Open Access Article
Open Access ArticleA study of SeqA subcellular localization in Escherichia coli using photo-activated localization microscopy†
Jacek T.
Mika‡
*a,
Aster
Vanhecke‡
a,
Peter
Dedecker
a,
Toon
Swings
b,
Jeroen
Vangindertael
a,
Bram
Van den Bergh
b,
Jan
Michiels
b and
Johan
Hofkens
a
aDepartment of Chemistry, KU Leuven, Celestijnenlaan 200F, Box 2404, 3001 Heverlee, Belgium. E-mail: jacek.mika@chem.kuleuven.be
bCentre of Microbial and Plant Genetics (CMPG), KU Leuven, Kasteelpark Arenberg 20, Box 2460, 3001 Leuven, Belgium
First published on 13th July 2015
Abstract
Escherichia coli (E. coli) cells replicate their genome once per cell cycle to pass on genetic information to the daughter cells. The SeqA protein binds the origin of replication, oriC, after DNA replication initiation and sequesters it from new initiations in order to prevent overinitiation. Conventional fluorescence microscopy studies of SeqA localization in bacterial cells have shown that the protein is localized to discrete foci. In this study we have used photo-activated localization microscopy (PALM) to determine the localization of SeqA molecules, tagged with fluorescent proteins, with a localization precision of 20–30 nm with the aim to visualize the SeqA subcellular structures in more detail than previously possible. SeqA–PAmCherry was imaged in wild type E. coli, expressed from plasmid or genetically engineered into the bacterial genome, replacing the native seqA gene. Unsynchronized cells as well as cells with a synchronized cell cycle were imaged at various time points, in order to investigate the evolution of SeqA localization during the cell cycle. We found that SeqA indeed localized into discrete foci but these were not the only subcellular localizations of the protein. A significant amount of SeqA–PAmCherry molecules was localized outside the foci and in a fraction of cells we saw patterns indicating localization at the membrane. Using quantitative PALM, we counted protein copy numbers per cell, protein copy numbers per focus, the numbers of foci per cell and the sizes of the SeqA clusters. The data showed broad cell-to-cell variation and we did not observe a correlation between SeqA–PAmCherry protein numbers and the cell cycle under the experimental conditions of this study. The numbers of SeqA–PAmCherry molecules per focus as well as the foci sizes also showed broad distributions indicating that the foci are likely not characterized by a fixed number of molecules. We also imaged an E. coli strain devoid of the dam methylase (Δdam) and observed that SeqA–PAmCherry no longer formed foci, and was dispersed throughout the cell and localized to the plasma membrane more readily. We discuss our results in the context of the limitations of the technique.
Introduction
SeqA is an Escherichia coli (E. coli) DNA-binding protein, that is involved in regulation of DNA replication initiation.1,2 During cell division E. coli replicates its DNA and there are mechanisms ensuring that each daughter cell receives one copy of the genome after cell division. In E. coli the DNA is usually methylated at GATC sites by the dam methylase. Newly replicated GATC sequences stay hemimethylated for a while until the dam enzyme gets the opportunity to methylate them. E. coli is able to recognize and inactivate new origins of replication because they are hemimethylated.3 In a process termed DNA sequestration, SeqA binds the new, freshly replicated origin of chromosomal replication (oriC), specifically at the hemimethylated GATC sequences that are highly abundant in the oriC region, whereby it prevents DnaA from starting replication initiation.1,2,4The subcellular localization of SeqA in E. coli is particular. It has been shown with immunofluorescence microscopy and GFP-tagging that the protein forms discrete foci inside E. coli that are predominantly localized on the bacterial nucleoid.5–7 The number of foci per cell varies between cells and also depends on growth condition (amount of chromosomes), but in general these studies found between 1 and 4 foci per cell under slow growth conditions.6,8,9 The nature of the foci is dynamic: as the cell grows a single focus divides into two foci that migrate in opposite directions within the cell.5 With conventional fluorescence microscopy approaches, these foci appear as single spots and it is possible that additional detail of these structures is obscured by the diffraction limit.
The size of bacterial cells (typically in the orders of micrometers) renders them difficult to study with conventional light microscopy. Due to the diffraction limit of visible light (±250 nm) only limited detail of the cellular substructure is revealed. In the last ten years, several new approaches have been developed that aim at increasing the resolution of fluorescence light microscopy.10 They are often referred to as superresolution microscopy techniques and give researchers a tool to look at structures of living cells with nanometer scale resolution.11–15 Superresolution microscopy has already contributed to an improved understanding of bacterial subcellular organization. To name a few examples, photo-activated localization microscopy PALM,11,12 has been used to visualize the chemotactic protein network,16 the spatial organization of the bacterial nucleoid and nucleoid associated proteins17–19 or to uncover features of the cytoskeletal structures formed by FtsZ and MreB20,21 proteins. The improved localization precision of this imaging technique, capable of imaging single proteins, revealed organizational features such as clustering and measured the sizes of structures that are below the diffraction limit on the tens of nanometers scale.22 Albeit with limitations originating from the nature of the fluorescent proteins (photophysics and maturation) PALM and other single molecule fluorescence microscopy techniques have been used to count or estimate protein copy numbers in E. coli cells.23–26
Inspired by the improved localization precision of PALM, we have employed this technique to visualize the subcellular localization of SeqA in E. coli with a spatial resolution that was previously not available to conventional fluorescence microscopy. To this end we have created E. coli strains where the native SeqA protein was replaced with a variant of the protein fused to fluorescent proteins (FPs: eYFP, mEos3.2 and PAmCherry) to investigate the protein at close to physiological expression levels. We have also performed PALM on cells overexpressing the SeqA-FP fusion from a plasmid in wild type cells and in cells devoid of the dam methylase. Using the stringent response, we have synchronized the cell cycle of the E. coli cells expressing SeqA–PAmCherry from the genome. Next, we looked into changes in subcellular localization of this protein and counted the protein copy numbers in the cells grown in rich medium at 37 °C and 30 °C using PALM. We found that while typically the SeqA protein localized to foci as reported previously, it was not the sole subcellular localization of the protein. A significant fraction of the SeqA–PAmCherry proteins localized outside of the foci and in some cells the protein was more enriched at the cell membrane or near the cell periphery. Additionally, we observed cells that did not display defined, large foci. In an attempt to answer the question whether SeqA had a fixed number of molecules per focus and a defined size we have counted the number of SeqA–PAmCherry molecules per focus and determined the sizes of SeqA clusters. The number of molecules displayed a broad distribution as did the sizes of the foci which led us to favor the possibility that SeqA–PAmCherry does not display a fixed number of molecules per focus. We discuss the visualization of SeqA subcellular localization with PALM and the protein number quantification attempt in light of the limitations of the technique.
Experimental procedures
Strains and growth
All microscopy experiments were performed on E. coli K-12 MG1655 and related strains. For experiments where SeqA tagged with a fluorescent protein was expressed from the genome, the native seqA gene was fused at its 3′ end to fluorescent protein genes, PAmCherry,27 eYFP21 or mEos3.2,28 separated with a linker (ASPPPGRSR) and including a kanamycin resistance marker. Recombineering29 was used to replace the native SeqA with the fusion genes (Table 1). The genomic knock-ins were validated with PCR and subsequently with DNA sequencing. In experiments where the SeqA–PAmCherry protein was overexpressed from a plasmid E. coli MG1655 or BW25113 Δdam were transformed with pBAD–SeqA–PAmCherry (this study).Cells were grown in LB medium (10 g l−1 Bacto Tryptone (Becton Dickinson), 5 g l−1 Yeast extract (Becton Dickinson) and 10 g l−1 NaCl (Merck)) with vigorous shaking in Erlenmeyer flasks at 37 °C or at 30 °C (expression from the genome in synchronized cells experiments) until mid-log phase (OD600 = 0.3–0.5) and harvested for microscopy, unless stated otherwise. For plasmid overexpression experiments cells were induced with 0.002% (w/vol) arabinose for 2–3 hours at 37 °C. Where necessary the cultures were supplemented with antibiotics: kanamycin (40 µg ml−1) for the genomic knock-ins and ampicillin (100 µg ml−1) for the strains harbouring pBAD–SeqA–PAmCherry.
Synchronization of cells
Where indicated, the cells expressing SeqA–PAmCherry from the genome were synchronized in their cell cycle using the stringent response.32,33 The synchronization was performed as described by Ferullo and co-workers.32 Briefly, E. coli cultures were grown to an OD600 = 0.2–0.3. Subsequently, 8 mM of the synchronizing agent serine hydroxamate (SHX, Sigma) was added to the cultures. After 90 min, when the cells have arrested their cell cycle and finished ongoing DNA replication rounds, SHX was removed from the cultures by centrifugation (5000g, 5 min). The cells were introduced into fresh LB medium and restarted growth, upon which a majority of the cells resumed their cell cycle in a synchronous fashion.Sample preparation for PALM microscopy
1 ml of culture was harvested after growth. To decrease the fluorescence background from the LB medium, cells were washed with NaPGCl buffer (NaPGCl = 95 mM sodium phosphate, pH 7.0, 50 mM glucose plus 125 mM sodium chloride), isoosmotic to the LB medium. The cells were then fixed with a solution of 4% paraformaldehyde in NaPGCl buffer, similarly to what was described by Greenfield and coworkers.16 To diminish cell motion the cells were spotted onto poly-L-lysine coated coverslips (poly-L-lysine 0.1% solution, Sigma-Aldrich) and imaging was performed.PALM acquisition
PALM microscopy was carried out on a home-built set up based on an epifluorescence inverted microscope (IX83, Olympus, Japan) with a 60× TIRF oil immersion objective lens (APON 60XOTIRF, NA 1.49, Olympus, Japan). Images were captured with an electron-multiplying CCD camera (ImagEM C9100-13, Hamamatsu Photonics, Hamamatsu, Japan) and recorded using Hokawo Imaging software V2.5 (Hamamatsu Photonics, Hamamatsu, Japan). For mEos3.2 and PAmCherry a 561 nm laser was used for excitation (constant illumination) and a 405 nm laser was used for photoconversion. The output power of the 405 nm laser was adjusted for each experiment based on the observed switching rate in order to obtain a low emitter density (i.e., less than one emitter per diffraction limited area in an individual cell in each frame) by use of neutral density filters. Fluorescence emission was collected with a 572 nm long pass and a 590/40 band pass filters. For eYFP a constant illumination of the 514 nm laser was used and the fluorescence emission was collected using a 530 nm long pass filter. The lasers used are from Coherent (Coherent, CA, USA) and the optical filters are from Chroma (Chroma technology, VT, USA). Samples were acquired with 100 ms exposure times. The duration of the PALM experiments was typically 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 to 15
000 to 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 frames. For the data sets with synchronized cells, where molecule counting was performed, the acquisition was continued until no more or hardly any photoactivation events could be detected upon visual inspection.
000 frames. For the data sets with synchronized cells, where molecule counting was performed, the acquisition was continued until no more or hardly any photoactivation events could be detected upon visual inspection.
PALM analysis
The PALM raw data was analyzed using the Localizer software34 in combination with Igor Pro (Wavemetrics, OR, USA). In the first step, each frame was analyzed to localize single emitters. Briefly, individual acquired images were segmented into emissive spots and noise, using the GLRT algorithm as described by Sergé and co-workers.35 Subsequently, the exact position of the emitter was estimated by fitting a 2D Gaussian distribution to the measured brightness profiles, using the Levenberg–Marquardt least squares algorithm. The coordinates of the maximum of the fitted 2D Gaussian distribution were then stored as the position of the emitter for that frame.Correction for multiple counting of identical emitters
To correct for multiple counting of the same fluorophores emitting over multiple frames or emitters reappearing after several frames of absence (e.g., due to blinking or emitting too few photons to be recognized by the fitting algorithm) we used an algorithm implemented in Localizer (termed “consolidation of identical emitters”). This algorithm groups localized emitters that were observed in close vicinity in time and space. Two parameters were required by this algorithm: the maximum allowed distance between two positions and the maximum allowed dark time (in frames) to be considered as coming from the same emitter. In order to determine the optimal parameters for this algorithm the approach of Annibale and co-workers36 was followed. The “consolidation of identical emitters” algorithm was repeated for different allowed jump distances and different allowed dark times. The total number of estimated emitters was plotted versus the maximum allowed dark times (Fig. S1 and Table S1†). The larger the maximum allowed dark time, the lower the number of estimated emitters. This decrease was composed of two regimes. The first one was the contribution of single emitters that remained dark/undetected for some frames in between two localization events, as a result of blinking or not being fitted by the algorithm. The second regime was the contribution of two different emitters that appeared close together in space and in time. The first regime should be steep and exponential, the second regime quasi linear. Both regimes were fitted using a procedure written in the Igor Pro analysis software based on the approach of Annibale and colleagues.36 Based on this fitting the optimal maximum allowed jump distance and the maximum allowed dark time values were obtained for each data set and consolidation of identical emitters was performed (Fig. S1 and Table S1†). The result of this analysis was also used to estimate the average localization precision of 21–25 nm for PAmCherry, depending on the dataset. The procedure did not take into account incomplete maturation efficiency nor the efficiency of photoactivation.27,37Calculating the sizes of SeqA foci
To obtain the sizes of individual SeqA–FP foci the L-clustering analysis was performed.18,38 In each analyzed cell large clusters that resemble the typical SeqA foci5,6,8,39 were selected upon visual inspection and a region of interest was selected around the area of the cluster/focus. Subsequently, L-clustering analysis was performed at this region and a distribution curve was obtained. The maximum of this curve was taken as the representative size of the cluster (SeqA focus). In the case where the L-clustering curve showed two clearly defined maxima, the maximum corresponding to the smaller cluster size was chosen. The rationale for that is that due to dynamic nature of the SeqA foci, a distribution with the two maxima is likely to represent a dividing SeqA focus. The small, often solitary proteins localized outside of the big typical SeqA foci were not analyzed.Results
PALM imaging reveals more information about SeqA structures in Escherichia coli
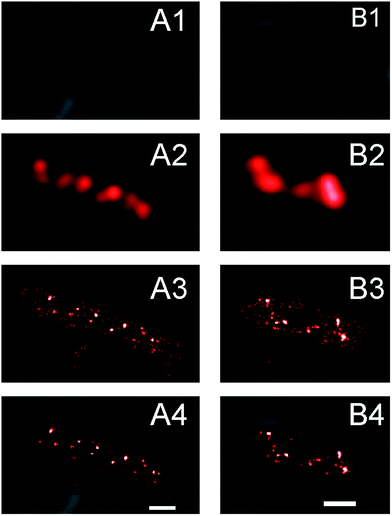
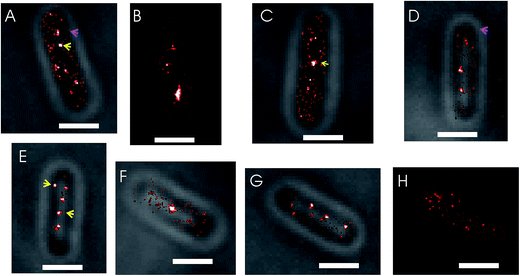
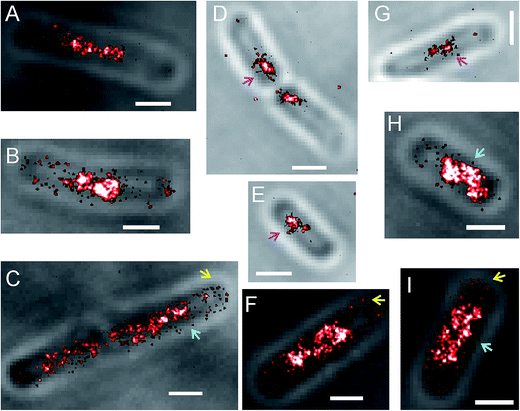
In conventional fluorescence microscopy the diffraction limit of light makes it difficult to resolve features of imaged objects that are smaller than 250 nm. It was shown extensively that SeqA forms discrete foci that appear as small, single spots in conventional fluorescence microscopy images.5–9 Driven by the increased localization precision of molecules enabled by PALM, we anticipated to find more detail of the SeqA foci properties using this technique. In Fig. 1 conventional fluorescence microscopy is compared with PALM imaging in two E. coli cells expressing SeqA-PAmCherry from the genome. The conventional fluorescence microscopy images (Fig. 1A2 and B2) were reconstructed from PALM data, where the localized molecules were rendered as diffraction limited spots (∼250 nm). In the PALM images (Fig. 1A3 and B3) the single localized SeqA–PAmCherry molecules are rendered as 25 nm spots according to their average localization precision. In Fig. 1 it can be observed that the PALM images contain more detail than the conventional fluorescence microscopy images. Moreover, while some of the SeqA–PAmCherry foci in conventional fluorescence microscopy images appear as single objects/spots, in PALM reconstructions it can actually be observed that they are composed of two foci or possess additional features.Upon visual inspection of the PALM images acquired in this study it was found that the structures formed by SeqA–PAmCherry in E. coli cells are heterogeneous (Fig. 2). As anticipated, the SeqA molecules did arrange into foci (Fig. 2, yellow arrows), as reported previously,5,6,8,39,40 however the foci were irregular in size and shape (compare Fig. 2A and C with Fig. 2B) and it was also possible to observe cells without clearly defined (large) SeqA foci (Fig. 2H, see also section Quantitative characterization of the amount of SeqA-PAmCherry molecules in Escherichia coli cells). The SeqA–PAmCherry molecules were not solely confined to (large) foci and in a vast majority of cells it was possible to find molecules outside of the (large) foci often as solitary proteins (see Fig. 2A and D, magenta arrows for examples). In a small subset of the data, we counted the total amount of proteins per cell and the amount of proteins that were localized in (large) foci. The foci were identified upon visual inspection arbitrarily as structures/clusters clearly resembling the foci reported in previous studies.5,6,8 The amount of proteins located within the (large) foci varied largely between cells (Fig. S2†) with on average only 30–34% of molecules located in the large foci. The purpose of this measurement is not to give absolute numbers of molecules that are within or outside the SeqA structures (as this may vary upon what one defines as a SeqA focus as well as on the maturation and detection efficiency of the fluorescent tags), but to provide example that SeqA molecules observed outside the large foci are abundant. Qualitatively, similar characteristics of SeqA foci were also observed for E. coli cells expressing SeqA–eYFP genomically (Fig. S3†). While it is possible that a portion of the SeqA–fluorescent protein fusion (SeqA–FP) molecules located outside of the foci are artifacts resulting from PALM reconstructions (e.g., sample impurities that were wrongly recognized during PALM reconstructions as SeqA–FP foci), we believe this localization is genuine. First, the small spots (putatively single molecule entities) outside of the foci were observed in most cells. Second, the fraction of molecules outside of the foci was high (Fig. S2†). And third, the single molecule-like localizations (small spots) were much more enriched inside the imaged cells compared to the area outside of the cells, which implies that these spots are not solely due to sample impurity (see below and Fig. S4†). It remains possible that a portion of the small spots are free PAmCherry molecules that were cleaved off from the SeqA–PAmCherry fusion protein.
As a test for false-positives, we have performed PALM reconstructions of wild type E. coli cells (MG1655) not expressing any FPs but imaged in the same way as the PAmCherry datasets. We found that the fraction of false-positive localizations in PALM reconstructions is negligible, as in the 120 imaged cells, 86 cells did not show any localizations and the cells that did, showed typically 1–5 localizations within the cell boundaries (Fig. S4†) giving on average 1.5 localizations per cell.
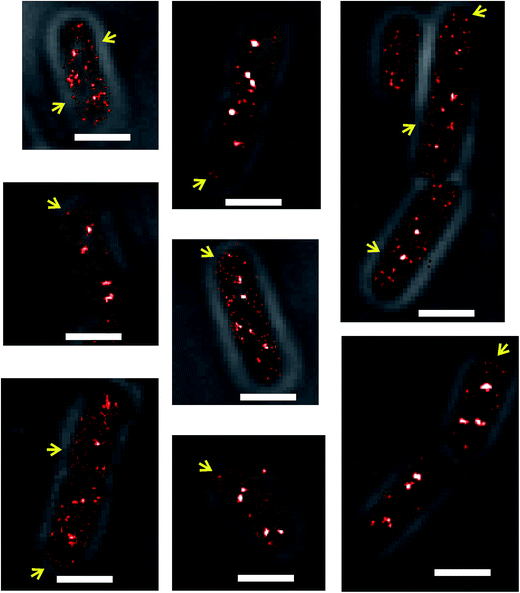
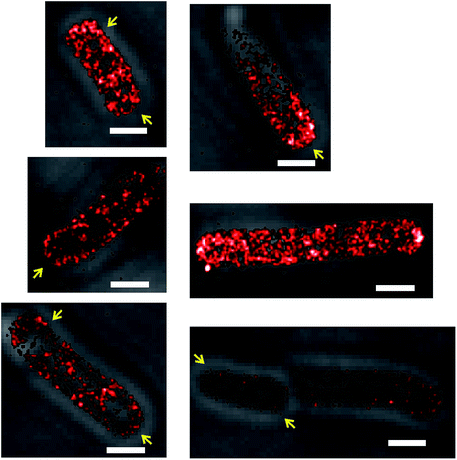
Some SeqA–PAmCherry molecules seem to localize at the membrane
Another detail of SeqA localization inside E. coli cells is that in some cells a fraction of the SeqA–PAmCherry molecules appeared to be localized or locally enriched at the membrane (Fig. 3, yellow arrows). This phenomenon was rare and upon visual inspection of the datasets for synchronized cells it was estimated to occur in about 12–18% of the cells (43 out of 336 cells, from 18 independent datasets, grown at 37 °C and 77 out of 419 cells, from 20 independent datasets, grown at 30 °C). It was also observed to occur in E. coli wild type strain and in the Δdam strain, both overexpressing SeqA–PAmCherry from the plasmid (pBAD–SeqA–PAmCherry, Fig. 9 and 10, respectively). The phenomenon was not observed in E. coli cells genomically expressing SeqA–YFP (Fig. S3†). A possible explanation for why the SeqA–eYFP molecules were not observed at the bacterial membrane might be the photophysical characteristics of eYFP.21 It is expected that only a small fraction of the eYFP molecules are actually detected during PALM acquisition. Therefore the membrane localization might be missed due to not enough eYFP molecules being detected. To the best of our knowledge the membrane localization of SeqA has not been reported previously by fluorescence microscopy but there are reports indicating SeqA or SeqA-interacting proteins are present in membrane fractions in cellular fractionation experiments.1,41–43Numbers of SeqA–PAmCherry molecules in Escherichia coli cells
PALM was used previously to count the number of molecules in living bacterial cells.23,44 Quantification of the number of molecules detected with PALM needs to be corrected for multiple counting of identical emitters that might result from the presence of the same emitter in subsequent frames, blinking and failure to recognize the emitter in subsequent frames by the PALM fitting algorithm. We corrected for this using the procedure outlined in the Correction for multiple counting of identical emitters section in the Experimental procedures. To facilitate the molecule counting and correction for multiple counting of identical emitters, it was crucial that the fluorophores did not move. For this aim cells were fixed with paraformaldehyde. Fixation with paraformaldehyde was performed for all the microscopy experiments in this study.E. coli cells grown in liquid media are not synchronized in their cell cycle. We performed synchronization of cells using the stringent response.19,32,33 To capture the evolution of SeqA foci during the cell cycle, cells were grown at 37 °C and 30 °C, synchronized with serine hydroxamate (SHX) and subsequently, after the synchronizing agent was removed and cells resumed growth, cells were harvested and prepared for microscopy (including fixation) at different time points (see the Synchronization of cells section in Experimental procedures for details). Fig. 4A and C show the results of SeqA–PAmCherry molecule counting in individual cells at different time-points after the synchronizing agent was removed and cells resumed growth. The amount of SeqA–PAmCherry molecules per cell showed a big spread between 17 and 302 molecules at 37 °C and between 15 and 195 molecules at 30 °C. Due to limited statistics (at 37 °C n = 10–24 cells at each time point and a total of 102 cells; at 30 °C n = 11–38 cells and a total of 182 cells) it is hard to draw conclusions to whether there are trends in changes of the amount of molecules as a function of the cell cycle. The data for all time points were combined (Fig. 4B and D) and there was an average of 97 ± 59.7 (standard deviation) and a median value of 74.5 SeqA–PAmCherry molecules per cell for cells grown at 37 °C and an average of 65 ± 33.5 and a median value of 58 molecules at 30 °C. To compensate for the fact that longer cells might have more molecules per cell the data for each cell was divided by the cell's length to obtain the number of molecules per 1 µm of cell. Similarly, the data showed a large spread (Fig. 4E and F) with cells grown at 37 °C having between 7 and 73 SeqA–PAmCherry molecules per 1 µm of cell and an average of 25 ± 13.3 molecules (median 21 molecules); cells at 30 °C (Fig. 4G and H) having between 5 and 62 molecules per 1 µm of the cell with an average of 21 ± 10.7 molecules (median 18 molecules). Cells grown at 30 °C also had on average fewer SeqA–PAmCherry molecules per cell than cells grown at 37 °C after the normalization for cell length. This observation might be rationalized by true differences in cell physiology resulting from growth at different temperatures but it cannot be excluded that at the lower temperature (30 °C) the maturation efficiency of PAmCherry in live cells is lower than for cells grown at 37 °C, which would affect the number of detected molecules.
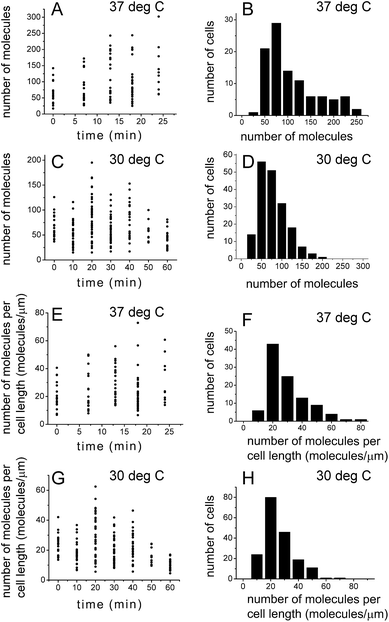 | ||
| Fig. 4 Quantification of the number of SeqA–PAmCherry molecules in Escherichia coli. E. coli cells were grown in LB at 37 °C and at 30 °C and synchronized in the cell cycle (see main text for details). At various time points after the growth was resumed, the number of localized SeqA–PAmCherry molecules was calculated in each cell and corrected for multiple counting of the same emitters using a method described by Annibale36 (see main text for details). (A and C) Number of molecules in each cell at a given time point after resumption of cell growth at 37 °C and at 30 °C, respectively. (B and D) Histograms of combined data from all time points in A and C. (E and G) The number of molecules per cell data (from A and C) was divided for each cell by the corresponding cell length, to correct for the possibility of larger cells having more molecules. (F and H) Histograms of combined data from all time points in E and G. (A, C, E and G) Each point represents a single cell. At 37 °C n = 10–24 cells at each time point and a total of 102 cells; at 30 °C n = 11–38 cells and a total of 182 cells. | ||
Numbers of SeqA–PAmCherry foci in Escherichia coli cells
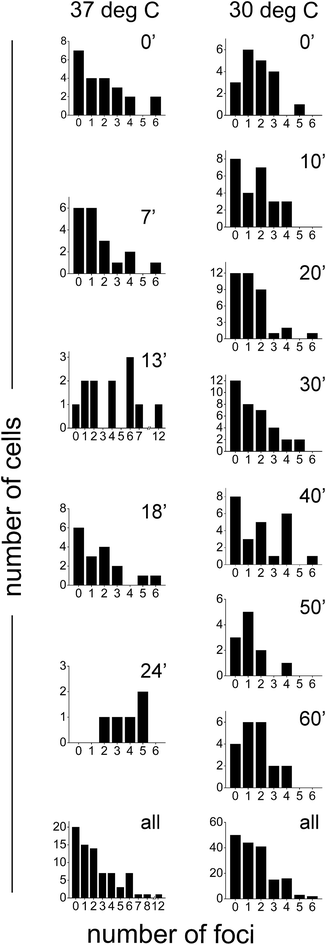
Since SeqA is known to organize into defined foci, upon visual inspection of the PALM images we determined the number of foci per cell in synchronized cells grown at 37 °C and 30 °C (see above for which structures were considered as large foci). Most cells showed between 1 and 4 foci per cell (Fig. 5). There was a significant fraction of cells that did not show defined (large) SeqA foci: 26% (20 out of 76 cells) and 29% (50 out of 171 cells) at 37 °C and 30 °C, respectively (Fig. 5, bottom panels). There were cells that showed more than 5 foci per cell with some cells having up to 12 foci. The larger foci numbers were much more prevalent for cells grown at 37 °C (13 out of 76 cells) than for cells grown at 30 °C (5 out of 171 cells).Numbers of molecules per SeqA focus
We also sought out to count the number of SeqA–PAmCherry molecules per focus, to test whether the SeqA foci have a defined/fixed molecule number in each focus. The numbers of molecules per focus were characterized by a broad distribution for the different temperatures and at all time points tested (Fig. 6), with cells grown at 37 °C displaying foci with up to 55 SeqA–PAmCherry molecules per focus. At 30 °C the focus with highest amount of SeqA–PAmCherry counted 36 molecules (Fig. 6). Under the limited statistical sampling of this study (at 37 °C n = 20–52 foci at a given time point and a total of 172 foci; at 30 °C n = 12–54 foci at a given time point and a total of 248 foci) the numbers of molecules per focus at different time points had mostly similar distributions with the exception of the dataset at 24 min at 37 °C (Fig. 6). For the combined data from all the time points on average the numbers of SeqA–PAmCherry molecules per focus were comparable for cells grown at 37 °C and 30 °C, with most cells having between 10 and 15 molecules per focus, with cells grown at 30 °C having a median of 9 SeqA–PAmCherry molecules per focus and an average of 10.5 ± 6 molecules and at 37 °C having a median of 12 SeqA–PAmCherry molecules per focus and an average of 14.5 ± 9.6 molecules (Fig. 6 and 7A). A question now was whether the SeqA foci had a defined number of molecules per focus. We have tried to model the number of SeqA–PAmCherry molecules per focus in cells grown at 30 °C (to this end we combined data for all time points from cells grown at 30 °C obtaining a data set with the highest number of foci) with a binomial distribution assuming a fixed number of SeqA–PAmCherry per focus and assuming different PAmCherry detection efficiencies (i.e. 10%, 45% and 60%). In Fig. 7B it can be observed that within a data set of this size (248 foci) and with the distribution of molecules observed in the data from the experiment (gray bars) none of the binomial distributions fit well to the experimental data, as the latter ones have a broader distribution, implying that SeqA–PAmCherry does not have a fixed number of molecules per focus.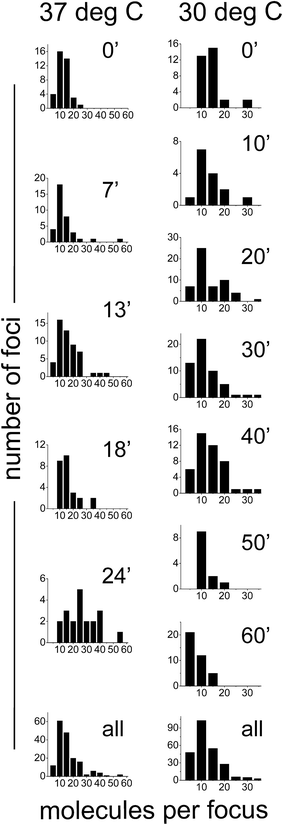 | ||
| Fig. 6 Quantification of the numbers of molecules of SeqA–PAmCherry per focus in Escherichia coli. E. coli cells were grown in LB at 37 °C and at 30 °C and synchronized in the cell cycle (see main text for details). At various time points after the growth was resumed, the number of localized SeqA–PAmCherry molecules for each focus was calculated in each cell and corrected for multiple counting of the same emitters using a method described by Annibale36 (see main text for details). Individual panels show the data for each given time point and temperature (left panels 37 °C and right panels at 30 °C). The bottom panels combine the data from all time points at each temperature. At 37 °C n = 20–52 foci at a given time point and a total of 172 foci; at 30 °C n = 12–54 foci at a given time point and a total of 248 foci. | ||
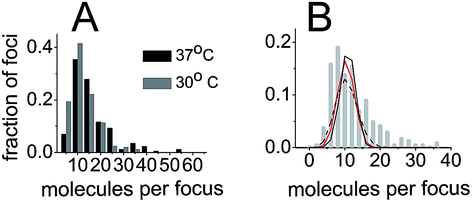 | ||
| Fig. 7 SeqA foci do not have a fixed number of molecules per focus. (A) Histogram of combined numbers of SeqA–PAmCherry molecules per focus from all time points (as in Fig. 6) for all cells at 37 °C and at 30 °C (black bars and gray bars, respectively), displayed with 5 molecule bins. (B) Histogram of combined numbers of SeqA–PAmCherry molecules per focus from all time points at 30 °C plotted with a higher frequency of bins (gray bars, 2 molecules bins) than in A. The lines show a fit of binomial distribution of molecule detection events at different molecule detection efficiencies (60% black line, 45% red line and 10% black dotted line), assuming that the SeqA–PAmCherry foci have a fixed number of molecules per focus. The actual data (gray bars) have a broader distribution than the fitted lines implying that SeqA–PAmCherry does not have a fixed number of molecules per focus. | ||
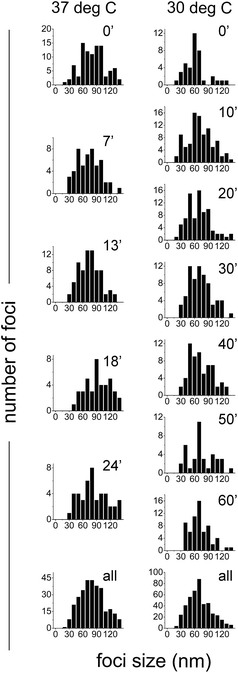
Quantification of the size of SeqA foci
To measure the sizes of the SeqA-FP foci, the PALM images were analyzed using the L-clustering algorithm (see Experimental procedures). We inspected the sizes of the foci in synchronized E. coli cells expressing SeqA–PAmCherry genomically at different time points after the cells had resumed growth. In Fig. 8 it can be observed that the SeqA foci are clearly smaller than the diffraction limit and that the sizes display a distribution between 30 and 130 nm with typical foci sizes in the 60–80 nm range. On average the clusters measured 76.9 ± 33 nm (median = 72.8 nm) for the combined data from cells grown at 37 °C (87 cells and 315 individual clusters, Fig. 8 and Table S2†) and 66.6 ± 30.2 nm (median = 62 nm) at 30 °C (174 cells, 456 individual clusters, Fig. 8 and Table S3†). In general we did not observe large changes in foci size at different timepoints after resumption of cell growth, with the exception of the timepoints at 18 min and 24 min at 37 °C, where the average foci size was 95.2 ± 44 and 90.1 ± 38.7 nm, respectively (Fig. 8 and Table S2†). These small changes are however close to the localization precision of PALM in our experimental set-up (21–25 nm). The foci sizes obtained for SeqA tagged with different FPs (PAmCherry, eYFP and mEos3.2) were very similar (Fig. S5A and Table S4†). The foci sizes obtained with data that were corrected for multiple counting of identical emitters were similar to the non-corrected ones (Fig. S5B and Table S5†), as were the foci sizes in synchronized cells compared to non-synchronized cells (Fig. S5C and Table S5†); the small differences were well within the localization precision.PALM imaging of SeqA–PAmCherry overexpressed from a plasmid
To compare SeqA localization in E. coli under conditions that resemble more closely the physiological expression levels of the protein (the SeqA–PAmCherry knock-in strain) with cells where SeqA–PAmCherry was overexpressed from a plasmid, we performed PALM imaging of E. coli harboring pBAD–SeqA–PAmCherry. The degree of overexpression varied between experiments and upon visual inspection it appeared that on average the cells overexpressing SeqA–PAmCherry had significantly more molecules, but the molecule numbers were not counted. As anticipated, we observed that SeqA–PAmCherry formed discrete foci (Fig. 9, magenta arrows) and in some cells the SeqA–PAmCherry molecules were also found at the membrane (Fig. 9, yellow arrows). The main difference in SeqA subcellular localization between the genomically expressed SeqA–PAmCherry (the knock-in strain) and the protein overexpressed from a plasmid (the strain harboring pBAD–SeqA–PAmCherry), was that in the latter case some cells showed regions of the cell with a high density of SeqA–PAmCherry molecules that appeared significantly larger than the SeqA foci. The non-uniform distribution of the molecules throughout the cell together with the fact that the proteins were localized mostly in the central region of the cell suggest that the SeqA–PAmCherry were residing at the nucleoid of the cells (Fig. 9, cyan arrows).PALM imaging of SeqA–PAmCherry in Δdam cells
The SeqA protein preferentially binds hemimethylated DNA.1,2 The E. coli DNA methyltransferase dam methylates newly synthesized DNA influencing the SeqA binding to DNA.45 We sought to investigate the subcellular localization of SeqA in cells lacking the dam methylase with PALM. The E. coli Δdam strain harboring pBAD–SeqA–PAmCherry was imaged. In the majority of the cells, the overexpressed SeqA–PAmCherry was dispersed throughout the cell instead of localizing into foci (Fig. 10). Upon visual inspection of PALM images only 5 out of 32 cells imaged showed structures that could be interpreted as foci. It was possible to observe that in some cells the SeqA protein seemed to localize to the membrane/cell periphery (Fig. 10, yellow arrows). In a previous study of SeqA localization in E. coli Δdam cells, where the wild type SeqA was imaged with immunofluorescence employing diffraction-limited conventional fluorescence microscopy, it was also found that SeqA no longer formed discrete foci and it was concluded that the protein is dispersed throughout the nucleoid6 or throughout the cell.5 Upon PALM imaging we observed, that while the SeqA–PAmCherry is not homogeneously distributed throughout the cell, it is not solely confined to the nucleoid (central region of the cell) as there are cells with a clearly non-nucleoid like localization (Fig. 10).Validation of the functionality of the SeqA–PAmCherry genomic knock-ins
We compared the growth rates of the wild-type strain (MG1655) expressing the native SeqA with those of the MG1655 SeqA–PAmCherry strain and we found that the genomic knock-in had no adverse effects on growth characteristics in LB under different temperatures tested (Fig. S6†). Western blots of the strains used in this study to perform microscopy show that qualitatively the expression level of SeqA in wild type cells is similar to the SeqA–FP levels in the genomic knock-ins (Fig. S8†). This is in good agreement with a previous report where it was demonstrated that a genomic knock-in of SeqA–eYFP has similar expression levels to those of SeqA in wild-type E. coli cells.39The biological functionality of the SeqA–FP fusions has been characterized extensively before.5,39 It has been shown that both the SeqA–GFP expressed from plasmid in the presence of native SeqA as well as SeqA–YFP expressed from the genome and replacing the native SeqA retain the ability to sequester the origins of replication, indicating that the fusion of SeqA to FPs does not compromise its biological function. Based on the similarity of the 3D structure between PAmCherry and eYFP (both are β-barrels of approximately similar molecular dimensions), the good qualitative agreement between the data obtained using the SeqA–eYFP and SeqA–PAmCherry genomic knock-ins (compare Fig. S3† with main text Fig. 1–3 and 8) and the fact that in the absence of dam methylase the SeqA–PAmCherry no longer organized into foci (Fig. 10) we conclude that there are strong indications to assume that the SeqA–PAmCherry can functionally replace the SeqA protein.
Discussion
SeqA is not solely localized into foci in Escherichia coli
The aim of this study was to obtain a more detailed view of SeqA subcellular localization, by employing a super-resolution microscopy technique – PALM. As shown previously SeqA localized into discrete foci. In some cases the improved localization precision of PALM allowed us to observe that the SeqA–PAmCherry structures that appeared as single discrete foci in diffraction limited microscopy were actually two foci in close vicinity (Fig. 1). A similar observation was reported by Helgesen and co-workers, who found that some foci that appear as single entities with conventional fluorescence microscopy are in fact composed of two clusters of SeqA molecules when reconstructed with super-resolution microscopy, using dSTORM imaging and SeqA labeled with fluorescent antibodies.46The number of these foci per cell varies in most cells between 1 and 4 foci per cell with some cells showing up to 12 foci (Fig. 5). The high foci number per cell is consistent with other reports, where E. coli growing rapidly in rich media showed up to 12 foci per cell, with typically between 4 and 8 foci.9 Yet, it was also possible to observe cells that did not display defined foci and under the experimental conditions of this study they made up to 26–29% of all the cells inspected. Cells that do not display SeqA foci have been observed previously with microscopy of SeqA–GFP fusions.8,40 For cells grown in minimal media (slow growth conditions) the cells without SeqA foci composed between 20 and 45% of the imaged cells. However, these studies do not report cells devoid of SeqA foci when E. coli is grown in rich media (LB or similar). In our study the cells were grown in a rich medium (LB, fast growth conditions) and we still observed a fraction of cells without foci in the case of the SeqA–PAmCherry expressed from the genome in cells synchronized using SHX. Intriguingly, in the non-synchronized cells expressing SeqA–eYFP from the genome we did not observe high fractions of cells without foci (Fig. S3†). This might imply that the fraction of cells without foci observed in the synchronized cells could result from changes in cell physiology and nucleoid arrangement upon SHX treatment or that they reflect cells where there is no ongoing replication. We do not know whether the stringent response influences the SeqA propensity to arrange into foci. But it has been demonstrated that the stringent response is dependent on the presence of SeqA in cells, that during this response there is limited replication at oriC and that nucleoids of E. coli cells are decondensed.33 We monitored cell growth after removal of the synchronizing agent (SHX) and we did observe resumption of growth, which indicates the end of the stringent response and release from the cell arrest (Fig. S7†). However, these are bulk measurements (optical density of the cultures) and it is possible that in a fraction of cells the cell cycle arrest is not relieved. We observed that foci were not the sole subcellular localization of SeqA–FP in E. coli as visualized by PALM. A fraction of SeqA–FP in cells expressing the fusion from the genome was localized as solitary or non-clustered proteins outside of the well defined large foci (Fig. 2 and S3†). As mentioned in the results section it is possible that the number of these molecules that are localized with PALM reconstructions outside of the foci might be due to the imperfections of the experimental set up used in this study (e.g., sample impurities wrongly recognized by the PALM algorithm or free FPs that were cleaved off from the SeqA-FP fusion). If one assumes however that these localizations are real, these molecules might reflect SeqA molecules traveling to or between the existing foci or the beginnings of new SeqA foci. Since these are mostly single molecule localizations (solitary proteins or non-clustered protein oligomers) it is likely they were not visible in studies employing conventional fluorescence microscopy. In cells overexpressing SeqA–PAmCherry from the plasmid, where the protein expression level was much higher than in the genomically expressing cells, SeqA formed structures that were much bigger than the typical SeqA foci (Fig. 9), implying that it is possible to disturb the SeqA localization into foci.
Membrane localization of SeqA
In a fraction of cells (12–18% in cells expressing SeqA–PAmCherry from the genome, depending on growth temperature) we observed that some SeqA–PAmCherry molecules seemed to be localized at the membrane or the cell periphery (Fig. 3). SeqA is a soluble protein47,48 and it has no domains that are expected to interact directly with the membrane. To the best of our knowledge membrane localization of SeqA with fluorescence microscopy has not been reported previously. There is, however, a great deal of reports of SeqA localization at the membrane coming from cell fractionation experiments. SeqA activity, measured as the binding of hemimethylated oriC DNA, was identified in membrane fractions41,49 and the presence of SeqA in membrane fractions was further confirmed with an anti-SeqA antibody.42,43 Shakibai and co-workers42 showed that another protein in membrane fractions was auxiliary for oriC binding; this protein was named SeqB. D'Alençon and co-workers demonstrated that membrane fractions isolated from synchronized cells had highest SeqA activity during initiation of replication,43 higher than membrane fractions from exponentially growing (non-synchronized) wild type cells. Moreover, Slater and co-workers report that SeqA is present but not enriched in membrane fractions as compared to the total cellular protein level of SeqA.1 This could rationalize why we observe SeqA–PAmCherry at the membrane only in a fraction of cells and only for a small total amount of all molecule localizations with PALM in a given cell. In exponentially growing (non-synchronized) SeqA–eYFP expressing from the genome we did not observe SeqA to clearly localize at the membrane. This phenomenon might be partially explained by the specific photophysics of eYFP,21 where only a small fraction of the total eYFP proteins are imaged (as mentioned in the results section) and some of the localizations might be missed by PALM. It is possible that the membrane localization of SeqA was not detected by conventional fluorescence microscopy previously as the much higher fluorescence intensity of the SeqA–FP foci might have overshadowed the membrane localized SeqA–FPs. Yet, in a single molecule localization microscopy technique such as PALM, it was possible to detect this phenomenon. This membrane localization was more common and more pronounced in cells overexpressing SeqA–PAmCherry from plasmid (Fig. 9), and even more so in the Δdam strain (Fig. 10). We speculate that as DNA binding sites get saturated or completely disappear, more SeqA molecules would be available to interact with SeqB or other putative interaction sites at the membrane, which makes the membrane localization easier to observe.Quantification of the numbers of SeqA–PAmCherry molecules in cells
We have used PALM to count the numbers of SeqA–PAmCherry molecules in E. coli where the protein was expressed from the genome in synchronized cells. Within the statistically limited amount of cells probed in this study at each given time point (10–38 cells) we did not observe clear trends to changes in the numbers of SeqA copy numbers per cell as a function of cell cycle (Fig. 4). In general there was a big spread of the number of proteins per cell (15 to 300 molecules per cell) with on average 97 SeqA–PAmCherry molecules per cell for cells grown at 37 °C and an average of 65 molecules for cells grown at 30 °C. The smaller amount of molecules counted at 30 °C might reflect true differences in cell physiology or could result from a lower maturation efficiency of PAmCherry at this temperature. By quantifying the expression level of SeqA with western blotting using the anti-SeqA antibody Slater and co-workers report that wild type E. coli grown in minimal media has on average a 1000 SeqA molecules per cell (between 847 and 1435 SeqA molecules per cell depending on the strain and western blotting technique), which is about ten times more than was determined for the SeqA–PAmCherry fusion used in this study.1 PAmCherry has been used previously for counting molecule numbers in microorganisms23,50 and it was concluded that the numbers obtained with this probe are likely underestimated due to incomplete fluorophore maturation, fixation and premature photobleaching. Additionally Durisic and co-workers measured that the (in vivo) photoactivation efficiency of PAmCherry is only as little as 45% in Xenopus oocytes,37 which will also contribute to an underestimation of the absolute molecule numbers. The photoactivation efficiency of PAmCherry is similar to those of other fluorescent proteins routinely used for PALM, which range between 39% (PA-GFP) and 61% (mEos2).37 Yet, a clear advantage of PA–mCherry over other fluorophores used for molecule counting with PALM is that it blinks less23,37 making the estimates of the amounts of unique emitters more reliable (Fig. S1†). Indeed, in this study we have corrected the numbers of localized molecules by employing the “consolidate identical emitters” procedure (see Quantitative characterization of the amount of SeqA–PAmCherry molecules in Escherichia coli cells in the Results section). We aimed to visualize all or nearly all PAmCherry molecules present in the samples as the data acquisition was continued until no more new photoactivation events could be detected upon visual inspection. Another possible source of error in the estimation of SeqA molecule numbers in cells, is the change of expression levels of the protein resulting from fusing it to the FPs. We have performed Western blots of wild type cells expressing SeqA and FP tagged expressing trains (eYFP, mEos3.2 and PAmCherry) and qualitatively we do not observe large changes in the expression level upon FP tagging as compared to the physiological level of SeqA (Fig. S8†). This is consistent with a previous report where it was shown that the genomically expressed SeqA–eYFP has a similar expression level to SeqA in wild-type cells.39 We therefore conclude that the likely underestimated SeqA molecule numbers are not due to changes in expression level as a result of tagging the protein with FPs.Broad distributions of SeqA molecule numbers per focus and foci sizes
Since SeqA forms discrete foci we sought to quantify the amount of SeqA–PAmCherry molecules per focus (Fig. 6). The number of molecules per focus was not observed to be dependent on the cell cycle, in the conditions of our cell cycle synchronization experiment. We observed a broad distribution of numbers of molecules per focus with on average 10–15 molecules per focus (for the largest dataset at 30 °C the average was 10.5 molecules per focus). Note that due to the properties of PAmCherry these numbers might be underestimated, as explained above. The modeling of the experimental distribution with a binomial distribution assuming a fixed amount of SeqA–PAmCherry molecules per focus and different fixed detection efficiencies as the only error source, did not fit well to the experimental data, which would be an indication that the foci do not have a fixed amount of SeqA–PAmCherry molecules per focus. However, peak widening may occur if there is more than one source of error.In previous studies employing conventional fluorescence microscopy SeqA foci appeared as single spots and no further detail of these structures was resolved. With the improved localization precision of PALM microscopy we were able to show that these structures are indeed smaller than the diffraction limit (Fig. 1 and 8). By sizing the SeqA foci using the L-clustering analysis, we have shown that these structures have a distribution of sizes between 30 and 130 nm with most SeqA foci ranging between 60 and 80 nm. In a recent study employing dSTORM, Helgesen and co-workers46 report the width of SeqA immunolabelled with Alexa647 to be 34 nm. The difference in reported foci sizes can partly be explained by the difference in localization precision or the sampling method employed (width of spots in the Helgesen study vs. L-clustering analysis in this study). The broad distribution of sizes of SeqA foci between different cells is similar to the distribution of numbers of molecules per focus, yet again implying that the foci do not have a fixed size.
It has been proposed that the SeqA complexes are dynamic in a threadmill-like fashion, i.e. growing in the leading end as SeqA molecules bind to newly replicated GATC sites and shrinking in the trailing end as dam methylates the older GATC sites.2 While the exact mechanism of SeqA dynamics (assembly/disassembly and migration) remains to be fully elucidated, our claim that the SeqA foci do not have fixed numbers of molecules per focus, supports the threadmilling model or a gradual growth. The growing of SeqA structures (e.g., resulting from the threadmilling) would rationalize the broad distributions of numbers of SeqA molecules per focus and the distribution of SeqA cluster sizes we observe.
Conclusions
In this study we have used super-resolution microscopy to study the subcellular localization of the E. coli DNA binding protein SeqA. This protein forms discrete foci in cells that have features sized below the diffraction limit. PALM enabled us to observe that some foci that can appear as single structures in conventional fluorescence microscopy are in fact composed of two foci. We have observed that not all of the SeqA-PAmCherry molecules are confined to foci, with some solitary molecules outside of the foci structures and in a fraction of cells some protein molecules localized to the membrane/cell periphery. While, to the best of our knowledge, the membrane localization of SeqA has not been demonstrated before with fluorescence microscopy, there were indications of this subcellular localization from cell fractionation studies. We thus believe the improved sensitivity of the technique used allowed us to observe details that were previously unavailable to diffraction-limited microscopy.We also performed quantitative analysis of the numbers of molecules in cells that were synchronized in their cell cycle using the stringent response process. We found that the numbers of SeqA–PAmCherry molecules showed a broad distribution between cells with on average 50–100 molecules per cell, but no correlation between this number and the stage of cell cycle was found. In order to answer the question on whether the structures formed by SeqA have a fixed amount of molecules per focus, we have quantified the number of SeqA-PAmCherry molecules per focus and similarly found a broad distribution with a single focus containing on average 10–15 molecules. Furthermore, the SeqA foci sizes were also characterized by a distribution with most foci in the 60–80 nm range. We are inclined to believe that the SeqA foci have a dynamic nature and do not have a fixed sized or amount of molecules per focus.
The possible sources of error in this study might be of two origins. First, the cell physiology (and thus the subcellular localization of SeqA) could have been altered as a result of treatment with the cell cycle synchronization agent serine hydroxamate. Second, the PALM technique, especially its quantitative aspect, has still some limitations and room for improvement. The molecule number counting data reported here are most likely underestimated as a result of the rather low detection efficiency (resulting from photo-activation efficiency) of PAmCherry.
Nevertheless, despite the possible uncertainties, PALM contributed to understanding new biology as shown in this study and by others (for examples see the review of Gahlmann and Moerner22). Quantitative biology is an emerging field that aims to assign actual numbers to the key cellular parameters, such as protein copy numbers, stoichiometries of macromolecular complexes, reaction rates and diffusion coefficients in vivo.51,52 To understand how proteins function in cells and to understand their regulatory mechanisms one needs to analyze molecules in their natural, cellular environment and not under idealized test tube conditions. Super-resolution imaging is undergoing constant development and new, brighter, monomeric, better-folding fluorescent proteins with better photophysical characteristics are made,15 which will provide better tools to study biological processes quantitatively in living cells.
Acknowledgements
We would like to thank Prof. Kirsten Skarstad and Anne Wahl for the kind gift of the SeqA antibody and suggestions on western blotting. J. T. M. would like to thank Geert van den Bogaart and Wim Vandenberg for helpful discussions. J. T. M. would like to acknowledge funding from NWO-Rubicon (project number 825.12.026) and the Marie Curie Actions—Intra-European Fellowship (IEF, project number 329018). J. M., T. S. and B. B would like to acknowledge funding from KU Leuven Research Council (PF/10/010; IDO/13/008) and IAP-BELSPO, FWO (G.0B25.15, G.0471.12).References
- S. Slater, et al., E. coli SeqA protein binds oriC in two different methyl-modulated reactions appropriate to its roles in DNA replication initiation and origin sequestration, Cell, 1995, 82, 927–936 CrossRef CAS.
- T. Waldminghaus and K. Skarstad, The Escherichia coli SeqA protein, Plasmid, 2009, 61, 141–150 CrossRef CAS PubMed.
- D. W. Russell and N. D. Zinder, Hemimethylation prevents DNA replication in E. coli., Cell, 1987, 50, 1071–1079 CrossRef CAS.
- M. Lu, J. L. Campbell, E. Boye and N. Kleckner, SeqA: a negative modulator of replication initiation in E. coli., Cell, 1994, 77, 413–426 CrossRef CAS.
- T. Onogi, H. Niki, M. Yamazoe and S. Hiraga, The assembly and migration of SeqA–Gfp fusion in living cells of Escherichia coli., Mol. Microbiol., 1999, 31, 1775–1782 CrossRef CAS.
- S. Hiraga, C. Ichinose, H. Niki and M. Yamazoe, Cell cycle-dependent duplication and bidirectional migration of SeqA-associated DNA-protein complexes in E. coli., Mol. Cell, 1998, 1, 381–387 CrossRef CAS.
- S. Hiraga, C. Ichinose, T. Onogi, H. Niki and M. Yamazoe, Bidirectional migration of SeqA-bound hemimethylated DNA clusters and pairing of oriC copies in Escherichia coli., Genes Cells, 2000, 5, 327–341 CrossRef CAS.
- T. Brendler, J. Sawitzke, K. Sergueev and S. Austin, A case for sliding SeqA tracts at anchored replication forks during Escherichia coli chromosome replication and segregation, EMBO J., 2000, 19, 6249–6258 CrossRef CAS PubMed.
- Morigen, I. Odsbu and K. Skarstad, Growth rate dependent numbers of SeqA structures organize the multiple replication forks in rapidly growing Escherichia coli., Genes Cells, 2009, 14, 643–657 CrossRef CAS PubMed.
- B. Huang, M. Bates and X. Zhuang, Super-resolution fluorescence microscopy, Annu. Rev. Biochem., 2009, 78, 993–1016 CrossRef CAS PubMed.
- E. Betzig, et al., Imaging intracellular fluorescent proteins at nanometer resolution, Science, 2006, 313, 1642–1645 CrossRef CAS PubMed.
- S. T. Hess, T. P. K. Girirajan and M. D. Mason, Ultra-high resolution imaging by fluorescence photoactivation localization microscopy, Biophys. J., 2006, 91, 4258–4272 CrossRef CAS PubMed.
- M. J. Rust, M. Bates and X. Zhuang, Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM), Nat. Methods, 2006, 3, 793–795 CrossRef CAS PubMed.
- T. A. Klar, S. Jakobs, M. Dyba, A. Egner and S. W. Hell, Fluorescence microscopy with diffraction resolution barrier broken by stimulated emission, Proc. Natl. Acad. Sci. U. S. A., 2000, 97, 8206–8210 CrossRef CAS.
- W. Vandenberg, M. Leutenegger, T. Lasser, J. Hofkens and P. Dedecker, Diffraction-unlimited imaging: from pretty pictures to hard numbers, Cell Tissue Res., 2015, 360, 151–178 CrossRef PubMed.
- D. Greenfield, et al., Self-organization of the Escherichia coli chemotaxis network imaged with super-resolution light microscopy, PLoS Biol., 2009, 7, e1000137 Search PubMed.
- W. Wang, G.-W. Li, C. Chen, X. S. Xie and X. Zhuang, Chromosome organization by a nucleoid-associated protein in live bacteria, Science, 2011, 333, 1445–1449 CrossRef CAS PubMed.
- S. F. Lee, M. A. Thompson, M. A. Schwartz, L. Shapiro and W. E. Moerner, Super-resolution imaging of the nucleoid-associated protein HU in Caulobacter crescentus, Biophys. J., 2011, 100, L31–L33 CrossRef CAS PubMed.
- C. Spahn, U. Endesfelder and M. Heilemann, Super-resolution imaging of Escherichia coli nucleoids reveals highly structured and asymmetric segregation during fast growth, J. Struct. Biol., 2014, 185, 243–249 CrossRef CAS PubMed.
- G. Fu, et al., In vivo structure of the E. coli FtsZ-ring revealed by photoactivated localization microscopy (PALM), PLoS One, 2010, 5, e12682 Search PubMed.
- J. S. Biteen, et al., Super-resolution imaging in live Caulobacter crescentus cells using photoswitchable EYFP, Nat. Methods, 2008, 5, 947–949 CrossRef CAS PubMed.
- A. Gahlmann and W. E. Moerner, Exploring bacterial cell biology with single-molecule tracking and super-resolution imaging, Nat. Rev. Microbiol., 2014, 12, 9–22 CrossRef CAS PubMed.
- U. Endesfelder, et al., Multiscale spatial organization of RNA polymerase in Escherichia coli, Biophys. J., 2013, 105, 172–181 CrossRef CAS PubMed.
- A. Badrinarayanan, R. Reyes-Lamothe, S. Uphoff, M. C. Leake and D. J. Sherratt, In vivo architecture and action of bacterial structural maintenance of chromosome proteins, Science, 2012, 338, 528–531 CrossRef CAS PubMed.
- R. Reyes-Lamothe, D. J. Sherratt and M. C. Leake, Stoichiometry and architecture of active DNA replication machinery in Escherichia coli, Science, 2010, 328, 498–501 CrossRef CAS PubMed.
- S. Bakshi, A. Siryaporn, M. Goulian and J. C. Weisshaar, Superresolution imaging of ribosomes and RNA polymerase in live Escherichia coli cells, Mol. Microbiol., 2012, 85, 21–38 CrossRef CAS PubMed.
- F. V. Subach, et al., Photoactivatable mCherry for high-resolution two-color fluorescence microscopy, Nat. Methods, 2009, 6, 153–159 CrossRef CAS PubMed.
- M. Zhang, et al., Rational design of true monomeric and bright photoactivatable fluorescent proteins, Nat. Methods, 2012, 9, 727–729 CrossRef CAS PubMed.
- K. A. Datsenko and B. L. Wanner, One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products, Proc. Natl. Acad. Sci. U. S. A., 2000, 97, 6640–6645 CrossRef CAS PubMed.
- F. R. Blattner, et al., The complete genome sequence of Escherichia coli K-12, Science, 1997, 277, 1453–1462 CrossRef CAS.
- T. Baba, et al., Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection, Mol. Syst. Biol., 2006, 2, 2006.0008 CrossRef PubMed.
- D. J. Ferullo, D. L. Cooper, H. R. Moore and S. T. Lovett, Cell cycle synchronization of Escherichia coli using the stringent response, with fluorescence labeling assays for DNA content and replication, Methods, 2009, 48, 8–13 CrossRef CAS PubMed.
- D. J. Ferullo and S. T. Lovett, The stringent response and cell cycle arrest in Escherichia coli, PLoS Genet., 2008, 4, e1000300 Search PubMed.
- P. Dedecker, S. Duwé, R. K. Neely and J. Zhang, Localizer: fast, accurate, open-source, and modular software package for superresolution microscopy, J. Biomed. Opt., 2012, 17, 126008 CrossRef PubMed.
- A. Sergé, N. Bertaux, H. Rigneault and D. Marguet, Dynamic multiple-target tracing to probe spatiotemporal cartography of cell membranes, Nat. Methods, 2008, 5, 687–694 CrossRef PubMed.
- P. Annibale, S. Vanni, M. Scarselli, U. Rothlisberger and A. Radenovic, Quantitative photo activated localization microscopy: unraveling the effects of photoblinking, PLoS One, 2011, 6, e22678 CAS.
- N. Durisic, L. Laparra-Cuervo, A. Sandoval-Álvarez, J. S. Borbely and M. Lakadamyali, Single-molecule evaluation of fluorescent protein photoactivation efficiency using an in vivo nanotemplate, Nat. Methods, 2014, 11, 156–162 CrossRef CAS PubMed.
- M. A. Kiskowski, J. F. Hancock and A. K. Kenworthy, On the use of Ripley's K-function and its derivatives to analyze domain size, Biophys. J., 2009, 97, 1095–1103 CrossRef CAS PubMed.
- S. Fossum-Raunehaug, E. Helgesen, C. Stokke and K. Skarstad, Escherichia coli SeqA structures relocalize abruptly upon termination of origin sequestration during multifork DNA replication, PLoS One, 2014, 9, e110575 Search PubMed.
- S. Adachi, T. Fukushima and S. Hiraga, Dynamic events of sister chromosomes in the cell cycle of Escherichia coli, Genes Cells, 2008, 13, 181–197 CrossRef CAS PubMed.
- G. B. Ogden, M. J. Pratt and M. Schaechter, The replicative origin of the E. coli chromosome binds to cell membranes only when hemimethylated, Cell, 1988, 54, 127–135 CrossRef CAS.
- N. Shakibai, et al., High-affinity binding of hemimethylated oriC by Escherichia coli membranes is mediated by a multiprotein system that includes SeqA and a newly identified factor, SeqB, Proc. Natl. Acad. Sci. U. S. A., 1998, 95, 11117–11121 CrossRef CAS.
- E. D'Alençon, A. Taghbalout, R. Kern and M. Kohiyama, Replication cycle dependent association of SeqA to the outer membrane fraction of E. coli., Biochimie, 1999, 81, 841–846 CrossRef.
- S.-H. Lee, J. Y. Shin, A. Lee and C. Bustamante, Counting single photoactivatable fluorescent molecules by photoactivated localization microscopy (PALM), Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 17436–17441 CrossRef CAS PubMed.
- T. Waldminghaus, C. Weigel and K. Skarstad, Replication fork movement and methylation govern SeqA binding to the Escherichia coli chromosome, Nucleic Acids Res., 2012, 40, 5465–5476 CrossRef CAS PubMed.
- E. Helgesen, S. Fossum-Raunehaug, F. Sætre, K. O. Schink and K. Skarstad, Dynamic Escherichia coli SeqA complexes organize the newly replicated DNA at a considerable distance from the replisome, Nucleic Acids Res., 2015, 43, 2730–2743 CrossRef CAS PubMed.
- A. Guarné, Q. Zhao, R. Ghirlando and W. Yang, Insights into negative modulation of E. coli replication initiation from the structure of SeqA-hemimethylated DNA complex, Nat. Struct. Biol., 2002, 9, 839–843 Search PubMed.
- A. Guarné, et al., Crystal structure of a SeqA–N filament: implications for DNA replication and chromosome organization, EMBO J., 2005, 24, 1502–1511 CrossRef PubMed.
- A. Chakraborti, S. Gunji, N. Shakibai, J. Cubeddu and L. Rothfield, Characterization of the Escherichia coli membrane domain responsible for binding oriC DNA, J. Bacteriol., 1992, 174, 7202–7206 CAS.
- D. Lando, et al., Quantitative single-molecule microscopy reveals that CENP-A(Cnp1) deposition occurs during G2 in fission yeast, Open Biol., 2012, 2, 120078 CrossRef PubMed.
- L. M. Gierasch and A. Gershenson, Post-reductionist protein science, or putting Humpty Dumpty back together again, Nat. Chem. Biol., 2009, 5, 774–777 CrossRef CAS PubMed.
- U. Moran, R. Phillips and R. Milo, SnapShot: key numbers in biology, Cell, 2010, 141, 1262–1262e1 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5fd00058k |
| ‡ The authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2015 |