. 3 we compare the linear trends inferred from binary analyses (aggregate control vs. talin-1 siRNA1) to the trends derived from an analogue sampling of the combined talin expression-level spectrum. These analogue trends are derived via a continuous fitting (see Materials and methods) based on median feature values calculated within each of ten cell subpopulations, defined as deciles (10% windows) of the talin expression spectrum.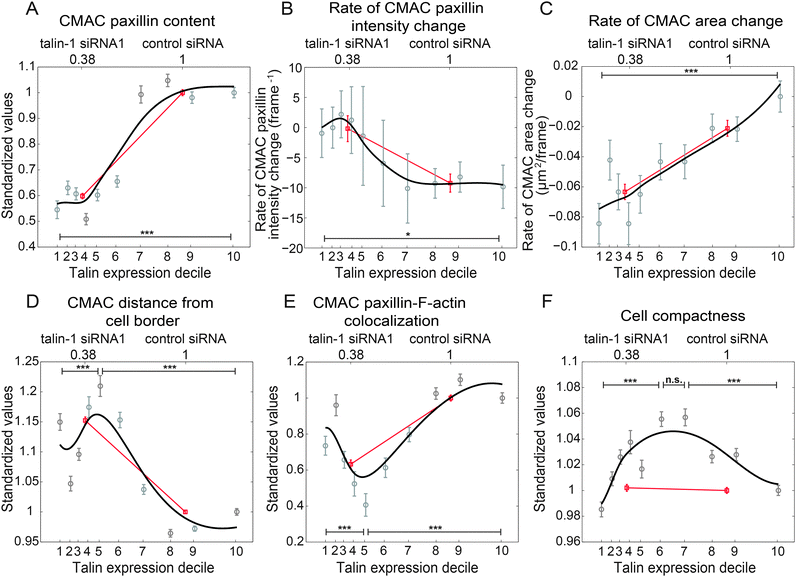 | ||
| Fig. 3 Analogue mapping of the talin expression spectrum reveals non-monotonic relationships between talin expression and cellular properties. Binary comparison of selected cellular properties between talin knockdown and control cells implies monotonic# dependencies (red lines) to talin expression-level (as described in Fig. S5, ESI†). In contrast, trends derived from decile-based stratification of talin expression-levels (black lines show smoothing spline fits based on median property values, per talin decile) reveal both monotonic (CMAC paxillin content (A), rate of CMAC paxillin intensity change (B), rate of CMAC area change (C)), and non-monotonic responses (CMAC distance from cell border (D), CMAC paxillin-F-actin colocalization (E), cell compactness (F)) associated with talin level changes. Normalized median talin expression values of control and talin knockdown cells are shown on the upper X-axes, while tick mark locations on the lower X-axes designate normalized median talin expression values of talin deciles. Y-axes in A and D–F display median property values in each talin decile, following standardization to the median of D10, while the Y-axes in B and C show absolute rates as indicated. In the binary comparisons, median property values of the knockdown cells were standardized to the median values of the control cells. Error bars indicate the 95% confidence intervals of the medians (details in Materials and methods). ***: p-values < 0.001, obtained from Wilcoxon rank-sum tests. Number of observations: control siRNA: 6684, talin siRNA: 8844; D1: 1425, D2: 1413, D3: 1514, D4: 1319, D5: 1426, D6: 1557, D7: 1532, D8: 1781, D9: 1736, D10: 1825. (Recall that deciles are based on the number of cells, not cell observations). # We apply the term “monotonicity” in the non-strict sense: i.e. a monotonically increasing trend may be both increasing and non-decreasing (flat). | ||
Fig. 3A–C display monotonic responses to progressively lower talin expression, including: a roughly sigmoidal reduction in CMAC paxillin content (suggesting a switch-like response with sensitivity at moderate talin levels, Fig. 3A); increasing rates of CMAC paxillin intensity change at moderate and low talin levels (with little response at high levels, Fig. 3B); and a nearly linear reduction in the rate of CMAC area change (Fig. 3C). Notably, the latter is the only one of the six displayed examples where binary analysis provides an accurate depiction of the true response pattern, with Fig. 3A and B describing distinctly non-linear relationships to talin expression. Even more striking are the non-monotonic response patterns observed in Fig. 3D–F, wherein: CMAC distance from cell border first increases then decreases with reducing talin (Fig. 3D); CMAC paxillin-F-actin colocalization first decreases then increases with reducing talin (Fig. 3E), and; cell compactness increases and then decreases with reducing talin (Fig. 3F). Note that when trends within control siRNA and talin-1 siRNA1 conditions are assessed independently, clear support is provided for the dependence of these responses on talin expression-level, rather than non-specific conditional effects (Fig. S8, ESI†). Critically, while linear inferences capture at least some minimal sense of the relationships depicted in Fig. 3D and E, the binary approach fails completely to describe, or even detect the existence of, the relationship between talin expression and cell compactness. Insensitivity to the symmetrical form of this relationship may explain the apparent inconsistency in binary inference results for this relationship when comparing two anti-talin siRNAs (Fig. S5H, ESI†) or assessing experimental reproducibility (Fig. S6H, ESI†). This is because the insensitivity of the binary method to the true signal increases its relative susceptibility to experimental noise. Overall, these results demonstrate an important proof-of-principle: the existence of non-monotonic responses to protein expression variation.
Probability distribution analyses reveal selective influences of talin on CMAC sub-populations
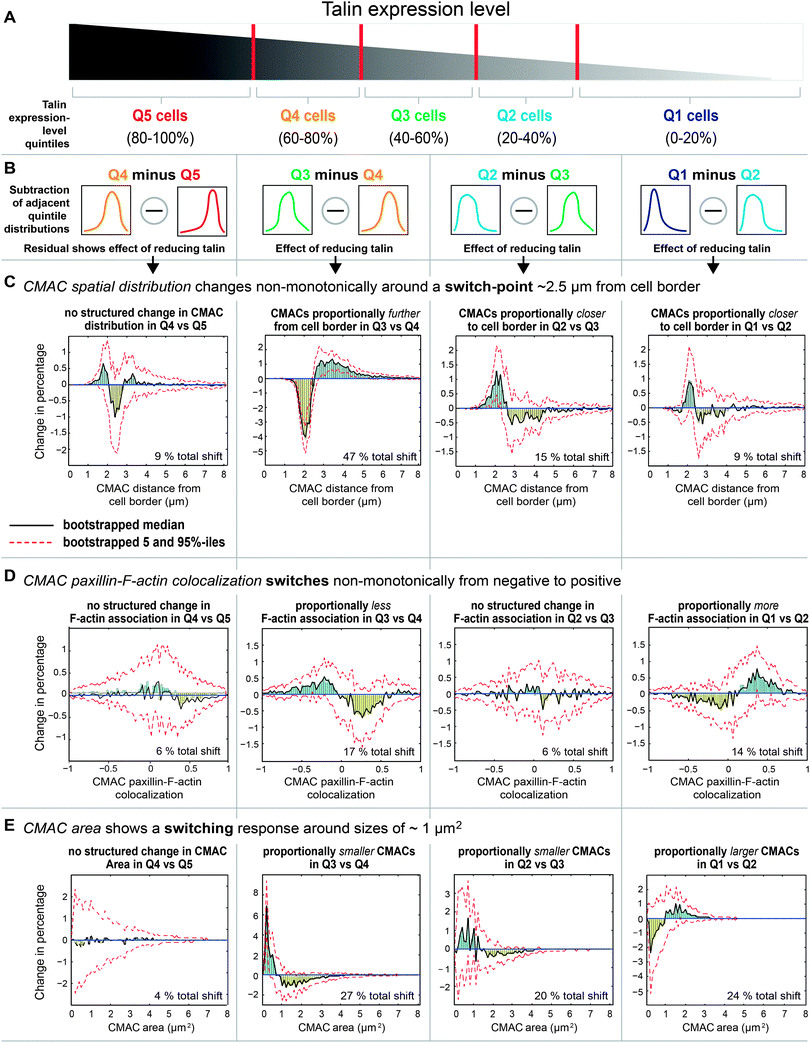
Having detected a range of complex talin-dependent alterations in cellular features, we next investigated whether talin expression-level variations may exert uniform or differential effects across CMAC populations. This is motivated by the existence of different CMAC sub-populations (e.g. nascent adhesions, focal complexes, focal adhesions31) within which the multi-functional talin protein may have alternate roles with distinctive concentration dependencies. To address this question, we stratified data for individual CMACs according to talin expression quintiles (Q1 [low] – Q5 [high], windows of 20%, Fig. 4A), calculated the probability distributions for properties within each quintile, and subtracted adjacent quintile probability distributions (Fig. 4B). This highlighted the selective influences on CMAC sub-populations of progressively lower cellular talin levels (Fig. 4C–E). Probability distributions were used in this setting to remove the influence of global changes in absolute adhesion number (which tended to decrease with lower talin levels).Using this approach, we observed complex changes in CMAC distance from cell border distributions as talin levels reduce from one quintile to the next (Fig. 4C), as follows: Q5 to Q4 – a small (9% total shift), noisy change in adhesion distribution (this reflects the limited median response to talin variation in this range); Q4 to Q3 – a large relative increase in central adhesions and decrease in peripheral adhesions (47% total shift, reflects increased CMAC distance from cell border in this talin range), with a switch-point in this effect ∼2.5 μm from the cell edge; Q3 to Q2 – inversion of the previous trend with relatively more peripheral adhesions detected (15% total shift, reflects a non-monotonic response and reduced CMAC distance from cell border in this talin range), with switch-point recurring at ∼2.5 μm from cell edge; Q2 to Q1 – a similar trend as Q3 to Q2 but weaker response (9% total shift, reflecting reduced CMAC distance from cell border in this talin range), with switch-point recurring at ∼2.5 μm from cell edge. Collectively, these results highlight the differential effects of talin level variation on CMAC frequencies at either side of a switch-point, or boundary, approximately 2.5 μm from the cell edge. Thus, the influence of talin expression-level variation is neither linear nor spatially uniform.
We next dissected the composition of the non-monotonic relationship between talin level and CMAC paxillin-F-actin colocalization (Fig. 4D). As talin levels are progressively reduced, we observed: Q5 to Q4 – a very small (6% total shift), noisy change in colocalization values (reflecting a limited aggregate response in this talin range); Q4 to Q3 – a moderately reduced proportion of adhesions having positive paxillin-F-actin colocalization (17% total shift, reflecting reduced aggregate colocalization in this talin range), switching around zero (from positive to negative); Q3 and Q2 – a very small (6% total shift), noisy change in colocalization values (reflecting a limited aggregate response in this talin range); Q2 to Q1 – a moderate increase (14% total shift) in the proportion of adhesions having positive colocalization (reflecting increased aggregate colocalization in this talin range), switching around zero (from negative to positive). Overall, this analysis details how reducing talin levels first decreases then increases the probability of EGFP-paxillin-RubyRed-LifeAct signal colocalization within CMACs by shifting the balance between spatial correlation and anti-correlation around the zero correlation boundary.
Given that adhesion localization (Fig. 4C) and adhesion-F-actin association (Fig. 4D) are both associated with CMAC maturation processes, we extended our probability distribution analysis to explore how heterogeneity in talin levels influences CMACs of different sizes (CMAC area, Fig. 4E). As talin levels progressively decrease, we observe: Q5 to Q4 – no coherent effect on the balance of CMAC area probabilities (4% total shift); Q4 to Q3 – a major reduction in the relative frequencies of large adhesions (27% total shift), with a switch-point at ∼1 μm2; Q3 to Q2 – continued reductions in the probability of large adhesions (20% total shift), with a switch point at ∼1.5 μm2; Q2 to Q1 – inversion of the previous trend, with increases in the probability of large adhesions (24% total shift, see example images in Fig. S9, ESI†), switching again at ∼1 μm2. This inverted response to decreasing talin arises entirely within the talin-1 siRNA1 data population and is therefore attributable to changes in talin expression-level, rather than potential non-specific conditional effects. Importantly, we note the highly structured differential influence of talin levels on the probabilities of large (area >1 μm2) versus small (area <1 μm2) adhesions across Q4–Q1, and the recurrence of the sign-change around the 1 μm2 value. This unexpected observation implies a precise and stable boundary delimiting talin's influence across the adhesion size spectrum. Mechanistically, this suggests talin's functional association with a switch-like event in CMAC maturation, occurring around the 1 μm2 boundary.
Talin levels influence the correlative relationships between cellular features
Above, we have established that variations in talin expression-level have a range of impacts on individual cellular properties. However, these analyses have not yet addressed the possibility that talin may also influence the relationships between cellular properties, such as how they are quantitatively and perhaps functionally correlated. Therefore, we next examined how talin expression-level variation impacts upon pair-wise correlations between an extended set of cellular properties linked to the processes of cell adhesion and migration.Continuous testing of the talin expression spectrum using partially overlapping sampling windows was used to assess talin level-dependent changes in Spearman‘s correlation coefficients between pairs of cellular properties (Fig. S10A and B, ESI†). We subsequently determined the median Spearman's correlation values within three talin expression-level groups, defined as low, moderate and high (Fig. S10C, ESI†). Each group covered an equal range of talin expression-levels, rather than equal cell numbers (as for deciles 1–10 in Fig. 3 or quintiles 1–5 in Fig. 4). Specifically, low talin spans the majority of the talin knockdown cell population, moderate talin contains mostly control siRNA-treated cells, and high talin corresponds to talin levels near and above the median of control cells. We applied a high stringency to identify statistically significant positive and negative correlations within these ranges, only selecting those where 95% confidence intervals excluded zero (Fig. S10C, ESI†).
The resulting inter-property relationships were arranged as correlative networks spanning features of the cell migration system,22 at the three levels of talin expression (Fig. 5A). Comparison of these networks highlighted talin level-dependent changes in network connectivity (Fig. 5B), as well as robust, talin level-independent relationships (Fig. 5C). For example, a positive correlation was detected between cell speed and cell compactness, consistent with our recent finding of a causal relationship between cell migration speed and cell shape.22 However, this relationship was only detectable in cells with high and moderate talin levels, but not at low talin levels. Thus, this correlation appears to be decoupled with talin depletion, an example similar to the type of “breaking” effect reported by Geiger and colleagues.17 Importantly though, we also observed that at low talin levels, higher cell speed correlated with increased CMAC distance to cell border (meaning that adhesions are located more centrally in faster cells), also consistent with our previous observations.22 Yet at moderate and high talin expression-levels (reflecting natural heterogeneity), the correlation between cell speed and CMAC positioning could not be detected. This, among other examples (Fig. 5B, moderate minus low), demonstrates that new inter-property relationships may become coupled via talin depletion, indicating adaptive responses rather than generalized decoupling.
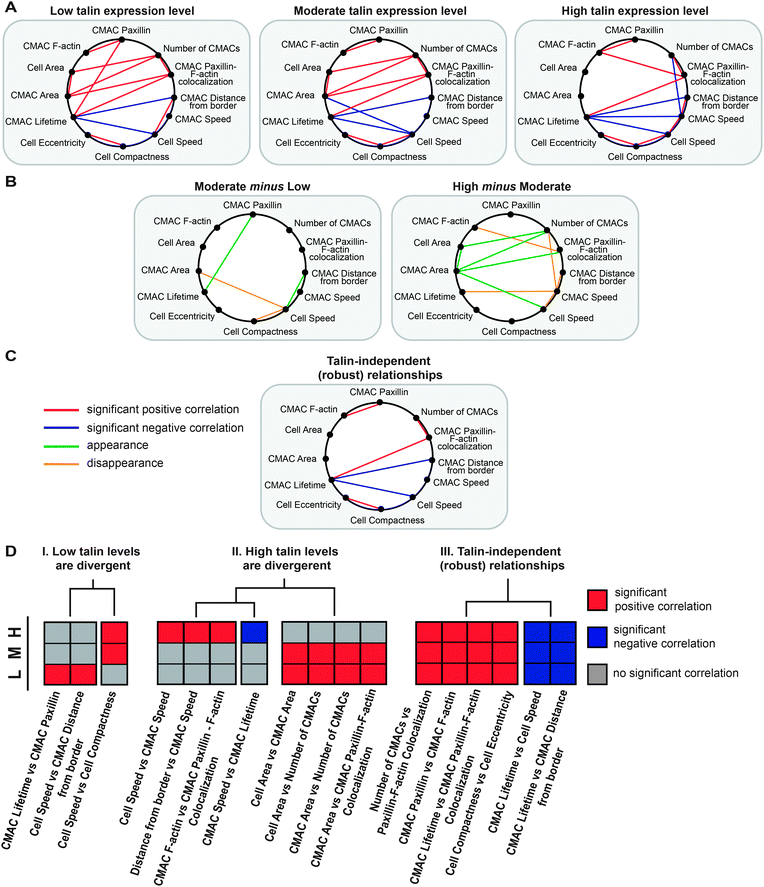 | ||
| Fig. 5 Talin level-dependent sensitivity in the correlations between cellular properties. (A) Given talin expression-level-stratified analyses of Spearman's correlations between pairs of cellular properties (described in Fig. S10, ESI†), significant Spearman's correlations detected at low, moderate and high levels of talin expression are displayed in inter-property correlative networks linking an expanded set of cellular properties. (B) “Subtraction” of these networks highlights how they change between low and moderate talin expression-levels, and between moderate and high talin expression-levels. (C) Correlative relationships that are robust to variation in talin expression-levels are also detected through analysis of networks in A. (D) All relationships depicted in the above networks were qualitatively clustered based on their sign and presence/absence at low, moderate and high talin levels. Clustering provides a systematic overview of relationships with similar dependencies. Rows denote the three levels of talin expression. The main clusters are: I. correlations divergent (only present or absent) at low talin levels; II. correlations divergent (only present and absent) at high talin levels; III. talin-independent, robust relationships. | ||
Some relationships are only detectable at high, but not at moderate or low talin levels. For example, cell speed was positively correlated with CMAC speed in cells with high talin levels, showing that when these cells move faster, CMACs also move faster. In contrast to the talin level-sensitive relationship between cell speed and CMAC speed, CMAC lifetime was negatively associated with cell speed at all talin expression ranges (Fig. 5C), showing that adhesion populations were more transient in faster moving cells, independent of talin levels.
Considering general patterns in this data, it is notable that most changes in the correlative networks arose between moderate and high talin ranges. Given that these groups are composed predominantly of control cells, this suggests that natural heterogeneity in talin may be sufficient to strongly influence the relationships between cellular features associated with control of the cell migration system.
Systematic analysis of talin level-sensitivity in inter-feature correlations
In order to systematically characterize the influence of talin levels on the correlative networks described in Fig. 5A, we qualitatively clustered the significant correlative relationships (based on their sign and their presence or absence) with respect to talin expression-levels (Fig. 5D). Given this clustering, three main relationship patterns were identified: (I) correlations absent or present only at low talin expression-levels. The positive correlation of CMAC lifetime and CMAC paxillin content was only detected at low talin expression-levels, showing that more stable adhesions tend to accumulate more paxillin when talin expression is low, but not when talin is high. This suggests changes in adhesion component recruitment kinetics dependent on talin levels, in line with the talin-dependent trend in the rate of net change in paxillin content, which became positive at low talin levels (Fig. 3B). In addition, the above-mentioned correlation between cell speed and cell compactness was detected in high and moderate talin cells, but not in cells expressing low levels of talin. (II) Correlations only present or only absent at high talin expression-levels reflect the regulatory influence of talin expression-level heterogeneity within the natural range, i.e. cell autonomous mechanisms. For example, if a correlation is only significant at high talin concentrations, it implies that stochastic or regulated cellular mechanisms would be sufficient to decouple that particular relationship. Notably, several such examples related to CMAC speed were found at high talin levels: CMAC speed was positively correlated with cell speed and CMAC distance from cell border, while being negatively correlated with CMAC lifetime. If, conversely, a correlation is absent only at high talin levels, it indicates decoupling above a talin expression-level threshold. In this cluster, we found properties associated with cell and CMAC size, as well as CMAC number: cell area vs. CMAC area; cell area vs. number of CMACs; CMAC area vs. number of CMACs; and CMAC area vs. CMAC paxillin-F-actin colocalization. This combination indicates that well spread cells have relatively plentiful and large adhesions in the context of low and moderate talin expression-levels (i.e. adhesion strength may be limiting for spreading), whereas in high talin expressing cells, spreading and CMAC features no longer correlate (perhaps because adhesion strength no longer limits spreading). (III) Correlations independent of talin-expression-levels are relationships robust to changes in talin levels. For example, the positive correlation between CMAC lifetime and CMAC paxillin-F-actin colocalization suggests that in cells with more stable adhesions, adhesions are more associated with F-actin. This relationship appears insensitive to talin expression-level variation.Discussion
In this study, we explored two types of responses associated with heterogeneity in talin expression-level. First we performed an analogue mapping of changes in individual cellular properties when conditioned on varying talin levels (focusing on several with conflicting published indications). Second, we explored how changes in talin levels influenced the correlations between cellular properties associated with cell adhesion and migration. In the first case, by stratifying cells according to talin expression-levels, we uncovered both linear and non-linear correspondences between talin expression and various cellular properties. Most significantly, several of these non-linear responses were in fact non-monotonic. This proof-of-principle finding advocates an important shift in how talin function, and perhaps protein function more generally, should be both assayed and conceived. In the second case, we detailed widespread and differential talin expression-level sensitivity in the correlative relationships between cellular properties, many of which are related to the process of cell migration. Overall, these findings highlight the complex and context-dependent nature of talin's influence on cellular properties. They also provide a basis to compare the types of inferences achievable when applying binary or analogue approaches to study protein function.As detailed earlier, the published literature provides an inconsistent picture of talin's influence over various cellular properties. For example, different responses to talin depletion have been published with respect to adhesion localization and F-actin association at the macromolecular scale, and with respect to cell shape at the cellular scale.13–17 Crucially, through our analogue assessment of talin expression, we present a framework within which many of these contradictory findings may be coherently integrated. This is based on the non-monotonic dose-dependence of talin correspondences with each of these features. This non-monotonicity means that responses to talin expression variation (e.g. experimental depletion) are contextually dependent on both the initial talin expression-level and the final talin level. This is in contrast to the linear inferences that can be drawn from binary analyses, which by their nature imply that only the strength, but not the direction (sign of change), of the response to expression perturbation is dependent on the starting and final values. The importance of differences in inferential capability between binary and analogue methods is best exemplified by the non-monotonic relationship between cell compactness and talin levels. This is because the particular form of this relationship (a roughly symmetrical “bell-shaped” non-monotonic dependence) is a worst-case scenario in terms of the sensitivity of binary analyses, which in such circumstances provide false negative indications, i.e. talin appears to have no functional relationship with cell shape. The potential for such false negatives to arise systematically in, for example, high-throughput RNAi or CRISPR-based depletion screens, may be significant. In positive terms, accurately mapping the concentration-dependent functional influences of proteins within cellular processes may enable substantial improvements in the ability to, for example, model and thereby understand the mechanisms underpinning complex cellular systems. In addition, when combined with perturbations of protein level (e.g. RNAi), analogue approaches may also help researchers to differentiate specific responses to protein level variation from non-specific conditional responses (such as RNAi off-target effects). This can be achieved by comparing expression-level dependence trends (for each recorded property) between control and knockdown conditions. Where expression-levels overlap between conditions, similar response trends support protein expression-level dependence. On the other hand, dissimilar trends at equivalent protein levels imply the influence of non-specific conditional effects. As applied in this study, where non-overlapping responses were excluded from further trend analysis, this comparison provides a mechanism to differentiate intended and unintended perturbation effects, thereby further improving data interpretation and inference.
How might non-monotonic talin expression-level dependencies arise? In general terms, this is most likely due to the interaction of several different yet interdependent mechanisms with which talin, a multi-functional protein, is associated. Indeed, as already noted, talin is known to play roles in integrin activation and clustering;5,32 adhesion reinforcement through F-actin recruitment;8,33 and adhesion disassembly.11,12 We hypothesize that these core mechanisms may have different sensitivities to talin expression-level, such that talin titration continually shifts the balance of their effects, producing complex outcomes observable at both adhesion and cellular scales.
Among the three core mechanisms noted above, the adhesion-F-actin linkage may be most prominent in defining non-monotonic talin dependencies. This is based not only on the observation that adhesion-F-actin association is itself non-monotonically related to talin levels, but also on our analysis of talin dose-dependent shifts in adhesion property distributions. In particular, the finding that talin titration differentially regulates small (<1 μm2) and large (>1 μm2) adhesions suggests selective influences of talin over CMACs in alternate states, such as focal complexes (typically <1 μm2) and focal adhesions (typically >1 μm2).34 Indeed, the recurrent boundary around 1 μm2 implies a switch-like transition, as has been proposed for focal complex to focal adhesion maturation. Critically, mechanisms reported to govern this switch revolve around the adhesion-F-actin linkage.35 These include, for example, competition for talin binding between RIAM and vinculin (an F-actin linker) and competition for integrin binding between talin and alternative F-actin linkers including α-actinin, filamin A and moesin.36–39 Mechanisms based on competition between talin and alternative integrin-F-actin linkers, such as α-actinin, filamin A or moesin, appear feasible since these proteins could be expected to outcompete the depleted talin population, thereby re-establishing associations between adhesions and F-actin. This corresponds with our findings that both paxillin-F-actin colocalization and adhesion area are increased given maximal talin depletion (compared to moderate depletion). The role for talin-sensitive regulation of the adhesion-F-actin linkage is also supported, though more indirectly, by the switch-like changes observed in adhesion localization probabilities at ∼2.5 μm from the cell edge. Given the coincidence between this probabilistic boundary and the spatial boundary between lamellipodial and lamellar domains in H1299 cells,40 it may be that the changes in actin filament organization, force generation and adhesion linkage that accompany this domain transition also influence the spatial sensitivity of adhesions to talin level variation. The lamellipodial-lamellar transition is indeed often associated with focal complex-to-focal adhesion switching, due to these F-actin-dependent changes.41 Thus, our data supports a key role for the adhesion-F-actin connection in shaping non-monotonic responses to talin titration. However, it is clear that effectively disentangling the contributions of the numerous molecules that likely comprise this non-linear mechanism will demand extensive and systematic approaches based on spatially and temporally resolved protein-concentration and -interaction data.
To fully understand talin's influence it is important to consider not only how expression-levels determine the values of cellular properties, but also how talin levels may alter relationships between these properties. Accordingly, we here demonstrate talin expression-level-dependent changes in networks of correlative inter-property relationships. Interestingly, these changes include both the decoupling and novel coupling of relationships in response to talin depletion, implying an adaptive response rather than only the disruption of correlative relationships.17 Notably, our analogue analysis allowed us to observe that the correlative relationships detected were dependent not just on the presence or absence of talin, but on specific talin levels. Indeed, most of the recorded relationships were sensitive to talin levels, but have significantly different sensitivity-thresholds (i.e. talin values at which the relationship appears/disappears). Interestingly, talin expression variation within the natural range had larger impacts on detected correlative networks than those recorded in the artificial (siRNA-depleted) range. Although this result may be contingent upon the particular properties explored, it suggests that endogenous talin level heterogeneity may have relatively strong impacts upon correlative connectivity between an array of cellular properties.
Conclusion
The key methodological advantages achieved herein derive from the integrated quantification of data capturing both talin expression-level and cellular properties-of-interest on a per cell basis. This is facilitated by correlative imaging of matched live and fixed, immunolabeled cells – an approach that is easily accessible to most researchers, requiring only immunofluorescence-compatible antibodies to the target protein. Notably though, the emergence of CRISPR/Cas9 editing technologies42,43 presents a highly attractive alternative approach, namely the creation of fluorescent protein fusions to endogenous genes. This enables the monitoring of target protein levels in real time, providing access to the spatial and temporal dynamics of target proteins, as well as permitting analyses over longer time frames – where our endpoint approach would become un-interpretable.Overall, the integrated single cell approach described here offers a method to avoid the pitfalls associated with aggregate descriptions of populations based on the “typical cell”.23 As a result, subsequent analogue analyses permitted the detection of non-monotonic dependencies on talin expression-level, and the mapping of extensive changes in correlative inter-property relationship networks over multiple talin expression-levels. These are novel outcomes in terms of talin-function analysis, but also in relation to general protein function analysis. Importantly, these approaches also grant improved physiological relevance to functional inferences due to the use of natural expression-level heterogeneity to infer patterns of protein influence,25,27 rather than relying solely on potentially extreme knockdown approaches. Nonetheless, the expansion of natural talin heterogeneity with RNAi, producing an extended yet continuous spectrum of talin expression, provided both enhanced sensitivity in our analysis, and a direct point of comparison to canonical approaches. Collectively, the combination of experimental design, correlative imaging and analogue data analysis has produced a number of proof-of-principle advances with wide applicability.
Materials and methods
Cell culture and siRNA transfection
H1299 (human non-small cell lung carcinoma, ATCC) cells, stably expressing EGFP-paxillin and RubyRed-LifeAct (H1299-P/L, details in ref. 22) were cultured in RPMI 1640 medium (Gibco) supplemented with 1 mM Glutamine and 10% fetal bovine serum (Gibco) and 400 μg ml−1 Geneticin (G-418 sulfate, Gibco) at 37 °C, 5% CO2. Oligonucleotides targeting human talin-1 with the following sequences: talin-1 siRNA1 (5′-GAA GAU GGU UGG CGG CAUU-3′); talin-1 siRNA2 (5′-GAA GAG AUA GGU UCC CAUA-3′) were synthesized by GenePharma (Shanghai, China). For siRNA transfection, 2 × 104 H1299-P/L cells per well were seeded into a 24-well plate. 24 h later, 20 pmol siRNA was transfected into H1299-P/L cells with 2 μl RNAiMAX (Invitrogen, USA), according to the manufacturer's protocol. After 48 h incubation, cells were harvested either for live cell imaging or for immunoblotting.Confocal cell fluorescence imaging and correlative microscopy
96-well glass-bottomed plates (0.17 mm optical glass, Matrical Bioscience, USA) were coated with purified fibronectin (5 μg ml−1) at 37 °C for 2 h followed by blocking with 1% heat denatured bovine serum albumin (Sigma-Aldrich) at 37 °C for 1 h. 2000 H1299 P/L cells were plated per fibronectin-coated well, and 2 h later, cells were subjected to live imaging with a Nikon A1 confocal microscope using an oil-immersion objective (PlanApo VC 60X/1.4 NA). Cells were maintained during imaging in normal culture medium without fetal bovine serum, at 37 °C and 5% CO2. Images of live cells were acquired at 5 min intervals for 6 h with a pixel resolution of 0.21 μm. Following live imaging, the cells were immediately fixed and immunolabeled for talin as described below. Next, using correlative microscopy, labeled cells were imaged again to acquire the information on talin expression-level of the same individual cells captured during live imaging. The talin-labeling protocol was optimized to retain cytoplasmic talin (details below). Thus, by using confocal microscope imaging, we capture the pool of talin available at the plane of the adhesion complexes. This quantity is recorded as the mean talin pixel intensity value per cell, reflecting the talin content of each cell and is referred to as talin expression-level.Automated image analysis
Acquired images were analyzed with the PAD software (v6.3) (Digital Cell Imaging Laboratories, Keerbergen, Belgium) as previously described.22 Briefly, CMACs with an area > 0.05 μm2 were segmented based on EGFP-paxillin signal, whereas cell boundaries were detected based on Ruby-LifeAct fluorescence. Segmented cells and CMACs were tracked over time based on nearest neighbor analysis (Fig. S4, ESI†). Only CMACs tracked continually for a minimum of 3 time-points were included in analyses, thereby minimizing the influence of image (e.g. shot) noise. Quantitative properties defining cells, per cell CMAC populations and individual CMACs were automatically extracted. CMAC intensities were background corrected by subtracting the local mean intensity (per channel, within 1 μm of each CMAC, excluding adjacent CMACs) from the mean intensity of each CMAC. In order to compare independent experiments (N = 7), intensity values from each channel (talin, RubyRed-LifeAct, EGFP-paxillin) were normalized to the median value per channel of the control siRNA-treated cell population, per experiment.Immunocytochemistry
Immunolabeling of control siRNA and talin-1 siRNA-transfected H1299-P/L cells for talin was performed with a liquid-handling robot (Model FREEDOM EVO, Tecan, Switzerland) to minimize the variance between experimental repeats. Briefly, the cells were fixed in 2% paraformaldehyde for 10 min at RT, washed once with PBS, and permeabilized with 0.1% Triton X-100 for 5 min. Following 4× washing, cells were blocked by incubation with 1% BSA/PBS. Then cells were incubated with an anti-talin antibody (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100, clone 8d4, Sigma-Aldrich) for 30 min, washed 4× with PBS, and incubated with Alexa 647-conjugated goat anti-mouse IgG (1
100, clone 8d4, Sigma-Aldrich) for 30 min, washed 4× with PBS, and incubated with Alexa 647-conjugated goat anti-mouse IgG (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300, Invitrogen, USA) for 30 min at RT. The cells were washed with PBS 4× before the plate was re-imaged at the microscope. Notably, immunofluorescent labeling of talin was optimized to retain the total cytoplasmic talin pool, as opposed to the canonical optimization for adhesion labeling. Such adhesion-specific labeling could also be achieved with the same antibody using a simultaneous fixation and permeabilization condition (Fig. S2A, ESI†). Specificity of the talin antibody was confirmed by comparison with secondary antibody (Alexa 647-conjugated goat anti-mouse IgG) labeling only (Fig. S2B and C, ESI†).
300, Invitrogen, USA) for 30 min at RT. The cells were washed with PBS 4× before the plate was re-imaged at the microscope. Notably, immunofluorescent labeling of talin was optimized to retain the total cytoplasmic talin pool, as opposed to the canonical optimization for adhesion labeling. Such adhesion-specific labeling could also be achieved with the same antibody using a simultaneous fixation and permeabilization condition (Fig. S2A, ESI†). Specificity of the talin antibody was confirmed by comparison with secondary antibody (Alexa 647-conjugated goat anti-mouse IgG) labeling only (Fig. S2B and C, ESI†).
Immunoblotting
Cells cultured in 24-well plates up to 90% confluence were washed with ice-cold PBS, then lysed with 100 μl cell lysis buffer (1 × PBS, 1% Triton-X 100, 0.5% sodium deoxycholate, 0.1% SDS, 1 mM EDTA, proteinase inhibitor cocktail (Roche Diagnostics)) on ice for 20 min. Whole cell lysates were collected and centrifuged at 4 °C, 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min. The resulting supernatants were collected and their protein concentrations were determined using a BCA protein quantification kit (Pierce/Thermo Scientific, USA). 20 μg of total protein from each sample was subjected to SDS-PAGE, followed by immunoblot analysis with primary antibodies against talin (1
000 rpm for 10 min. The resulting supernatants were collected and their protein concentrations were determined using a BCA protein quantification kit (Pierce/Thermo Scientific, USA). 20 μg of total protein from each sample was subjected to SDS-PAGE, followed by immunoblot analysis with primary antibodies against talin (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500, 8d4, Sigma-Aldrich), paxillin (1
500, 8d4, Sigma-Aldrich), paxillin (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200, 5H11, Invitrogen), vinculin (1
200, 5H11, Invitrogen), vinculin (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 150
150![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, V9131, Sigma-Aldrich), filamin-A (1
000, V9131, Sigma-Aldrich), filamin-A (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500, MAB1680, Millipore), FAK (1
500, MAB1680, Millipore), FAK (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500, BD610087, BD), zyxin (1
500, BD610087, BD), zyxin (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500, Z4751, Sigma-Aldrich), β-pix (1
500, Z4751, Sigma-Aldrich), β-pix (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500, MAB3829, Millipore), ERK (1
500, MAB3829, Millipore), ERK (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000, 9102, Cell Signaling Technology), β1-integrin (1
1000, 9102, Cell Signaling Technology), β1-integrin (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3000, BD610087, BD), α-actinin (1
3000, BD610087, BD), α-actinin (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500, A5044, Sigma-Aldrich), tubulin (1
500, A5044, Sigma-Aldrich), tubulin (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000, DM1A, Fisher Scientific) and the corresponding HRP-labeled secondary antibodies (anti-mouse: 1
2000, DM1A, Fisher Scientific) and the corresponding HRP-labeled secondary antibodies (anti-mouse: 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3000, Jackson Immunoresearch Laboratories, Inc, anti-rabbit: 1
3000, Jackson Immunoresearch Laboratories, Inc, anti-rabbit: 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3000, Jackson Immunoresearch Laboratories, Inc) using an enhanced chemiluminescence detection system (Pierce/Thermo Scientific).
3000, Jackson Immunoresearch Laboratories, Inc) using an enhanced chemiluminescence detection system (Pierce/Thermo Scientific).
Statistical comparison of different talin siRNAs and experimental repeats
The selected variables describing cell adhesion and migration of talin siRNA1 and talin siRNA2 treated cells were standardized to the median values of their corresponding controls (control siRNA-treated cells) per experiment. Following this standardization, the data from the controls were pooled (see Fig. S5, ESI†). In the comparison of the seven different experiments, the same type of standardization was performed (Fig. S7, ESI†). Medians of talin siRNA and control siRNA-treated cells were compared using two-sided Wilcoxon rank sum test. P-values were adjusted for multiple comparisons applying Bonferroni correction. We chose a significance threshold of p < 0.001. Statistical analyses were performed using custom-written functions in MATLAB R2013b (The MathWorks, Inc., Natick, Massachusetts, United States).Data extraction
The data set was constructed using a combination of variables extracted on different spatial (cells and CMACs) and temporal scales (live cell recordings correlated with talin antibody labeling). To combine these scales, individual trajectories of cells and CMACs were combined using MATLAB and each cell was categorized according to its mean talin intensity at the point of fixation and staining. CMAC statistics were calculated based on the trajectories of individual CMACs while cell statistics were calculated per cell trajectory. Each of the statistically calculated variables was finally sorted by the talin concentration of the corresponding fixed and stained cell.Data stratification and analysis of univariate trends
The total data set consisted of quantitative cellular properties extracted on cell and CMAC levels from 275 cells. Based on talin expression-level, ten cell sub-populations were defined (deciles; D1: low talin levels; D10: high talin levels), wherein each decile contains data from 27 or 28 cells. When performing comparisons using the deciles, the selected variables were standardized to the median values of the decile with the highest talin expression value (D10).In order to estimate trends in the dependence of each variable on talin expression-level smoothing splines were fitted to each variable as a function of talin expression-decile. These splines where calculated using De Boor's algorithm with a smoothing parameter of 0.999.
The 95% confidence intervals (CI) for the medians were calculated consistently based on MATLAB's “boxplot” function as follows: lower CI: 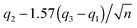 , upper CI:
, upper CI: 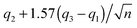 , where q2 is the median (50th percentile), q1 and q3 are the 25th and 75th percentiles, respectively, and n is the number of observations. Note that the concept of monotonicity was applied in the non-strict sense: i.e. a monotonically increasing trend may be both increasing and non-decreasing (flat).
, where q2 is the median (50th percentile), q1 and q3 are the 25th and 75th percentiles, respectively, and n is the number of observations. Note that the concept of monotonicity was applied in the non-strict sense: i.e. a monotonically increasing trend may be both increasing and non-decreasing (flat).
Importantly, trend data was parsed by excluding from further trend analysis any variables for which there was poor overlap between trends at equivalent talin expression-levels in control siRNA and talin-1 siRNA1 conditions (as assessed in Fig. S8, ESI†).
Calculation of differential probability distributions
We defined five talin expression quintiles, each containing data from 55 cells (Q1: low talin expression, Q5: high talin expression). Histograms, consisting of the same bin size and interval between minima and maxima of CMAC level variables in each talin quintile were calculated. To account for the different number of adhesions within talin expression quintiles, we calculated probability distributions within bins by dividing the values of each variable with the total number of CMAC observations within that quintile (Fig. 4). The proportional changes between adjacent talin expression quintiles (e.g. Q1 and Q2) were calculated by subtracting the bootstrapped proportional distributions (resampling with replacement) of adjacent quintiles.Continuous sampling of talin expression-levels
Per cell median values of each property were sorted according to increasing talin expression-levels of individual cells. Using a moving window sampling (window size: 25, overlap: 20), we calculated Spearman's rank correlation between any two variables within the sampling windows. We have chosen Spearman's rank correlation over the Pearson's product-moment correlation because we have variables measured on different scales. In addition, Spearman is suitable to detect non-linear relationships. By calculating the bootstrapped Spearman's correlation (using the MATLAB function “bootci”) within the overlapping sample windows, we obtained the median correlation value and the corresponding 95% confidence interval. Repeating this sampling on the permutation of talin expression-levels showed no significant correlations, indicating our ability to detect true correlations. A stringent filter was applied when selecting correlations represented within the networks of Fig. 5: we considered only significant correlations, wherein the confidence intervals did not contain the 0 value (meaning the majority of the bootstraps produced a correlation value other than 0), regardless of the exact value of the correlation coefficient. In order to reduce the bias for responses rather associated with talin knockdown than with per se talin expression-levels, only continuous correlative trends across knockdown and control conditions were evaluated.Acknowledgements
The authors thank Pablo Hernandez-Varas, Ulrich Berge and Gabriela Imreh for their comments and discussion on the manuscript. This work was supported by grants to SS from the EU-FP7 – Systems Microscopy NoE (Grant No. HEALTH-F4-2010-258068), the Center for Innovative Medicine at KI, the Swedish Research Council (Grants No. 340-2012-6001 and 521-2012-3180), and the Swedish Cancer Society. Imaging occurred at the live cell-imaging unit at the Department of Biosciences and Nutrition at KI, supported by grants from the Knut and Alice Wallenberg Foundation, the Swedish Research Council, the Centre for Innovative Medicine, the Jonasson donation to the School of Technology and Health, and the Royal Institute of Technology, Stockholm, Sweden. HAS was supported by a scholarship by the Higher Education Commission of Pakistan. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.References
- R. Horwitz and D. Webb, Curr. Biol., 2003, 13, R756–R759 CrossRef CAS PubMed.
- J. G. Lock, B. Wehrle-Haller and S. Stromblad, Semin. Cancer Biol., 2008, 18, 65–76 CrossRef CAS PubMed.
- D. A. Calderwood, Biochem. Soc. Trans., 2004, 32, 434–437 CrossRef CAS PubMed.
- B. T. Goult, M. Bouaouina, P. R. Elliott, N. Bate, B. Patel, A. R. Gingras, J. G. Grossmann, G. C. Roberts, D. A. Calderwood, D. R. Critchley and I. L. Barsukov, EMBO J., 2010, 29, 1069–1080 CrossRef CAS PubMed.
- S. Tadokoro, S. J. Shattil, K. Eto, V. Tai, R. C. Liddington, J. M. de Pereda, M. H. Ginsberg and D. A. Calderwood, Science, 2003, 302, 103–106 CrossRef CAS PubMed.
- F. Margadant, L. L. Chew, X. Hu, H. Yu, N. Bate, X. Zhang and M. Sheetz, PLoS Biol., 2011, 9, e1001223 CAS.
- J. D. Humphries, P. Wang, C. Streuli, B. Geiger, M. J. Humphries and C. Ballestrem, J. Cell Biol., 2007, 179, 1043–1057 CrossRef CAS PubMed.
- A. Carisey, R. Tsang, A. M. Greiner, N. Nijenhuis, N. Heath, A. Nazgiewicz, R. Kemkemer, B. Derby, J. Spatz and C. Ballestrem, Curr. Biol., 2013, 23, 271–281 CrossRef CAS PubMed.
- B. Wehrle-Haller, Curr. Opin. Cell Biol., 2012, 24, 569–581 CrossRef CAS PubMed.
- D. R. Critchley, Annu. Rev. Biophys., 2009, 38, 235–254 CrossRef CAS PubMed.
- C. Huang, Z. Rajfur, N. Yousefi, Z. Chen, K. Jacobson and M. H. Ginsberg, Nat. Cell Biol., 2009, 11, 624–630 CrossRef CAS PubMed.
- S. J. Franco, M. A. Rodgers, B. J. Perrin, J. Han, D. A. Bennin, D. R. Critchley and A. Huttenlocher, Nat. Cell Biol., 2004, 6, 977–983 CrossRef CAS PubMed.
- X. Zhang, G. Jiang, Y. Cai, S. J. Monkley, D. R. Critchley and M. P. Sheetz, Nat. Cell Biol., 2008, 10, 1062–1068 CrossRef CAS PubMed.
- P. Wang, C. Ballestrem and C. H. Streuli, J. Cell Biol., 2011, 195, 499–513 CrossRef CAS PubMed.
- H. Priddle, L. Hemmings, S. Monkley, A. Woods, B. Patel, D. Sutton, G. A. Dunn, D. Zicha and D. R. Critchley, J. Cell Biol., 1998, 142, 1121–1133 CrossRef CAS.
- P. M. Kopp, N. Bate, T. M. Hansen, N. P. Brindle, U. Praekelt, E. Debrand, S. Coleman, D. Mazzeo, B. T. Goult, A. R. Gingras, C. A. Pritchard, D. R. Critchley and S. J. Monkley, Eur. J. Cell Biol., 2010, 89, 661–673 CrossRef CAS PubMed.
- S. E. Winograd-Katz, S. Itzkovitz, Z. Kam and B. Geiger, J. Cell Biol., 2009, 186, 423–436 CrossRef CAS PubMed.
- G. Giannone, G. Jiang, D. H. Sutton, D. R. Critchley and M. P. Sheetz, J. Cell Biol., 2003, 163, 409–419 CrossRef CAS PubMed.
- B. Nieves, C. W. Jones, R. Ward, Y. Ohta, C. G. Reverte and S. E. LaFlamme, J. Cell Sci., 2010, 123, 1216–1226 CrossRef CAS PubMed.
- D. H. Kim and D. Wirtz, FASEB J., 2013, 27, 1351–1361 CrossRef CAS PubMed.
- S. Sen, W. P. Ng and S. Kumar, J. R. Soc., Interface, 2012, 9, 1311–1317 CrossRef CAS PubMed.
- J. G. Lock, M. J. Mamaghani, H. Shafqat-Abbasi, X. Gong, J. Tyrcha and S. Stromblad, PLoS One, 2014, 9, e90593 Search PubMed.
- S. J. Altschuler and L. F. Wu, Cell, 2010, 141, 559–563 CrossRef CAS PubMed.
- J. W. Cotari, G. Voisinne and G. Altan-Bonnet, Curr. Opin. Biotechnol., 2013, 24, 760–766 CrossRef CAS PubMed.
- S. Farkash-Amar, A. Zimmer, E. Eden, A. Cohen, N. Geva-Zatorsky, L. Cohen, R. Milo, A. Sigal, T. Danon and U. Alon, PLoS Genet., 2014, 10, e1004176 Search PubMed.
- C. J. Ku, Y. Wang, O. D. Weiner, S. J. Altschuler and L. F. Wu, Cell, 2012, 149, 1073–1083 CrossRef CAS PubMed.
- A. Sigal, R. Milo, A. Cohen, N. Geva-Zatorsky, Y. Klein, Y. Liron, N. Rosenfeld, T. Danon, N. Perzov and U. Alon, Nature, 2006, 444, 643–646 CrossRef CAS PubMed.
- S. Farkash-Amar, E. Eden, A. Cohen, N. Geva-Zatorsky, L. Cohen, R. Milo, A. Sigal, T. Danon and U. Alon, PLoS One, 2012, 7, e48722 CAS.
- Z. Li, J. G. Lock, H. Olofsson, J. M. Kowalewski, S. Teller, Y. Liu, H. Zhang and S. Stromblad, Mol. Biol. Cell, 2010, 21, 3317–3329 CrossRef CAS PubMed.
- J. G. Lock and S. Stromblad, Exp. Cell Res., 2010, 316, 1438–1444 CrossRef CAS PubMed.
- B. Geiger and K. M. Yamada, Cold Spring Harbor Perspect. Biol., 2011, 3, a005033 Search PubMed.
- D. A. Calderwood, R. Zent, R. Grant, D. J. Rees, R. O. Hynes and M. H. Ginsberg, J. Biol. Chem., 1999, 274, 28071–28074 CrossRef CAS PubMed.
- H. S. Lee, R. M. Bellin, D. L. Walker, B. Patel, P. Powers, H. Liu, B. Garcia-Alvarez, J. M. de Pereda, R. C. Liddington, N. Volkmann, D. Hanein, D. R. Critchley and R. M. Robson, J. Mol. Biol., 2004, 343, 771–784 CrossRef CAS PubMed.
- M. L. Gardel, I. C. Schneider, Y. Aratyn-Schaus and C. M. Waterman, Annu. Rev. Cell Dev. Biol., 2010, 26, 315–333 CrossRef CAS PubMed.
- M. Vicente-Manzanares, C. K. Choi and A. R. Horwitz, J. Cell Sci., 2009, 122, 199–206 CrossRef CAS PubMed.
- H. S. Lee, P. Anekal, C. J. Lim, C. C. Liu and M. H. Ginsberg, Mol. Biol. Cell, 2013, 24, 1354–1362 CrossRef CAS PubMed.
- T. Kiema, Y. Lad, P. Jiang, C. L. Oxley, M. Baldassarre, K. L. Wegener, I. D. Campbell, J. Ylanne and D. A. Calderwood, Mol. Cell, 2006, 21, 337–347 CrossRef CAS PubMed.
- P. Roca-Cusachs, A. del Rio, E. Puklin-Faucher, N. C. Gauthier, N. Biais and M. P. Sheetz, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, E1361–E1370 CrossRef CAS PubMed.
- P. Vitorino, S. Yeung, A. Crow, J. Bakke, T. Smyczek, K. West, E. McNamara, J. Eastham-Anderson, S. Gould, S. F. Harris, C. Ndubaku and W. Ye, Nature, 2015, 519, 425–430 CrossRef CAS PubMed.
- J. Ahn, P. Truesdell, J. Meens, C. Kadish, X. Yang, A. H. Boag and A. W. Craig, Mol. Cancer Res., 2013, 11, 952–963 CrossRef CAS PubMed.
- U. S. Schwarz and M. L. Gardel, J. Cell Sci., 2012, 125, 3051–3060 CrossRef CAS PubMed.
- P. D. Hsu, E. S. Lander and F. Zhang, Cell, 2014, 157, 1262–1278 CrossRef CAS PubMed.
- B. L. Oakes, D. C. Nadler and D. F. Savage, Methods Enzymol., 2014, 546, 491–511 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c4ib00291a |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2015 |