Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Gene expression as an indicator of the molecular response and toxicity in the bacterium Shewanella oneidensis and the water flea Daphnia magna exposed to functionalized gold nanoparticles†
T. A.
Qiu‡
a,
J. S.
Bozich‡
b,
S. E.
Lohse
c,
A. M.
Vartanian
c,
L. M.
Jacob
c,
B. M.
Meyer
a,
I. L.
Gunsolus
a,
N. J.
Niemuth
b,
C. J.
Murphy
c,
C. L.
Haynes
a and
R. D.
Klaper
*b
aDepartment of Chemistry, University of Minnesota, 207 Pleasant St SE, Minneapolis, MN 55455, USA
bSchool of Freshwater Sciences, University of Wisconsin Milwaukee, 600 E. Greenfield Ave, Milwaukee, WI 53204, USA. E-mail: rklaper@uwm.edu; Fax: +414 382 1705; Tel: +414 382 1713
cDepartment of Chemistry, University of Illinois at Urbana-Champaign, 600 S. Mathews Ave, Urbana, IL 61801, USA
First published on 4th August 2015
Abstract
Nanoparticle (NP) physiochemical properties have been shown to be important determinants of NP interactions with biological systems. Due to both nanomaterial diversity and environmental complexity, a mechanistic understanding of how physiochemical properties affect NP/organism interactions will greatly aid in the accurate assessment and prediction of current and emerging NP-induced environmental impacts. Herein, we investigated key biological apical endpoints, such as viability, growth, and reproduction and the expression of genes associated with related molecular pathways in response to exposure to gold nanoparticles (AuNPs) functionalized with either positively charged ligands, polyallyamine hydrochloride, or negatively charged ligands, mercaptopropionic acid, in two model organisms, the bacterium Shewanella oneidensis MR-1 and the water flea Daphnia magna. By linking changes in molecular pathways to apical endpoints, potential biomarkers for functionalized AuNP impacts were identified in both organisms. Specifically, act was identified as a potential biomarker in D. magna and 16S as a potential biomarker in S. oneidensis. We also revealed that changes in molecular pathways induced by ligand–NP combination were strongly dependent upon the type of ligand on the NP surface, and the effects from their respective ligands alone might predict these effects for the ligand–NP combination, but only in some cases. Lastly, we revealed that it is possible to identify similar pathways provoked upon NP exposure across organisms. This study shows that molecular pathways will help elucidate mechanisms for NP toxicity that are predictive of adverse environmental outcomes.
Nano impactBased on the great diversity of organisms in the environment and possible engineered nanomaterials, fundamental understanding of nanoparticle-biological interactions will be critical for accurate assessment and prediction of nanoparticle environmental impacts. Herein, we investigated key biological apical endpoints and molecular pathways in response to functionalized Au nanoparticles in two environmentally relevant model organisms, the bacterium Shewanella oneidensis and the water flea Daphnia magna. We identified some specific molecular responses that may serve as potential biomarkers for nanoparticle impacts, revealed that changes in apical endpoints and molecular pathways in both organisms strongly depend on ligand-nanoparticle combination, and uncovered shared pathways provoked upon nanoparticle exposure. This study facilitates a better understanding of nanoparticle-organism interaction mechanisms, building toward prediction of meaningful environmental outcomes. |
Introduction
Engineered nanoparticles (NPs) are being produced to enhance a wide range of societally beneficial applications, from energy storage capacities and material durability to medical therapeutics and water treatment devices.1–3 These applications are possible because of the novel physiochemical properties NPs display, such as high surface area and reactivity as well as distinct surface chemistries, compositions and size distributions. It is, however, these same size-dependent physiochemical properties that may influence their biocompatibility.4–8 For example, size, shape and core composition have been thought to mediate receptor–ligand binding rates, cellular phagocytosis, exocytosis and cytotoxicity.9–12 Other studies suggest that NP surface charge is the main determinant of biological interactions, with positively charged particles being more toxic than negatively or neutrally charged particles.13–18These classifications of critical features that determine biological impact all focus on the NPs themselves. The differences in response across organisms or cell types are less often considered despite the fact that toxicological evaluations of the biological impacts caused by engineered NPs have revealed a wide range in responses across cell types or organisms considered.19–23 For example, Sohaebuddin et al. (2010)24 demonstrated that cell type determines the extent of response to nanomaterials with different compositions and sizes. In another study using ZnO NPs, the EC50 differed by orders of magnitude for V. fischeri, D. magna and T. platyurus.25 Variation across cell systems and organisms makes it difficult to develop a common understanding of the properties of nanomaterials that may determine toxicity. Even for well-studied chemicals, such as pesticides, models that use general acute endpoint data to predict impacts often inaccurately estimate concentrations that cause effects across similar chemicals and rarely are applicable across organisms.26,27 These studies have shown that a more mechanistic understanding of the impacts of chemicals at sublethal doses provides a more accurate description of impacts and better data for modeling these effects across species. The goal of this project is to achieve a more mechanistic understanding of NP/organism interactions to facilitate efficient prediction of the impact nanotechnology will have on environmental health. Linking specific molecular mechanisms that are impacted by NPs across organisms to apical endpoints will not only greatly aid in assessing the potential environmental impact of these materials but is also crucial to informing NP design for safe and sustainable development of nanotechnologies.
Currently, the major proposed molecular mechanism for NP toxicity is oxidative stress.4,28–31 However, the exposures that produce oxidative stress in many studies are well above what is estimated to be the current or future environmental concentrations; long-term low dose exposures are the more likely scenario.32,33 In addition, the molecular mechanisms responsible for coping with oxidative stress are triggered upon exposure to a wide range of chemical species34,35 and are a natural biological response that does not necessarily lead to an adverse outcome.36 The focus on oxidative stress and lethal dose exposures makes it difficult to uncover other mechanisms that may have a greater predictive power for the environmental impact of NPs. Sublethal concentration-based exposures allow the cell to have a more natural perturbation by the contaminant that triggers subtle, but potentially specific, molecular responses.37,38 It is these more realistic exposure scenarios that will uncover more mechanism-based information to predict meaningful impacts across species.
Molecular biomarkers provide a sensitive indicator of the response of an organism to stressors such as exposure to a toxicant in addition to providing information on the mechanisms that are impacted by exposure.37,38 Mechanistic information that can be tied to larger impacts on reproduction for example enhances the possibility of predicting negative outcomes where standardized toxicological tests, although valuable, have limited ability to accurately predict the impact of emerging contaminants. Overreliance on these methods has led to risk assessment failures.39 Developing such candidate biomarkers for NP toxicity will greatly aid in the rapid assessment and impact prediction for current and emerging nanomaterials across a wide range of organisms. Previously developed biomarkers, for example, vitellogenin, have been used for the successful determination of adverse outcomes of some classes of endocrine disruptors and their impacts on vertebrate reproduction.40–42 Metallothioneins are used to detect metal ion exposure, and they are expressed in response to a wide range of metal-based contaminants associated with environmental pollution.43 Heat shock proteins, indicative of proteotoxic stressors, indicate subtlethal cellular damage and respond in a dose-dependent manner to environmental stressors.44 Biomarkers that provide mechanistic insight into nanoparticle-organism interactions, especially if they apply to effects seen across species, would provide a way to group nanomaterials by their molecular level effects. Furthermore, they may indicate both nonspecific and specific modes of action as well as underlying mechanisms for toxicity of NPs with particular physiochemical properties.
In this study, we examined several candidate biomarkers in two model species, the bacterium Shewanella oneidensis and the invertebrate Daphnia magna, that are associated with pathways of importance in these two species and determined how their expression related to the biological impacts of exposure to gold NPs (AuNPs) with positively or negatively charged surfaces. Shewanella oneidensis (MR-1) is an environmentally beneficial Gram-negative bacterium with a unique metal-reducing capability to respire heavy metals; S. oneidensis plays an important role in the cycling of metal elements in the ecosystem as well as the bioremediation of toxic elements.45Daphnia magna is a designated toxicology and toxicogenomics model organism by multiple agencies (OECD, NIH and EPA) and is an environmentally relevant freshwater invertebrate that composes an integral part of freshwater food webs.46 AuNPs were chosen as a model NP in this study due to the chemical inertness of the gold core and our ability to readily control size,47 shape48 and surface functionalization.49 Two ligands were used for AuNP functionalization, positively charged polyallylamine hydrochloride (PAH) and negatively charged mercaptopropionic acid (MPA).
We explored genes in various molecular pathways in our two model organisms. Pairs of genes selected from each organism were to represent pathways encoding for similar cellular functions in the two organisms, including oxidative stress, xenobiotic detoxification, protein folding, cellular electron transport, and cellular maintenance. In addition, genes in pathways related to reproduction in D. magna and to cell division, DNA repair and extracytoplasmic stress in S. oneidensis were also investigated. The goal was to determine: 1) how the exposure to NPs with differing surface properties impacted each organism and how this differed from their respective ligand controls; 2) if gene expression for these pathways were an indication of impacts seen in each organism; 3) if exposure duration altered effects and gene expression measurements and if acute measurements of gene expression would provide an indication of chronic impacts; and 4) if gene expression for similar pathways across organisms would provide biomarkers that were predictive across species. The NPs used in this study were quantitatively and qualitatively characterized prior to and after exposure to assay media to aid us in understanding how alterations in NP physical properties may impact molecular pathways. Overall, this work aims to link molecular pathways and apical endpoints to NP characteristics in two distinct environmentally relevant organisms.
Methods
Functionalized AuNP synthesis and characterization
All materials were used as received, unless otherwise noted. Gold tetrachloroaurate trihydrate (HAuCl4·3H2O), sodium borohydride (NaBH4), trisodium citrate, 3-mercaptopropionic acid (MPA) and polyallylamine hydrochloride (PAH; Mw 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g mol−1) were obtained from Sigma Aldrich. Ultrapure deionized water was prepared using a Barnstead NANOPURE water filtration system. PALL Minimate tangential flow filtration capsules for AuNP purification with 50 kDa pore size was obtained from VWR. Transmission electron microscopy grids were obtained from PELCO (SiO on copper mesh).
000 g mol−1) were obtained from Sigma Aldrich. Ultrapure deionized water was prepared using a Barnstead NANOPURE water filtration system. PALL Minimate tangential flow filtration capsules for AuNP purification with 50 kDa pore size was obtained from VWR. Transmission electron microscopy grids were obtained from PELCO (SiO on copper mesh).
The 4.7 (±1.5) nm-diameter PAH–AuNPs were prepared by polyelectrolyte wrapping of ~4 nm-diameter citrate-coated AuNPs. The (4.3 ± 1.3) nm-diameter MPA–AuNPs were prepared by direct synthesis. After synthesis, measuring and counting using TEM images determined size distributions. Detailed descriptions of the AuNP syntheses are given below.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 Mw PAH (10.0 mg mL−1) dissolved in 1.0 mM NaCl was then added. The wrapping solution was briefly mixed at vortex briefly and left to stand for 16 h. The PAH–AuNPs were subsequently purified by centrifugation and washing (55 min at 18
000 Mw PAH (10.0 mg mL−1) dissolved in 1.0 mM NaCl was then added. The wrapping solution was briefly mixed at vortex briefly and left to stand for 16 h. The PAH–AuNPs were subsequently purified by centrifugation and washing (55 min at 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 894 rcf), in ultrapure deionized water. The purified PAH–AuNPs were then concentrated in a diafiltration membrane.50
894 rcf), in ultrapure deionized water. The purified PAH–AuNPs were then concentrated in a diafiltration membrane.50
AuNP characterization and analysis
Synthesized functionalized AuNPs were characterized in Milli-Q water, bacteria growth medium, and Daphnia medium using various analytical techniques, including TEM for absolute sizes (JEOL 2100 Cryo TEM), dynamic light scattering for hydrodynamic diameter (Brookhaven ZetaPALS), zeta-potential for surface charge (Brookhaven ZetaPALS), and UV-vis localized surface plasmon resonance (LSPR) spectroscopy for particle concentration and aggregation (Mikropack DH-2000 UV-vis-NIR spectrometer).Free ligand suspensions
Free ligands, MPA and PAH (Mw 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g mol−1), were obtained from Sigma Aldrich. MPA and PAH ligands are readily soluble in water and do not require a co-solvent for dispersion. The ligands were dissolved into Milli-Q water at a maximum concentration of 50 mg L−1 and diluted accordingly for free ligand toxicity control experiments.
000 g mol−1), were obtained from Sigma Aldrich. MPA and PAH ligands are readily soluble in water and do not require a co-solvent for dispersion. The ligands were dissolved into Milli-Q water at a maximum concentration of 50 mg L−1 and diluted accordingly for free ligand toxicity control experiments.
Shewanella oneidensis MR-1 cultivation and cell oxygen uptake assay
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 into the growth medium supplied with NP/ligands in reaction vessels that were kept in a 32 °C water bath. Exposures of PAH–NPs were conducted at 30, 100, and 5000 μg L−1, and exposures of PAH ligand were 30, 100, 300, 600, 1000, 2000 and 5000 μg L−1. In all subsequent experiments and comparisons, a ten-fold mass concentration of PAH free ligand and an equivalent mass concentration of MPA free ligand were used as ligand controls, which was calculated to be an overestimate of possible total free ligand present in the suspension (see ESI†).53 A tube filled with 1 mL 30% (w/w) KOH solution was inserted into each reaction vessel to absorb carbon dioxide generated from cell respiration. The consumption of oxygen was compensated by continuous oxygen injection to keep the pressure constant in the headspace of the reaction vessels. Oxygen uptake was recorded every 10 minutes automatically for 24–48 hours by the instrument. The first derivative of oxygen uptake was plotted to identify the maximal oxygen uptake rate and the time of that maximum. To represent each oxygen uptake trace with a single value, the ratio of maximal oxygen uptake rate to the time when it reached maximal rate was calculated following the equation below:
10 into the growth medium supplied with NP/ligands in reaction vessels that were kept in a 32 °C water bath. Exposures of PAH–NPs were conducted at 30, 100, and 5000 μg L−1, and exposures of PAH ligand were 30, 100, 300, 600, 1000, 2000 and 5000 μg L−1. In all subsequent experiments and comparisons, a ten-fold mass concentration of PAH free ligand and an equivalent mass concentration of MPA free ligand were used as ligand controls, which was calculated to be an overestimate of possible total free ligand present in the suspension (see ESI†).53 A tube filled with 1 mL 30% (w/w) KOH solution was inserted into each reaction vessel to absorb carbon dioxide generated from cell respiration. The consumption of oxygen was compensated by continuous oxygen injection to keep the pressure constant in the headspace of the reaction vessels. Oxygen uptake was recorded every 10 minutes automatically for 24–48 hours by the instrument. The first derivative of oxygen uptake was plotted to identify the maximal oxygen uptake rate and the time of that maximum. To represent each oxygen uptake trace with a single value, the ratio of maximal oxygen uptake rate to the time when it reached maximal rate was calculated following the equation below:The ratio was then normalized to the average of control groups and represented as a percentage, where 100% indicates no inhibition of cell oxygen uptake.
Daphnia magna cultivation and biological assays
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8 light/dark cycle as designated by EPA protocols.54Daphnia breeding populations were held at a concentration of 14 adult Daphnia per 1 L of media in glass beakers and were discarded once adults reached 28 days old. Daphnia were fed 50 mL freshwater algae (Pseudokirchneriella subcapitata) at an algal density of 400
8 light/dark cycle as designated by EPA protocols.54Daphnia breeding populations were held at a concentration of 14 adult Daphnia per 1 L of media in glass beakers and were discarded once adults reached 28 days old. Daphnia were fed 50 mL freshwater algae (Pseudokirchneriella subcapitata) at an algal density of 400![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 algal cells per mL and 15 mL of dissolved alfalfa (Medicago sativa). Alfalfa stock was prepared by suspending 405 mg of Alfalfa in 50 mL Milli-Q water after 15 minutes of stirring and 5 minutes of centrifugation at 3829 RCF.
000 algal cells per mL and 15 mL of dissolved alfalfa (Medicago sativa). Alfalfa stock was prepared by suspending 405 mg of Alfalfa in 50 mL Milli-Q water after 15 minutes of stirring and 5 minutes of centrifugation at 3829 RCF.
Reproductive exposures adhered to the mortality and reproduction guidelines designated by the OECD (OECD guidelines 1998). Daphnids were kept at a concentration of 5 daphnids per 100 mL, and results were normalized to controls (i.e. daphnia exposed to only MHRW) to account for changes in reproduction and body size as these replicate exposures took place over a period of several months.
Gene expression exposures and RNA preservation
Gene expression exposures were performed in parallel with bacterial oxygen uptake and D. magna survival and reproduction assays.RNA extraction, reverse transcription and real-time quantitative PCR
A Direct-zol™ RNA MiniPrep kit (Zymo Research) was used for total RNA isolation and purification by spin columns. The manufacturer's recommended protocol was followed using a centrifugation speed of 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g with an on-column DNase I treatment at 30 °C for 15 minutes. RNA was finally eluted from the column at 16
000 × g with an on-column DNase I treatment at 30 °C for 15 minutes. RNA was finally eluted from the column at 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 1 minute. Total RNA was characterized using a Thermo Scientific NanoDrop 8000 and an Agilent 2100 Bioanalyzer for quality control.
000 × g for 1 minute. Total RNA was characterized using a Thermo Scientific NanoDrop 8000 and an Agilent 2100 Bioanalyzer for quality control.
Total RNA was reverse transcribed into cDNA following the manufacturer's protocols. Briefly, 100 ng of total RNA were incubated in the presence of either random primers (Promega) for S. oneidensis or oligo(dT)15 primer (Promega) for D. magna at 65 °C for 5 minutes. After cooling on ice for 1 minute, the SuperScript III reverse transcriptase, DTT, and RNaseOUT™ recombinant ribonuclease inhibitor (Life Technologies) were added into the mixture followed by incubation at 25 °C for 5 minutes (this step was only for random primers), 50 °C for 60 minutes, and 70 °C for 15 minutes for primer extension. Once synthesized, cDNA were stored at −20 °C.
Target genes were chosen for both S. oneidensis and D. magna. Four pairs of genes in similar pathways related to stress response in the two organisms were selected, including gst (S. oneidensis)/gst (D. magna, same order for the following pairs) in xenobiotic detoxification, nqrF/nadh for electron transport, katB/cat for oxidative stress attenuation, and ibpA/hsp70 for heat shock response. To link to apical endpoints, the vtg gene for D. magna reproduction and ftsK for bacterial cell division were also examined. Genes for actin (act) in D. magna and for 16S ribosomal RNA (16S) and RNA polymerase (rpoA) in S. oneidensis were monitored to consider NP/ligand impacts on basic organism machinery. In addition, stress response genes including pspB for extracytoplasmic stress, sodB for oxidative stress, and radA for DNA repair were also examined in S. oneidensis. Table 1 shows a full list of genes along with their corresponding functions.
| Shewanella oneidensis MR-1 | ||||
|---|---|---|---|---|
| Target gene | Forward primer (5′–3′) | Reverse primer (5′–3′) | Function | Accession number |
| Glutathione S-transferase (gst) | GCA AAG CAT TCC AGC AAT TT | GAC CTT CTT GCG TTT TGA GC | Xenobiotic detoxification | NP_720213.1 |
| Na-translocating NADH-quinone reductase subunit F (nqrF) | CGC TTA CTC GAT GGC TAA CTA C | GCA AGG CAG CGT CAA ATT AC | Mitochondrial electron transport NADH to ubiquinone | NP_716734.1 |
| Double-stranded DNA translocase (ftsK) | TAC GAG TCG TGT TGC GAT AAA | AAG GGC TGA CAC TGG AAT AAA | Cell division | NP_717901.1 |
| Catalase HPII (katB) | GGC ATT GAT CCT GAT TCT TCT C | TCC AAC GAG GGA AGT TAC CA | Catalase activity; response to oxidative stress | NP_716697.1 |
| 16 kDa heat shock protein A (ibpA) | GCA ACT CAG GTT ATC CTC CAT AC | CGC TAC TGA TCT CAA GCT CTT C | Response to heat; chaperone activity | NP_717873.1 |
| 16S ribosomal RNA (16S) | TCA AGT CAT CAT GGC CCT TAC | TAC GAC GAG CTT TGT GAG ATT AG | Component of prokaryotic ribosomes | NR_074798.1 |
| RNA polymerase alpha subunit (rpoA) | TCG CAT CCT ATT GTC GTC TAT G | CTT CTT GTA CGC CTT CCT TAC T | DNA-directed RNA polymerase activity | NP_715896.1 |
| ATP-dependent protease (radA) | TTC GGC AAT TTT CCT CTC C | ACA CCA CCA TGA CCA AGG AT | DNA repair | NP_716849.1 |
| Phage shock protein B (pspB) | TTG ATT GCG AAA GCC GAT A | ATC AAG AAT CGC CTC TAA GGT TT | Extracytoplasmic stress | NP_717416.1 |
| Fe/Mn superoxide dismutase (sodB) | GCA ATG TTC GCC CTG ACT AC | CCT GCG AAG TTT TGG TTC AC | Removal of superoxide radicals | NP_718453.1 |
| Daphnia magna | ||||
|---|---|---|---|---|
| Target gene | Forward primer (5′–3′) | Reverse primer (5′–3′) | Function | |
| Glutathione S-transferase (gst) | CAA CGC GTA TGG CAA AGA TG | CTA GAC CGA AAC GGT GGT AAA | Xenobiotic detoxification | AF448500.1 |
| Dehydrogenase (nadh) | GCA GGA AAC AAT AAG GCA AAC C | GGT GGC ACA GAC CAT TTC TTA | Mitochondrial electron transport and energy production | DQ340845.1 |
| Vitellogenin (vtg) | CTG TTC CTC GCT CTG TCT TG | CCA GAG AAG GAA GCG TTG TAG | Reproduction, sexual maturation and general stress | AB252737.1 |
| Catalase (cat) | CAG GAT CAT CGG CAG TTA GTT | CTG AAG GCA AAC CTG TCT ACT | Oxidative stress attenuation | GQ389639.1 |
| Heat shock protein 70 (hsp70) | CCT TAG TCA TGG CTC GTT CTC | TCA AGC GGA ACA CCA CTA TC | Response to heat; protein folding | EU514494.1 |
| β-Actin (act) | CCA CAC TGT CCC CAT TTA TGA A | CGC GAC CAG CCA AAT CC | Cytoskeleton production and cell maintenance | AJ292554.1 |
Primers for real-time quantitative PCR were designed by the PrimerQuest Tool (Integrated DNA Technologies). Two sets of primers were designed for each gene, and the one with efficiency closest to 1 was chosen to be the primer for subsequent real-time PCR. Table 1 includes a full list of primers used in this study.
Real-time quantitative PCR (qPCR) was performed on a StepOnePlus™ Real-Time PCR System (Life Technologies) using SYBR Green as the fluorescent intercalating dye (iTaq™ Universal SYBR® Green Supermix, Bio-Rad). For each qPCR reaction, cDNA and primers were mixed with the fluorescence dye following the manufacturer's protocol. Starting with an initial 10 min denaturation at 95 °C, real-time PCR repeated 40 cycles of amplification, each of which was 15 s at 95 °C followed by 30 s at 60 °C. Fluorescence of SYBR Green was detected at the end of each cycle. All qPCR experiments were done in technical duplicates.
NORMA-Gene analysis of qPCR data
Real-time quantitative PCR data were processed by the Miner55 program and NORMA-Gene algorithm.56 Miner applies an objective analysis scheme to obtain the dynamic fluorescence threshold (R), threshold cycle number (Ct), and efficiency (E) for each qPCR reaction, instead of using the same threshold for all reactions. Data for the normalized reporter signal (Rn) versus cycle number were extracted from amplification data exported from the StepOnePlus™ software as the input to the Miner program to obtain R, Ct, and E values for each reaction. R0, the initial fluorescent reporter signal, was calculated based on the equation below:| R0 = R × (1 + E)−Ct |
Statistical analysis
The normalized ratios from oxygen uptake traces were further subjected to statistical analysis. No normality and outliers were considered within this data set due to the limited sample size (N < 5). The two-tailed student's t-test was performed on treated samples versus their respective control group with α = 0.05. GraphPad Prism (GraphPad Software, Inc.) was used for statistical analysis.Data from Daphnia acute studies failed to meet the assumptions of normality. Therefore, the effects of NP and free ligand exposures on Daphnia survival, were compared to controls using the nonparametric Mann–Whitney U test for two-independent samples (N > 3). Impacts on daphnid reproduction and body size were assessed using one-way ANOVA with Tukey's multiple comparison tests after normality and variance homogeneity were determined (N > 3). One round of statistically determined outliers was removed, and treatments were deemed significantly different than controls at probability value <0.05. SPSS (IBM 2013) was used to interpret data.
The relative fold change values of S. oneidensis gene expression were log2-transformed followed by the combination of control groups. Outliers were identified and excluded from the data set (ROUT algorithm, Q = 1.0%, Prism GraphPad), and post hoc Tukey's tests after ANOVA were performed to determine statistical significance among different treatments at one time point and one gene of interest. For the 16S and sodB genes upon 100 μg L−1 PAH–AuNP exposure, as there was only one treatment, an unpaired t-test was used instead of ANOVA. Again, normality was not tested due to the limited sample size (N < 6). GraphPad Prism was used to perform statistical analysis.
The relative gene expression data from Daphnia short-term and long-term gene exposures were normalized to controls and log2 transformed to fit a normal distribution. Outliers were removed prior to statistical analysis. Significant differences in relative expression were determined using one-way ANOVA with Tukey's multiple comparison tests after normality and variance homogeneity were determined (p < 0.05) (N > 3). SPSS (IBM 2013) was used to interpret data.
Results
Nanoparticle characterization
TEM analysis of absolute size showed that the two AuNPs had very similar core size, while the hydrodynamic diameter of PAH–AuNPs in water was larger than MPA–AuNPs, possibly due to the polyelectrolyte wrapping (Table 2). It was notable that MPA–AuNPs showed an increased hydrodynamic diameter and a peak shift in UV-vis extinction upon resuspension in growth medium, indicating the aggregation of MPA–AuNPs, though the MPA–AuNPs still retained a negative surface charge in the growth medium (Table 2). The aggregation may result from elevated ionic strength in the growth medium or the pH change from slightly acidic Milli-Q water (pH ~ 6.3) to neutral growth medium (pH ~ 7.2). This aggregation might lead to altered NP toxicity, as previous studies have revealed.19,57 Similar behavior was not observed for PAH–AuNPs, indicating more stability of PAH–AuNPs than MPA-AuNPs in growth medium (Table 2).| PAH–AuNPs | MPA–AuNPs | |||
|---|---|---|---|---|
| S. oneidensis media | D. magna media | S. oneidensis media | D. magna media | |
| *Based on TEM analysis. See Fig. S1 for TEM images.a Localized surface plasmon resonance (LSPR) wavelength of maximum peak value (λmax). Errors are represented by standard deviations. | ||||
| LSPR λmax (nm) (in H2O)a | 528 | 515 | ||
| LSPR λmax (nm) (in medium) | 530 | 530 | 555 | 575 |
| d core (nm)* | 4.7 ± 1.5 (N ≥ 250) | 4.3 ± 1.3 nm (N = 501) | ||
| D h (nm) (in H2O) | 200.2 ± 3.5 | 126.4 ± 3.7 | ||
| D h (nm) (in medium) | 159.5 ± 0.6 | 79.43 ± 1.9 | 339.6 ± 21.9 | 364 ± 34.2 |
| ζ-Potential (mV) (in H2O) | +68.5 ± 1.6 | −17.3 ± 0.6 | ||
| ζ-Potential (mV) (in medium) | +24.57 ± 5.6 | +10.5 ± 4.8 | −24.28 ± 3.2 | −29.8 ± 1.3 |
Shewanella oneidensis oxygen uptake
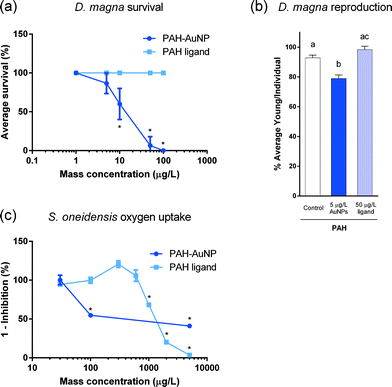
PAH–AuNPs significantly affected bacterial oxygen uptake at 100 μg L−1 (unpaired t-test, t = 9.895, df = 5, p < 0.05) while its corresponding free ligand control, 1 mg L−1 of PAH free ligand elicited similar inhibition compared to control groups (unpaired t-test, t = 4.222, df = 6, p < 0.05) (Fig. 1(c)). A concentration of 30 μg L−1 of PAH–AuNPs was chosen as the sublethal dose as this concentration produced no inhibition; this NP dose was paired with the 10-fold dose, 300 μg L−1, as the corresponding PAH free ligand control. MPA–AuNPs did not inhibit bacterial oxygen uptake at the highest dose tested (5 mg L−1), while the respective 5 mg L−1 of MPA free ligand demonstrated oxygen uptake inhibition (t = 9.713, df = 2, p < 0.05) (Fig. S2†).As the oxygen uptake reflects bacterial population growth, the doubling time of bacterial growth at the exponential phase was calculated based on oxygen uptake traces (see ESI†). Results showed that S. oneidensis had an average doubling time between 2 and 3 hours in the growth medium used in this study; thus, 1 hour was chosen as a time point for short-term exposure and 6 hour for long-term exposure in the subsequent gene expression studies.
S. oneidensis gene expression response
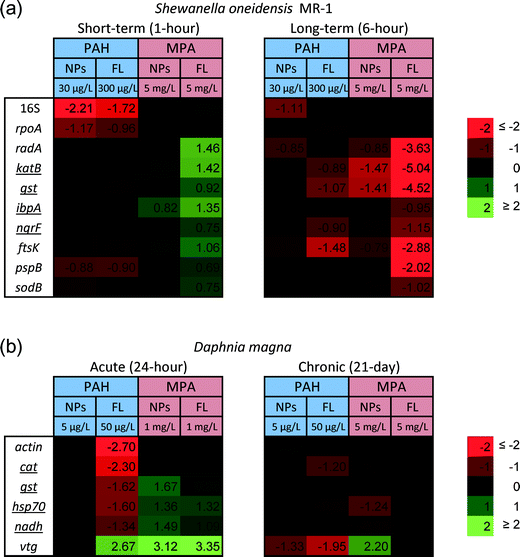
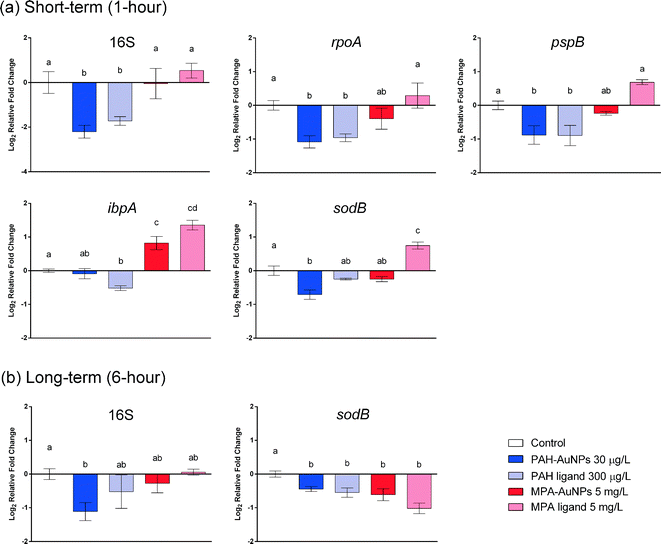
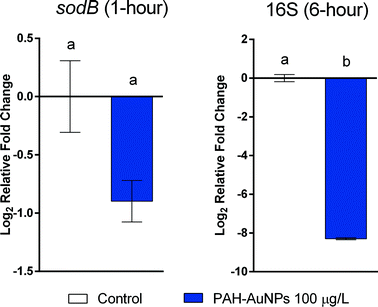
At the sublethal exposure dosages, differential expression levels of ten genes in S. oneidensis were observed at both 1 hour and 6 hour time points when comparing treatment and control. The general pattern of gene expression is summarized in the heat map (Fig. 2(a)).In all cases, the differences in gene expression appear to be dominated by ligand rather than NP exposure. All changes in gene expression induced by ligand–NP combination were accompanied by the changes in their respective free ligand control, including 16S (PAH, F = 18.33, df = 22, p < 0.0001), rpoA (PAH, F = 8.177, df = 31, p = 0.0001), pspB (PAH, F = 8.198, df = 22, p < 0.0003), and ibpA (MPA, F = 36.92, df = 22, p < 0.0001) at 1 hour exposure (Fig. 3(a)), and sodB (PAH and MPA, F = 10.06, df = 22, p < 0.0001) at 6 hour exposure (Fig. 3(b)). Exceptions are two NP-specific effects that were observed in sodB (PAH, F = 7.543, df = 22, p < 0.05) at 1 hour exposure and 16S (PAH, F = 3.238, df = 22, p < 0.05) at 6 hour exposure, where the free ligand control did not elicit similar effects as NPs when compared to control. For these two genes, S. oneidensis was exposed to a higher dosage (100 μg L−1) of PAH–AuNPs to explore the link to inhibition of oxygen uptake (Fig. 4). The 16S gene expression decreased upon 100 μg L−1 PAH–AuNP exposure at 6 hour exposure (unpaired t-test, t = 38.67, df = 7, p < 0.0001), while sodB gene expression did not show a significant difference compared to the control group at 1 hour exposure.
The difference in ligand–NP combination appears to be important in determining the differential gene expression pattern at 1 hour exposure, as only down-regulation was observed in PAH–AuNP exposure but only up-regulation was observed in MPA–AuNP exposure (Fig. 2(a)). However, upon 6 hour exposure, the ligand–NP combination did not determine the gene expression pattern, as only down-regulation was observed for all treatments, regardless of the type of ligand (Fig. 2(a)).
Time frame is also an important factor in terms of gene expression response, as differential gene expression responses were observed at different time points. In response to PAH–AuNP/ligand exposure, effects that were observed in the rpoA and pspB genes at 1 hour exposure diminished by the 6 hour exposure timepoint. More interestingly, for MPA–AuNP/ligand exposure, the expression level compared to control at the 6 hour exposure appeared to be opposite of the response in the 1 hour exposure, especially for MPA ligand exposure.
Daphnia magna acute toxicity
NP surface functionalization played an important role in acute toxicity in the form of daphnid survival, with positively charged PAH–AuNPs being orders of magnitude more toxic than the negatively charged MPA–AuNPs (Fig. 2(a)). PAH–AuNPs significantly affected daphnid mortality, eliciting 40% mortality at 10 μg L−1 (U = 0, p < 0.05).13 MPA–AuNPs did not significantly affect daphnid survival at the highest concentration tested, 25 mg L−1 (data not shown) (p > 0.05).13 The free ligands used in NP functionalization had no impact on daphnid survival at any concentration tested.Daphnia magna chronic toxicity
Ligand–NP combination is also important in governing the chronic impacts on daphnid reproduction. Of the two NPs tested, PAH–AuNPs significantly decreased daphnid reproduction over the 21 day chronic exposure (Fig. 2(b)) while MPA–AuNPs did not (data not shown). PAH–AuNPs significantly decreased daphnid reproduction by 15% at the highest concentration tested, 5 μg L−1 (F = 14.751, df = 23, p < 0.05). In comparison, PAH free ligand caused a statistically insignificant increase in daphnid reproduction at 50 μg L−1. As previously reported, 5 mg L−1 MPA free ligand increased daphnid reproduction by 14% (U = 4, p < 0.05, data not shown).13Daphnia magna acute gene expression response
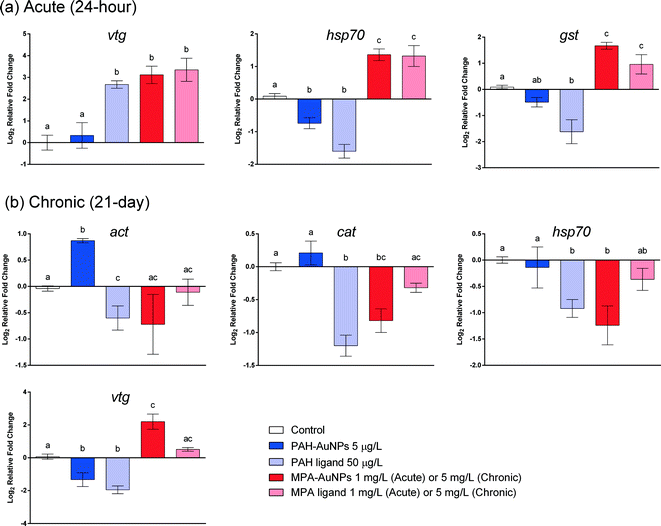
After a 24 h acute exposure, NP functionalization is also an important factor in determining Daphnia response at the gene level when exposed to PAH and MPA–AuNPs, resulting in different gene expression patterns for cat, nadh, vtg, gst and hsp70 (Fig. 2). For Daphnia exposed to PAH–AuNPs, there was a significant 0.74 fold decrease in the relative expression of hsp70 (F = 31.799, df = 49, P < 0.05) compared to controls. Daphnia exposed to MPA–AuNPs caused a significant 1.36 fold increase for hsp70 (F = 31.799, df = 49, P < 0.05), 1.49 fold increase for nadh (F = 29.066, df = 55, p < 0.05), 1.67 fold increase for gst (F = 23.116, df = 53, p < 0.05) and 3.12 fold increase for vtg (F = 11.556, df = 47, p < 0.05) over controls. MPA–AuNP-exposed Daphnia had significantly different gene expression patterns than Daphnia exposed to PAH–AuNPs for nadh, vtg, gst and hsp70 (Fig. 2 and 5). Notably, PAH–AuNPs caused a 0.33 fold increase in relative expression of vtg while MPA–AuNPs elicited a 3.12 fold increase in relative expression of vtg (F = 11.556, df = 47, p < 0.05).The impacts of free ligands used in particle functionalization closely follow the gene expression patterns observed for their respective functionalized NPs at 24 h (Fig. 2). Daphnia exposed to the PAH ligand showed no statistical difference compared to Daphnia exposed to PAH–AuNPs for all genes tested except cat (F = 8.640, df = 55, p < 0.05) and vtg (F = 11.556, df = 47, p < 0.05). Each gene that showed a significant positive fold change in relative expression for Daphnia exposed to MPA–AuNPs also showed a significant fold change in relative expression for the MPA free ligand treatment and did not significantly differ between the two.
Daphnia magna chronic gene expression response
Similar to the 24 h acute exposure, AuNP surface functionalization played an important role in determining gene expression levels in Daphnia chronically exposed to AuNPs (Fig. 2 and 5). For Daphnia exposed to PAH–AuNPs, there was a significant 1.33 fold decrease in the relative expression of vtg (F = 16.592, df = 42, p < 0.05) and a significant 0.87 fold increase in the relative expression of act (F = 9.68, df = 42, p < 0.05) over controls (Fig. 5). MPA–AuNPs elicited a significant 1.24, 0.82 and 0.93 fold decreases in the relative expression of hsp70 (F = 9.294, df = 42, p < 0.05), cat (F = 18.128, df = 44, p < 0.05) and nadh (F = 14.9, df = 44, p < 0.05), respectively, compared to controls (Fig. 2 and 5). Notably, for this treatment, there was a significant 2.2 fold increase in the relative expression of vtg (F = 16.592, df = 42, p < 0.05) over controls (Fig. 2). A significantly different gene expression response was observed for several genes when AuNP treatments were compared. PAH–AuNP treatment elicited a positive fold change in the relative expression of cat, nadh and act while MPA–AuNP elicited a negative fold for these same genes. The greatest difference between these two treatments was observed for vtg.NP-specific impacts were observed in Daphnia chronically exposed functionalized AuNPs versus their respective PAH and MPA ligands as reflected in the gene expression patterns (Fig. 2 and 5). PAH–AuNP and PAH ligand caused a similar relative expression pattern in Daphnia for genes gst, hsp70, vtg and nadh, as no significant difference was observed among these conditions (Fig. 2). However, PAH ligand caused 0.6 fold decrease in relative expression for act compared with the PAH–AuNP treatment that elicited a 0.98 fold increase in relative expression for act (F = 9.68, df = 42, p < 0.05) (Fig. 5). There were no significant differences between MPA ligand and MPA–AuNP treatments on Daphnia expression for any genes tested.
Discussion
Ligand–NP combinations differed in the extent of organismal apical endpoint impacts. Both model organisms were impacted to a greater extent by positively charged ligand–NP combinations with differential sensitivities. PAH–AuNPs were determined to be 2–3 orders of magnitude more toxic than the MPA–AuNPs for both S. oneidensis and D. magna. MPA–AuNPs caused no acute mortality in D. magna or inhibition on S. oneidensis oxygen uptake at the highest concentration tested (25 mg L−1 for daphnids and 5 mg L−1 for bacteria) (Fig. S2†).13 PAH–AuNPs elicited mortality in D. magna at concentrations as low as 10 μg L−1 and decreases in reproduction at 5 μg L−1 (Fig. 1(a)),13 while S. oneidensis started to show respiratory inhibition at 100 μg L−1 (Fig. 1(c)). Electrostatic interactions could largely drive the differences in sensitivity to the differently charged particles, as both eukaryotes and prokaryotes have cell surfaces that are negatively charged.58,59 It is thought that, due to electrostatic interactions, positively charged NPs are more likely to interact with cell surfaces than negatively charged NPs. Goodman et al. (2004)60 observed similar differences in the toxicity of AuNPs functionalized with cationic and anionic side chains when exposed to mammalian cell lines and bacterial cells, and Feng et al.61 demonstrated a similar correlation between toxicity and NP-cell association where increased NP-cell association was found for positively charged NPs compared to negatively charged NPs in bacteria. In addition to electrostatic interactions, the low toxicity of MPA–AuNPs may be potentially explained by the high degree of aggregation of MPA–AuNPs experienced in both organisms' exposure media (Table 2), thus reducing bioavailability. Overall, similar differential toxicity of the two functionalized NPs were observed in our study in both model organisms, indicating these organisms may follow the same electrostatic mechanism for interacting with NPs despite having distinct membrane surface chemistry and that the general response of whole organism may be extrapolated from the response of cell lines, although they differ in sensitivity.In some cases the toxicity of select NPs may not be determined by their respective ligand alone, which demonstrates NP-specific organismal impacts. However, this NP specific effect was only true for D. magna, where the impacts to S. oneidensis could largely be attributed to the ligand itself and only at much higher concentrations. The differences in sensitivity observed for these two model organisms exposed to PAH–AuNPs may be due to the distinct differences in the cell surface chemistry of Gram-negative bacteria and the aquatic eukaryotes. Besides the cytoplasmic membrane, which are found in both bacterial and Daphnia cells, the Gram-negative S. oneidensis bacterial cell also has an envelope that consists of a peptidoglycan–lipoprotein complex, periplasmic zone, and an outer membrane layer.58 The outer membrane layer is the first barrier that NPs would encounter, and this lipid bilayer retains various amounts of embedded lipopolysaccharides (LPS).58 LPS are high molecular weight molecules with a basal lipid anchored in the lipid bilayer and a long negatively charged chain of polysaccharide. Recent work using S. oneidensis demonstrated that LPS is an important binding site for AuNPs.62 Compared to the animal cell membrane, the complex structure of the cell envelope in S. oneidensis may provide extra protection when NPs are in proximity to the cells, thus desensitizing bacterial cells to NP exposures. In addition, studies demonstrate that eukaryote cells have many more mechanisms for supramolecular and colloidal particle internalization (e.g. receptor mediated endocytosis, pinocytosis and phagocytosis) for both nano- and macro-sized particles, while very few studies show plausible evidence of internalization of nanomaterials into bacterial cells.63–65 Furthermore, the manner by which multi- and single-cellular organisms interact with NPs may also contribute to the difference in sensitivity. Daphnia actively accumulate NPs internally while bacteria only passively interact with NPs through random encounters on the surface. The difference in how NP interact and accumulate in two organisms may also result in the NP-specific effect observed in D. magna but not in S. oneidensis. PAH–AuNPs resulted in a decrease in Daphnia survival (10 μg L−1) while the respective PAH free ligand control (100 μg L−1) did not show any mortality (Fig. 1(a)). However, when PAH–AuNPs elicited inhibition to bacterial oxygen uptake at 100 μg L−1, the respective ligand control (1 mg L−1) displayed a similar inhibition (Fig. 1(c)). These biological differences and impacts of NP surface functionalization and free ligand type are further addressed by the presented gene expression study.
Gene expression revealed insight into potentially molecular pathways that may be impacted upon exposure to NPs and may explain the differences in toxicity across different ligand–NP combinations and supported the mortality and respiration results indicating a particle- specific impact in Daphnia versus Shewanella. In both acute and chronic assays, Daphnia exposed to PAH–AuNPs elicited a significantly different gene expression pattern compared with Daphnia exposed to MPA–AuNPs, despite the two NPs having the same gold core. These differences were notable in the 24 h acute exposure for hsp70, gst, vtg and nadh and in the 21 day chronic assay for hsp70, vtg, nadh, cat and act. Amongst the genes that responded, a positive relative fold change for act was unique to the PAH–AuNP treatment in the 21 day assay with respect to the ligand control. Actin (act) encodes for a protein important to cytoskeleton and muscle fibril production as well as other cell functions. Studies have linked an increase in protein concentration of actin as a compensatory mechanism to maintain muscular and cellular performance in times of environmental stress.66 In addition, studies have indicated a high binding affinity of microparticles for actin67 and have shown that multiple NP types damage actin filaments in vitro.68–70 PAH–AuNPs could be potentially damaging muscle fibrils and cellular structure over long-term exposures in Daphnia. The relationship of this gene with apical endpoints impacted in Daphnia within this study remains unclear.
Daphnia exposed to MPA–AuNPs only uniquely responded to the treatment with an increase in the relative fold change of gst at 24 h. This gene encodes for an enzyme glutathione S-transferase and is an important enzyme in xenobiotic detoxification as it conjugates compounds with glutathione and may be elevated in times of oxidative stress. Our previous studies observed gst induction in Daphnia dependent upon NP functionalization of fullerenes but only at concentrations that elicited significant mortality (>5 mg L−1).9 Like MPA–AuNPs, these NPs exhibited a high degree of aggregation and exhibited low toxicity in Daphnia. This may demonstrate an acute whole organismal response to a high amount of negatively charged NPs. Our more recent previous study examined adult daphnid guts exposed to 4 nm PAH and MPA–AuNPs and their ligands at low concentrations (<0.05 mg L−1).18 Here, we showed that significant amounts of ROS were produced for both MPA and PAH AuNPs and their respective ligands at the same concentrations. This leads us to believe that ROS production does not fully explain the adverse outcomes observed in our acute and chronic studies. Therefore, other mechanisms may be responsible for the observed impacts as Daphnia responded differently to MPA and PAH AuNPs but had similar amounts of ROS detected upon exposure to these treatments at the same concentrations. However, in our current study and the previous, gene expression patterns were different for the two ligand–NP combinations. These results suggest that pathways affected by NPs are strongly dependent upon NP surface properties.
For S. oneidensis, gene expression assays were again indicative of the observed apical endpoint impacts. Most of the gene expression responses for S. oneidensis were provoked equally by the free ligand exposure and ligand–NP combination at both time points. While MPA–AuNPs did not show any impact that was specific to NPs, the decrease in expression of 16S at 6 hour exposure and sodB gene at 1 hour exposure were unique to the PAH–AuNPs but not to PAH free ligand. The sodB gene encodes for one of the superoxide dismutases (SODs) that protect cells from deleterious reactions with reactive oxygen species;71 it has been previously reported that the sodB gene was up-regulated upon S. oneidensis exposure to chromium(VI).72 More related, a previous study using 60 nm amino-functionalized polystyrene nanomaterial (PS-NH2-NPs) on E. coli single-gene deletion mutants showed that the ΔsodB mutant was more sensitive to the exposure of PS-NH2-NPs compared to the parent strain.73 As PAH–AuNPs have a similar surface-functionalization of amine groups with PS-NH2-NPs, these results suggests that the sodB gene plays an essential role in bacterial cell response to amine-functionalized nanomaterials, making it possible to use sodB as a biomarker for this specific NP surface functionalization. 16S ribosomal RNA (rRNA) is one of the three rRNAs, which are components of prokaryotic ribosomes. rRNA transcription is the rate-limiting step in ribosome synthesis, and thus, directly correlates to protein synthesis and cell growth.74 Previous research has reported that rRNA degradation occurs during environmental stress, including oxidative stress and starvation.75–77 Notably, it was also reported that rRNA is degraded due to a change in cell membrane permeability, potentially leading to the entry of RNase I, an endoribonuclease, from the periplasmic space into the cytoplasm.78,79 Extensive cell membrane damage can also result in the efflux of RNA due to the loss of plasma membrane integrity.80 Previous research has shown the disruption of membrane integrity in S. oneidensis cells upon PAH–AuNP exposure,61 correlating with the decrease in the expression of 16S. It should be noted that at 1 hour exposure, the respective PAH ligand control also elicited decrease in 16S expression, while at 6 hour exposure only PAH–AuNPs showed the effect; thus, the potential of 16S to be used as a biomarker that is specific for PAH–AuNPs is limited to long-term exposures. In effort to link 16S and sodB gene response to the apical biological endpoints, the gene expression level of these two genes was examined at a higher dosage (100 μg L−1) that also caused inhibition in bacterial oxygen uptake (Fig. 1(c)). While the sodB gene at 1 hour exposure did not elicit change in gene expression, 16S at 6 hour exposure showed a similar decrease upon 100 μg L−1 PAH–AuNP exposure (Fig. 4), proving that 16S can be potentially used as a biomarker for the impact of PAH–AuNPs on bacterial oxygen uptake; future work will explore the adverse outcome pathway from the decrease in 16S rRNA expression to the inhibition of bacterial oxygen uptake, and we postulate that the inhibition is mediated via reduced activity in protein synthesis. MPA–AuNPs did not induce a similar response of 16S rRNA expression, or any other NP-specific response, indicating a distinction between the same AuNP cores functionalized with different surface ligands.
Length of exposure had an impact on the effects seen in both species and on both gene expression and apical endpoints measured. Short-term exposures for both D. magna and S. oneidensis revealed that functionalized NP impacts on certain molecular pathways might be predicted by their respective ligand alone. Out of all S. oneidensis regulated genes, three genes stand out as potential predictors of NP impacts based on the ligand alone. These genes are pspB and rpoA for PAH–AuNP/ligand and ibpA for MPA–AuNP/ligand at 1 hour exposure, as they were influenced similarly upon exposure to both the ligand-bound AuNPs and the respective free ligand. For D. magna, three genes were most notable; these genes were hsp70 and vtg for PAH–AuNP/ligand and hsp70, vtg and nadh for MPA–AuNP/ligand. These results suggest that NP impacts on specific molecular pathways may be predicted based on response to the ligand alone. This finding is especially important for ligands or functional groups that are commonly used to achieve desired physiochemical properties for NPs. However, as demonstrated with our study, ligand–NP combinations did alter several genes that the ligand alone did not, and the concentrations of NPs that impacted apical endpoints, in particular PAH–AuNPs, differed from that of the ligand. This diminishes the potential ability to use ligand information alone as a predictor for NP toxicity; rather, the overall NP characteristics, including charge or size, may be more informative.
Our study revealed that gene expression in acute exposures was not predictive of long-term impacts or differences among treatments with respect to ligand versus ligand–NP combinations. In addition, long-term exposure to NPs resulted in gene expression patterns that could not be predicted based on gene expression patterns from short-term exposures. Upon exposure to MPA–AuNP/ligand, both S. oneidensis and D. magna showed decreases in gene expression during short-term exposure and that this response flipped to mostly an increase in gene expression upon long-term exposure. Exceptions to this finding were observed in the decrease of 16S and sodB expression upon PAH–AuNP exposure in S. oneidensis and the increase of vtg gene expression upon MPA–AuNP exposure in D. magna, which show similar response in gene expression levels at both time points. Our results indicate that, although it is possible to predict long-term gene expression impacts based on short-term impacts, it is limited to selected genes, which may downplay the significance of this finding.
Gene expression responses across organisms provide an indication of how organisms are similar or different in their response to NP exposures. A notable signature shared across two organisms was the up-regulation of ibpA/hsp70 induced by MPA–AuNP and ligand for short-term exposures. Both ibpA and hsp70 encode for heat shock protein in S. oneidensis and D. magna, respectively. Heat shock proteins (Hsp) are a large family of proteins that help unfolded or misfolded proteins to fold correctly in vivo and are widely considered to be good indicators of proteotoxic stress.81,82 The up-regulation of heat shock protein induced by MPA–AuNPs and ligands potentially indicates the disruption of membrane proteins, provoking pathways that help adapt to change in chemical environment caused by introduction of NPs or ligands. This feature, shared by both organisms, potentially indicates a universal stress-response to negatively charged NPs, making the genes encoding for heat shock protein a good candidate for predicting the effect of NPs based on the response to their respective ligands. However, MPA–AuNPs did not lead to any adverse outcomes at the concentrations we tested, which makes understanding the importance of this pathway within the context of our study difficult.
Conclusion
Molecular studies have the ability to tease out distinct modes of action for NP toxicity and help to develop biomarkers for assessing NP impacts on environmentally relevant endpoints. Using standard toxicological and gene expression assays, we revealed that: 1) the ligand–NP combinations determine the extent of impacts on apical endpoints and the toxicity of select NPs may not be determined by their respective ligand alone; 2) depending on the organism considered, exposure to ligand–NP combinations may impact unique molecular pathways that differ from the ligand alone; 3) short-term exposures reveal that ligand–NP impacts on certain molecular pathways might be predicted by their respective ligand alone but the ability to predict long-term impacts may be minimal; and 4) examining gene expression responses across organisms may provide an indication of how organisms are similar or different in their response to NP exposures. Lastly, this study reveals that there are mechanisms other than oxidative stress for NP toxicity and that these may be elucidated using molecular level experiments and exposures that consider sublethal concentrations.Acknowledgements
This work was funded by the National Science Foundation Center for Chemical Innovations grant CHE-1240151: Center for Sustainable Nanotechnology and a Research Experience for Undergraduates fellowship supported by the National Science Foundation Center for Chemical Innovation Program (CHE-1240151) awarded to Ky G. Christenson. We thank Ky G. Christenson for his help in gene expression work. We wish to thank the UWM Great Lakes Genomics Center for the use of equipment and assistance by staff. Lastly, we want to thank Dr. Y. Hayashi for access to the NORMA-Gene workbook.References
- A. S. Arico, P. Bruce, B. Scrosati, J.-M. Tarascon and W. van Schalkwijk, Nat. Mater., 2005, 4, 366–377 CrossRef CAS PubMed.
- J. Xie, G. Liu, H. S. Eden, H. Ai and X. Chen, Acc. Chem. Res., 2011, 44, 883–892 CrossRef CAS PubMed.
- M. M. Pendergast and E. M. V. Hoek, Energy Environ. Sci., 2011, 4, 1946–1971 CAS.
- A. Nel, T. Xia, L. Madler and N. Li, Science, 2006, 311, 622–627 CrossRef CAS PubMed.
- Y. Pan, S. Neuss, A. Leifert, M. Fischler, F. Wen, U. Simon, G. Schmid, W. Brandau and W. Jahnen-Dechent, Small, 2007, 3, 1941–1949 CrossRef CAS PubMed.
- A. Albanese, P. S. Tang and W. C. Chan, Annu. Rev. Biomed. Eng., 2012, 14, 1–16 CrossRef CAS PubMed.
- D. A. Arndt, M. Moua, J. Chen and R. D. Klaper, Environ. Sci. Technol., 2013, 47, 9444–9452 CrossRef CAS PubMed.
- R. Klaper, D. Arndt, K. Setyowati, J. Chen and F. Goetz, Aquat. Toxicol., 2010, 100, 211–217 CrossRef CAS PubMed.
- R. Klaper, J. Crago, J. Barr, D. Arndt, K. Setyowati and J. Chen, Environ. Pollut., 2009, 157, 1152–1156 CrossRef CAS PubMed.
- B. D. Chithrani, A. A. Ghazani and W. C. Chan, Nano Lett., 2006, 6, 662–668 CrossRef CAS PubMed.
- H. Yang, C. Liu, D. Yang, H. Zhang and Z. Xi, J. Appl. Toxicol., 2009, 29, 69–78 CrossRef CAS PubMed.
- K. L. Aillon, Y. Xie, N. El-Gendy, C. J. Berkland and M. L. Forrest, Adv. Drug Delivery Rev., 2009, 61, 457–466 CrossRef CAS PubMed.
- J. S. Bozich, S. E. Lohse, M. D. Torelli, C. J. Murphy, R. J. Hamers and R. D. Klaper, Environ. Sci.: Nano, 2014, 1, 260–270 RSC.
- A. M. El Badawy, R. G. Silva, B. Morris, K. G. Scheckel, M. T. Suidan and T. M. Tolaymat, Environ. Sci. Technol., 2010, 45, 283–287 CrossRef PubMed.
- C. He, Y. Hu, L. Yin, C. Tang and C. Yin, Biomaterials, 2010, 31, 3657–3666 CrossRef CAS PubMed.
- N. M. Schaeublin, L. K. Braydich-Stolle, A. M. Schrand, J. M. Miller, J. Hutchison, J. J. Schlager and S. M. Hussain, Nanoscale, 2011, 3, 410–420 RSC.
- A. Verma and F. Stellacci, Small, 2010, 6, 12–21 CrossRef CAS PubMed.
- G. A. Dominguez, S. E. Lohse, M. D. Torelli, C. J. Murphy, R. J. Hamers, G. Orr and R. D. Klaper, Aquat. Toxicol., 2015, 162, 1–9 CrossRef CAS PubMed.
- A. Albanese and W. C. W. Chan, ACS Nano, 2011, 5, 5478–5489 CrossRef CAS PubMed.
- N. Lewinski, V. Colvin and R. Drezek, Small, 2008, 4, 26–49 CrossRef CAS PubMed.
- N. Khlebtsov and L. Dykman, Chem. Soc. Rev., 2011, 40, 1647–1671 RSC.
- C.-W. Lam, J. T. James, R. McCluskey, S. Arepalli and R. L. Hunter, Crit. Rev. Toxicol., 2006, 36, 189–217 CrossRef CAS PubMed.
- E. Fröhlich, Int. J. Nanomed., 2012, 7, 5577–5591 CrossRef PubMed.
- S. K. Sohaebuddin, P. T. Thevenot, D. Baker, J. W. Eaton and L. Tang, Part. Fibre Toxicol., 2010, 7, 22 CrossRef PubMed.
- M. Heinlaan, A. Ivask, I. Blinova, H.-C. Dubourguier and A. Kahru, Chemosphere, 2008, 71, 1308–1316 CrossRef CAS PubMed.
- H. Sanderson and M. Thomsen, Bull. Environ. Contam. Toxicol., 2007, 79, 331–335 CrossRef CAS PubMed.
- D. R. Moore, R. L. Breton and D. B. MacDonald, Environ. Toxicol. Chem., 2003, 22, 1799–1809 CrossRef CAS PubMed.
- A. A. Shvedova, A. Pietroiusti, B. Fadeel and V. E. Kagan, Toxicol. Appl. Pharmacol., 2012, 261, 121–133 CrossRef CAS PubMed.
- B. J. Marquis, S. A. Love, K. L. Braun and C. L. Haynes, Analyst, 2009, 134, 425–439 RSC.
- A. Manke, L. Wang and Y. Rojanasakul, BioMed Res. Int., 2013, 2013, 942916 Search PubMed.
- E. J. Park and K. Park, Toxicol. Lett., 2009, 184, 18–25 CrossRef CAS PubMed.
- R. Klaper, D. Arndt, J. Bozich and G. Dominguez, Analyst, 2014, 139, 882–895 RSC.
- F. Gottschalk, T. Sonderer, R. W. Scholz and B. Nowack, Environ. Sci. Technol., 2009, 43, 9216–9222 CrossRef CAS PubMed.
- A. Valavanidis, T. Vlahogianni, M. Dassenakis and M. Scoullos, Ecotoxicol. Environ. Saf., 2006, 64, 178–189 CrossRef CAS PubMed.
- R. Franco, R. Sanchez-Olea, E. M. Reyes-Reyes and M. I. Panayiotidis, Mutat. Res., Genet. Toxicol. Environ. Mutagen., 2009, 674, 3–22 CrossRef CAS PubMed.
- T. Finkel and N. J. Holbrook, Nature, 2000, 408, 239–247 CrossRef CAS PubMed.
- J. Ryan and L. Hightower, Stress-Inducible Cellular Responses, Springer, 1996, pp. 411–424 Search PubMed.
- C. G. Kilty, J. Keenan and M. Shaw, Expert Opin. Drug Saf., 2007, 6(2), 207–215 CrossRef CAS PubMed.
- C. Eason and K. O'Halloran, Toxicology, 2002, 181, 517–521 CrossRef.
- N. J. Niemuth, R. Jordan, J. Crago, C. Blanksma, R. Johnson and R. D. Klaper, Environ. Toxicol. Chem., 2014, 34(2), 291–296 CrossRef PubMed.
- G. T. Ankley, K. M. Jensen, M. D. Kahl, J. J. Korte and E. A. Makynen, Environ. Toxicol. Chem., 2001, 20, 1276–1290 CrossRef CAS.
- S. M. Johns, M. D. Kane, N. D. Denslow, K. H. Watanabe, E. F. Orlando, D. L. Villeneuve, G. T. Ankley and M. S. Sepúlveda, Environ. Toxicol. Chem., 2009, 28, 873–880 CrossRef CAS PubMed.
- J. S. Garvey, Preprints of Papers Presented at National Meeting, Division of Water, Air and Waste Chemistry, American Chemical Society, (USA), 1988 Search PubMed.
- M. Fischbach, E. Sabbioni and P. Bromley, Cell Biol. Toxicol., 1993, 9, 177–188 CrossRef CAS.
- H. H. Hau and J. A. Gralnick, Annu. Rev. Microbiol., 2007, 61, 237–258 CrossRef CAS PubMed.
- R. Gulati, Hydrobiologia, 1978, 59, 101–112 CrossRef.
- N. R. Jana, L. Gearheart and C. J. Murphy, Langmuir, 2001, 17, 6782–6786 CrossRef CAS.
- T. K. Sau and C. J. Murphy, J. Am. Chem. Soc., 2004, 126, 8648–8649 CrossRef CAS PubMed.
- D. A. Giljohann, D. S. Seferos, W. L. Daniel, M. D. Massich, P. C. Patel and C. A. Mirkin, Angew. Chem., Int. Ed., 2010, 49, 3280–3294 CrossRef CAS PubMed.
- S. F. Sweeney, G. H. Woehrle and J. E. Hutchison, J. Am. Chem. Soc., 2006, 128, 3190–3197 CrossRef CAS PubMed.
- J. A. Yang, S. E. Lohse and C. J. Murphy, Small, 2014, 10, 1642–1651 CrossRef CAS PubMed.
- C. J. Ackerson, P. D. Jadzinsky and R. D. Kornberg, J. Am. Chem. Soc., 2005, 127, 6550–6551 CrossRef CAS PubMed.
- M. D. Torelli, R. A. Putans, Y. Tan, S. E. Lohse, C. J. Murphy and R. J. Hamers, ACS Appl. Mater. Interfaces, 2015, 7, 1720–1725 CAS.
- EPA, Standard operating procedure for moderately hard reconstituted water, SoBran, Dayton, OH, USA, 2003 Search PubMed.
- S. Zhao and R. D. Fernald, J. Comput. Biol., 2005, 12, 1047–1064 CrossRef CAS PubMed.
- L.-H. Heckmann, P. Sorensen, P. Krogh and J. Sorensen, BMC Bioinf., 2011, 12, 250 CrossRef PubMed.
- S. Wang, W. Lu, O. Tovmachenko, U. S. Rai, H. Yu and P. C. Ray, Chem. Phys. Lett., 2008, 463, 145–149 CrossRef CAS PubMed.
- P. A. Rice and B. H. Iglewski, Rev. Infect. Dis., 1988, 10, S277–S278 CrossRef PubMed.
- A. Martínez-Palomo, International Review of Cytology, Academic Press, 1970, vol. 29, pp. 29–75 Search PubMed.
- C. M. Goodman, C. D. McCusker, T. Yilmaz and V. M. Rotello, Bioconjugate Chem., 2004, 15, 897–900 CrossRef CAS PubMed.
- Z. V. Feng, I. L. Gunsolus, T. A. Qiu, K. R. Hurley, L. H. Nyberg, H. Frew, K. P. Johnson, A. M. Vartanian, L. M. Jacob, S. E. Lohse, M. D. Torelli, R. J. Hamers, C. J. Murphy and C. L. Haynes, Chem. Sci., 2015 10.1039/C5SC00792E.
- K. H. Jacobson, I. L. Gunsolus, T. R. Kuech, J. M. Troiano, E. S. Melby, S. E. Lohse, D. Hu, W. B. Chrisler, C. J. Murphy, G. Orr, F. M. Geiger, C. L. Haynes and J. A. Pedersen, Environ. Sci. Technol., 2015 DOI:10.1021/acs.est.5b01841.
- Y. Y. Zhao, Y. Tian, Y. Cui, W. W. Liu, W. S. Ma and X. Y. Jiang, J. Am. Chem. Soc., 2010, 132, 12349–12356 CrossRef CAS PubMed.
- S. Dalai, S. Pakrashi, R. S. Kumar, N. Chandrasekaran and A. Mukherjee, Toxicol. Res., 2012, 1, 116–130 RSC.
- A. Kumar, A. K. Pandey, S. S. Singh, R. Shanker and A. Dhawan, Chemosphere, 2011, 83, 1124–1132 CrossRef CAS PubMed.
- S. Schwerin, B. Zeis, T. Lamkemeyer, R. J. Paul, M. Koch, J. Madlung, C. Fladerer and R. Pirow, BMC Physiol., 2009, 9, 8 CrossRef PubMed.
- M. Ehrenberg and J. L. McGrath, Acta Biomater., 2005, 1, 305–315 CrossRef PubMed.
- P. Ruenraroengsak and A. T. Florence, J. Drug Targeting, 2010, 18, 803–811 CrossRef CAS PubMed.
- V. G. Walker, Z. Li, T. Hulderman, D. Schwegler-Berry, M. L. Kashon and P. P. Simeonova, Toxicol. Appl. Pharmacol., 2009, 236, 319–328 CrossRef CAS PubMed.
- T. Mironava, M. Hadjiargyrou, M. Simon, V. Jurukovski and M. H. Rafailovich, Nanotoxicology, 2010, 4, 120–137 CrossRef CAS PubMed.
- S. B. Farr and T. Kogoma, Microbiol. Rev., 1991, 55, 561–585 CAS.
- K. Chourey, M. R. Thompson, J. Morrell-Falvey, N. C. VerBerkmoes, S. D. Brown, M. Shah, J. Zhou, M. Doktycz, R. L. Hettich and D. K. Thompson, Appl. Environ. Microbiol., 2006, 72, 6331–6344 CrossRef CAS PubMed.
- A. Ivask, E. Suarez, T. Patel, D. Boren, Z. Ji, P. Holden, D. Telesca, R. Damoiseaux, K. A. Bradley and H. Godwin, Environ. Sci. Technol., 2012, 46, 2398–2405 CrossRef CAS PubMed.
- B. J. Paul, W. Ross, T. Gaal and R. L. Gourse, Annu. Rev. Genet., 2004, 38, 749–770 CrossRef CAS PubMed.
- D. R. Crawford, Y. H. Wang, G. P. Schools, J. Kochheiser and K. J. A. Davies, Free Radical Biol. Med., 1997, 22, 551–559 CrossRef CAS.
- G. N. Basturea, M. A. Zundel and M. P. Deutscher, RNA, 2011, 17, 338–345 CrossRef CAS PubMed.
- D. Hsu, L.-M. Shih and Y. C. Zee, J. Bacteriol., 1994, 176, 4761–4765 CAS.
- M. P. Deutscher, Nucleic Acids Res., 2006, 34, 659–666 CrossRef CAS PubMed.
- M. Kuwano, H. Taniguchi, M. Ono, H. Endo and Y. Ohnishi, Biochem. Biophys. Res. Commun., 1977, 75, 156–162 CrossRef CAS.
- Z. Darzynkiewicz, S. Bruno, G. Delbino, W. Gorczyca, M. A. Hotz, P. Lassota and F. Traganos, Cytometry, 1992, 13, 795–808 CrossRef CAS PubMed.
- S. D. Brown, M. R. Thompson, N. C. VerBerkmoes, K. Chourey, M. Shah, J. Z. Zhou, R. L. Hettich and D. K. Thompson, Mol. Cell. Proteomics, 2006, 5, 1054–1071 CAS.
- K. Chourey, M. R. Thompson, J. Morrell-Falvey, N. C. VerBerkmoes, S. D. Brown, M. Shah, J. Z. Zhou, M. Doktycz, R. L. Hettich and D. K. Thompson, Appl. Environ. Microbiol., 2006, 72, 6331–6344 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5en00037h |
| ‡ These two authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2015 |