Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Centrifugal microfluidic platforms: advanced unit operations and applications
O.
Strohmeier†
ab,
M.
Keller†
ab,
F.
Schwemmer†
b,
S.
Zehnle†
a,
D.
Mark
ab,
F.
von Stetten
ab,
R.
Zengerle
abc and
N.
Paust
*ab
aHahn-Schickard, Georges-Koehler-Allee 103, 79110 Freiburg, Germany. E-mail: Nils.Paust@Hahn-Schickard.de; Tel: +49 761 203 73245
bLaboratory for MEMS Applications, IMTEK – Department of Microsystems Engineering, University of Freiburg, Georges-Koehler-Allee 103, 79110 Freiburg, Germany
cBIOSS Centre for Biological Signalling Studies, University of Freiburg, Schaenzlestr. 18, 79104 Freiburg, Germany
First published on 2nd June 2015
Abstract
Centrifugal microfluidics has evolved into a mature technology. Several major diagnostic companies either have products on the market or are currently evaluating centrifugal microfluidics for product development. The fields of application are widespread and include clinical chemistry, immunodiagnostics and protein analysis, cell handling, molecular diagnostics, as well as food, water, and soil analysis. Nevertheless, new fluidic functions and applications that expand the possibilities of centrifugal microfluidics are being introduced at a high pace. In this review, we first present an up-to-date comprehensive overview of centrifugal microfluidic unit operations. Then, we introduce the term “process chain” to review how these unit operations can be combined for the automation of laboratory workflows. Such aggregation of basic functionalities enables efficient fluidic design at a higher level of integration. Furthermore, we analyze how novel, ground-breaking unit operations may foster the integration of more complex applications. Among these are the storage of pneumatic energy to realize complex switching sequences or to pump liquids radially inward, as well as the complete pre-storage and release of reagents. In this context, centrifugal microfluidics provides major advantages over other microfluidic actuation principles: the pulse-free inertial liquid propulsion provided by centrifugal microfluidics allows for closed fluidic systems that are free of any interfaces to external pumps. Processed volumes are easily scalable from nanoliters to milliliters. Volume forces can be adjusted by rotation and thus, even for very small volumes, surface forces may easily be overcome in the centrifugal gravity field which enables the efficient separation of nanoliter volumes from channels, chambers or sensor matrixes as well as the removal of any disturbing bubbles. In summary, centrifugal microfluidics takes advantage of a comprehensive set of fluidic unit operations such as liquid transport, metering, mixing and valving. The available unit operations cover the entire range of automated liquid handling requirements and enable efficient miniaturization, parallelization, and integration of assays.
1. Introduction
Microfluidics enables the miniaturization, integration, and automation of laboratory processes ranging from basic operations to complex biochemical assays. Obviously, an increase in the research activities in this field has been accompanied by a much slower conversion of microfluidic approaches into products. The reasons for this tardy technology transfer have been extensively discussed in previous studies,1,2 stating for instance a lack of flexibility of the microfluidic implementations, which allow for a very limited number of applications for a single microfluidic device. All of the research, development, and certification expense would have to be paid off by these very limited number of applications developed for a small market segment.As one possible solution, microfluidic platform-based approaches have been suggested.3,4 A microfluidic platform provides a set of microfluidic unit operations such as liquid transport, metering, mixing and valving. The unit operations are validated, scalable, and standardized, and can be combined in an easy and consistent manner. In some cases, it might be possible that a fixed set of unit operations is implemented within a generic disposable cartridge, in which different applications can be processed, simply by adjusting chemistry. In general, the key advantage of using platforms is the possibility to make use of building blocks from existing solutions to implement new applications with reduced effort and risk, and to address an increased market, which can be as large as the number of applications implemented within a platform.
The company Cepheid impressively demonstrated platform based automation of biochemical analysis. An application specific cartridge was introduced, but the cartridge is capable of performing analysis for many different targets by changing the analysis chemistry. Thus, a single cartridge covers a large range of products for nucleic acid-based sample-to-answer testing with high market penetration (e.g., $411 million annual turnover by Cepheid, 2014).5 Based on one cartridge format, 22 different tests are currently available, covering applications in the fields of healthcare-associated infections, critical infectious diseases, sexual health, and oncology. In dependency of the desired throughput, processing devices for 1, 2, 4, or 16 cartridges in parallel are available.5 Another success story for in vitro diagnostics testing at the point-of-care is the handheld device and the microfluidic cartridges from Abbott's i-STAT system, for which more than 35 million tests were sold in 2014.6 Cartridges are available for measuring blood chemistries and electrolytes, hematology, blood gases, coagulation, or cardiac markers.6 It has been predicted that the market for microfluidic automation will continue to grow. The market for microfluidic devices for point-of-care applications alone is expected to grow from US$200 million today to a US$800 million turnover in 2019.7 In order to be successful, a microfluidic platform has to fully cover the functionalities from sample input to data analysis for the desired range of applications. Several recent publications e.g. by Mark et al., Sin et al. or Madou et al., provide criteria to select an appropriate microfluidic platform.8–10
This review intends to deepen the understanding of platform-based microfluidic automation. It focuses exclusively on platforms making use of centrifugal microfluidics in order to provide detailed insight into this obviously emerging technology. When compared to other microfluidic platforms, centrifugal microfluidics has several strengths: the centrifugal propulsion mechanism allows for a closed fluidic system, free of any interfaces to external pumps. The removal of any bubbles that may interfere with the proper performance of an assay is particularly simple due to the scalable buoyancy in the centrifugal gravity field. In addition, residual liquids that may be trapped due to surface forces can be removed from channels, chambers and sensor matrixes, again, simply by adjusting the volume forces by rotation. The strength of centrifugal microfluidics is reflected by an enormous breadth of available unit operations and initiated an increase in research activity on the one hand and an increasing commitment by major diagnostic companies on the other hand. Panasonic, Roche, Samsung, 3M, and Abaxis already have centrifugal microfluidic-based products on the market and a considerable number of additional companies are currently evaluating the use of centrifugal microfluidics for their applications.
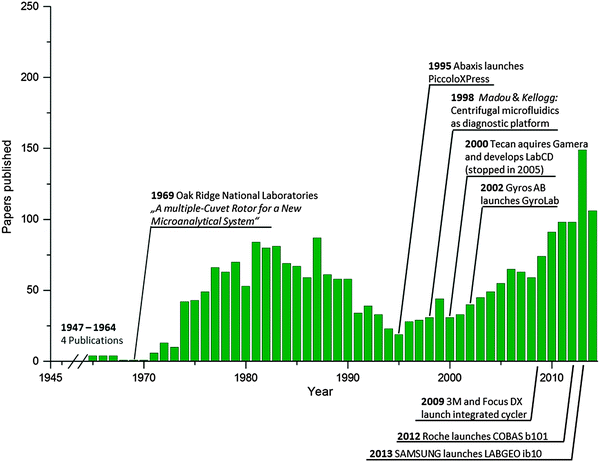
The last published comprehensive review on centrifugal microfluidics focused on the history and individual biomedical applications.11 Since then, more than 300 papers have been published on centrifugal microfluidics. An overview of the scientific journal publications and selected milestones in technology transfer is depicted in Fig. 1. Among the scientific publications, a clear trend toward the full integration of a complex sample-to-answer analysis can be observed. In addition, ground breaking novel unit operations have been developed that have the potential of making significant contributions to the field in the near future. Consequently, our review highlights these recent innovations. Special focus is directed towards the process of translating the assay step by step into a microfluidic layout, particularly the method used for combining unit operations to facilitate the miniaturization, integration, and automation of laboratory processes on centrifugal microfluidic platforms. Whereas basic fluidic functionalities are called unit operations, for a concatenation of such basic functionalities representing a laboratory workflow, we introduce the term “process chain.” In this context, we propose to standardize fluidic unit operations for the implementation of basic stand-alone functionalities such as metering, valving, and mixing. For the integration of frequently applied complete laboratory workflows, process chains should be standardized to allow for their efficient implementation without the need to deal with the basic functionalities. Examples of process chains are chemical cell lysis, nucleic acid purification and amplification, blocking to avoid unspecific binding, washing, immunocapture, etc. The terms used to describe the centrifugal microfluidic platform-based approach are defined in Table 1. Application examples for the hierarchy of a fluidic layout using process chains are depicted in the respective application chapter. Throughout this review, wherever suitable, we attempt to explain the implemented centrifugal microfluidic applications using the categories “process chains” and the underlying “unit operations.”
| Term | Definition |
|---|---|
| Microfluidic platform | A microfluidic platform provides a set of validated fluidic unit operations, which are designed for easy combination within a standardized fabrication technology.8 The platform approach enables efficient implementation of various laboratory workflows and/or applications. |
| Microfluidic chip/microfluidic cartridge | A microfluidic chip, which is often referred to as a microfluidic cartridge, is a substrate that provides structures like chambers, channels, etc. for the hardware implementation of the fluidic unit operations. For most applications, microfluidic chips are disposed of after use to avoid cross contamination and/or save regeneration cost. |
| Fluidic unit operations | …are basic fluidic functionalities such as the following: |
| • sample and reagent supply | |
| • reagent pre-storage and release | |
| • liquid transport | |
| • valving and switching | |
| • metering and aliquoting | |
| • mixing | |
| • separation | |
| • droplet generation | |
| • detection | |
| • … | |
| Processing device | The processing device (often also called the “instrument”) is a piece of reusable hardware that provides additional means to operate the microfluidic chip. This may comprise the main actuator (e.g., spinning drive) to control the fluids, as well as external means such as temperature control and/or magnetic, electric, optic, pneumatic, or mechanical features, including a means for detection/read-out. |
| Process chains | …are assemblies of fluidic unit operations and external means that represent laboratory workflows on a higher level of integration. Examples of process chains are… |
| • blood plasma separation | |
| • cell lysis | |
| • nucleic acid purification | |
| • nucleic acid amplification | |
| • immunocapture | |
| • washing | |
| • blocking | |
| • … |
This review is structured as follows. First, the physics of centrifugal microfluidics is briefly outlined, followed by a comprehensive review of the established and recently proposed centrifugal microfluidic unit operations. Based on the review of microfluidic unit operations, we reach conclusions about how some of the described developments will foster the integration of more complex applications. Subsequently, we review centrifugal microfluidic implementations of nucleic acid-based analysis; immunodiagnostics; clinical chemistry; and the analysis of food, water, and soil. Specific embodiments of centrifugal microfluidic systems, e.g., specific platforms using centrifugal microfluidics that are commercially available or under development are briefly outlined thereafter. Finally, we summarize the strengths and limitations and identify and discuss future trends.
1.1 Physics of centrifugal microfluidics
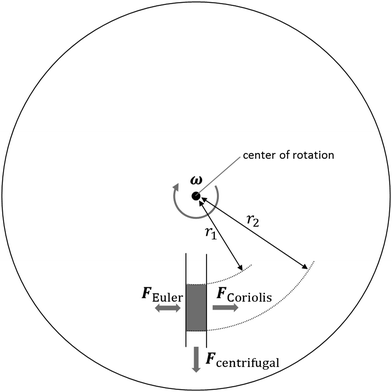
In order to understand the unit operations used in centrifugal microfluidics, we hereby introduce the forces that are exploited on this platform, as illustrated in Fig. 2. In general, we differentiate between intrinsic forces—sub-classified into pseudo-forces and non-pseudo forces—that are induced merely by the presence or absence of centrifugation, and extrinsic forces resulting from the use of external means.| Fc = −mω × (ω × r) | (1) |
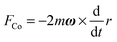 | (2) |
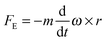 | (3) |
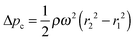 | (4) |
Non-pseudo forces are present in rotating systems, as well as in non-rotating systems. Hence, they are not limited to centrifugal platforms, but still play a major role in many centrifugal unit operations. The most dominant and most exploited non-pseudo forces and their corresponding differential pressures are the viscous force (Δpv) (eqn (5)), pneumatic force (Δppneu) (eqn (6)) exerted by a pressurized gas, capillary force (Δpcap) (eqn (7)), and fluidic inertia (Δpi) (eqn (8)).
| Δpv = −Rhydq | (5) |
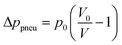 | (6) |
| Δpcap = σκ | (7) |
| Δpi = −ρla | (8) |
In the case of particle transport in fluids, such as in sedimentation processes, the particles are subject to a viscous force: the drag force (Fd). It is given by
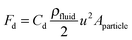 | (9) |
| Fs = 6πμru. | (10) |
Paramagnetic beads are commonly used in suspensions and attracted by external magnets on- or off-chip. The magnetic force Fmag acting on a spherical paramagnetic bead exposed to a magnetic flux density B is given by
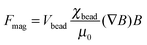 | (11) |
Electric forces can be applied in centrifugal systems via electrodes, which are preferably integrated into the microfluidic cartridge. This ensures the permanent and proximal exposure of samples to an electric field to perform electrolysis, dielectrophoresis, and other separation processes. The use of an external pneumatic pressure in centrifugal microfluidics can be realized in a non-contact fashion such as by directing a pressurized gas jet at certain openings of a rotating platform. Thus, the impact pressure of the gas is applied to the microfluidic network.12
2. Unit operations
A microfluidic platform provides a set of validated fluidic unit operations, which are designed for easy combination within a standardized fabrication technology.8 Unit operations are defined as the basic fluidic functionalities of a microfluidic platform. Examples of unit operations include sample and reagent supply, reagent pre-storage and release, liquid transport, valving and switching, metering, aliquoting, mixing, and detection. Assemblies of unit operations enable the efficient implementation of various process chains, which are laboratory workflows and/or applications on a higher level of integration. Examples of such process chains include blood plasma separation, cell lysis, nucleic acid purification, nucleic acid amplification, immunocapture, washing, and blocking. In the following, prominent unit operations are introduced and discussed in the light of their applications.2.1 Sample and reagent supply
It is inherently necessary to load the sample material and certain reagents for sample processing and analysis into the centrifugal microfluidic cartridge, either prior to or during processing. In more advanced applications and commercially available products, reagents are typically prestored in the cartridge to facilitate handling. Despite their importance, sample supply and reagent prestorage are seldom considered in academic publications. The following section will give an overview of the relevant concepts for sample loading and prestorage and the release of reagents in centrifugal microfluidic cartridges.The direct uptake of whole blood via a cartridge-integrated capillary was demonstrated by Rombach et al.19 An integrated capillary primes upon contact with a fingerprick blood sample and fills up with a defined volume. Subsequently, the blood is centrifuged to downstream processing chambers and directly processed by the cartridge to detect cholesterol. The uptake of whole blood by capillary forces was also integrated into the Roche Cobas b 101 system.20
The prestorage of liquid reagents allows complete hands-off automation obviating the need for manual reagent addition during processing. The diverse nature of chemical and biochemical reagents, including alcohols, solvents, aqueous solutions, e.g., with a high salt concentration24 or proteins and enzymes, renders their long-term stable prestorage extremely challenging. Alcoholic reagents evaporate easily and therefore need to be prestored in materials with low vapor transmission rates. Solvents and aqueous solutions might chemically interact with the surrounding material. Proteins and enzymes can degrade over time, with a loss in activity or change in concentration in the solution as a result of adsorption to the cartridge and container material.
The concepts for the prestorage of liquid reagents can be roughly divided into two groups: (1) prestorage in suitable containers that are placed in the cartridge or (2) prestorage directly in microfluidic chambers on the cartridge. The prestorage of reagents in additional containers might be a superior way to reduce physical and chemical interactions between the reagent and the cartridge material (mainly polymers) and is less critical with respect to swelling, water uptake, and vapor transmission.24 However, the required technologies for container fabrication and the mechanisms for releasing the reagents from the containers into the fluidic networks are more complex. Because of its advantages, commercially available centrifugal microfluidic systems like the Abaxis Piccolo Xpress25 or Roche Cobas b 10120 use reagent prestorage in additional containers.
The long-term stable prestorage of liquid reagents for DNA extraction has been demonstrated by Hoffmann et al.24 Washing- and elution-buffers were encapsulated in glass ampoules, which were placed in the cartridge. To release the reagents into the microfluidic structures, the glass ampoules were crushed manually prior to processing. Ethanol and water have been prestored for time periods of up to 300 days without any noticeable losses. Glass ampoules have further been used to prestore rehydration buffer for lyophilized polymerase pellets (Fig. 3b).26 A prestorage concept with a release mechanism that solely relies on centrifugal forces was presented by van Oordt et al. Liquid reagents were packed in miniature stick packs, which were fabricated from vapor-tight aluminum composite foil. Liquid was released via a peelable seal27 on the outer side of the stick pack by exceeding a defined centrifugal force (Fig. 3a). A 250 μL quantity of 10% v/v isopropanol in water did not show any significant evaporation after storage at 70 °C for 21 days, which corresponded to 18 months of storage at room temperature.28 This concept has later been used by Czilwik et al. for prestorage and on-demand release of a rehydration buffer for PCR reagents.29 The reagent release by centrifugation would furthermore enable the handling of highly wetting reagents, such as alcoholic buffer solutions, which could cause unwanted capillary priming of the microfluidic channel network if loaded to the disk in absence of centrifugal forces. The prestorage of highly reactive bromine water in inert Teflon or glass tubes sealed by ferrowax plugs was demonstrated by Hwang et al. The reagent release was controlled by melting the wax plugs via laser irradiation allowing the bromine to diffuse out while the diffusion was stopped after resolidification of the wax. This principle allowed the release of reagents in small increments depending on the progress of the chemical reaction.30 Kawai et al. presented a rotatable reagent cartridge that was placed in a centrifugal microfluidic disk. Different reagents for an enzymatic L-lactate assay with volumes between 230 nL and 10 μL were sequentially released by rotating the container, and thereby connecting the respective compartment with the microfluidic channel network. The recovery of more than 96% of the prestored reagents was reported.31
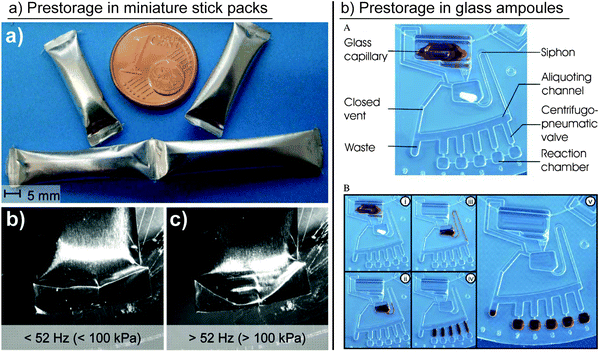 | ||
| Fig. 3 Different concepts for liquid reagent prestorage in containers. (a) Prestorage of liquids in miniature stick packs and release via pealable seal.27,28 (Reproduced with permission from The Royal Society of Chemistry.) (b) Prestorage of liquid in glass ampoules and release by crushing the ampoules.26 (Reproduced with permission from The Royal Society of Chemistry.) | ||
Liquid reagent prestorage directly within a cyclic olefin polymer (COP) cartridge has been demonstrated with fluid reservoirs connected to the microfluidic system via optofluidic valves. Prestorage without noticeable fluid loss was demonstrated for a period of one month.32,33 The prestorage of tetramethyl benzidine (TMB), washing buffer, and detection antibody solution directly in the cartridge was demonstrated by Kim et al. The single reservoirs were connected to the microfluidic network via ferrowax valves that were opened by laser irradiation.34 A similar concept was used to connect the prestored liquids to the microfluidic channel network via wax valves with different melting temperatures, thereby making it possible to sequentially release liquids into the network by melting the valves using infra-red heating.35
Recently, the LabTube was introduced as a new concept for centrifugal microfluidics based on stacked microfluidic elements.36 A centrifugally actuated ballpen mechanism enables the simultaneous axial and rotatory movement of the stacked elements “revolvers” relative to each other. A first revolver comprises cavities for the storage of reagents with pierceable aluminum foil. A second revolver is equipped with lancing structures. The serial release of reagents is controlled by the ballpen mechanism, which lances the reagent cavities either in parallel or one after the other.
The prestorage of dry reagents is mostly conducted by drying reagents to the surface or placing dry/lyophilized pellets or functional beads into microfluidic chambers during fabrication. Drying of reagents directly onto the cartridge surface has successfully been demonstrated for polymerase chain reaction (PCR) primers and probes37–40 and genomic DNA.41 In another work, dry enzyme pellets for the detection of nitrite and hexavalent chromium were prestored in microfluidic chambers on the cartridge. After a storage period of 31 days in a desiccator, the relative standard deviation of the concentration adjusted absorbance was 7.91%.42 The prestorage of lyophilized enzymes for DNA amplification was demonstrated by Lutz et al.26 and Strohmeier et al.43
2.2 Transport of liquids
A fundamental unit operation in centrifugal microfluidics is the transport of liquids within a fluidic network of channels and chambers. Typically, centrifugal forces, created by a defined rotation, have been exploited to transport fluids from a radially inward position (high level of potential energy) to a radially outward position (low level of potential energy). Because of the flow directed from the cartridge center radially outward, the number of cascadable unit operations and process chains is limited by the radius of the cartridge. In many cases, the available radius may not be large enough for the integration of all the process chains that are needed for a desired application. As a consequence, alternatives to the use of centrifugal forces to drive liquid transport in any direction—particularly radially inward—have been required and have recently been developed to enable the integration of larger and more complex fluidic networks.A straightforward approach for pumping liquids radially inward was demonstrated by Kong et al., and involved directing an external gas stream through orifices into a rotating microfluidic cartridge.12 At closely defined spinning frequencies and gas flow rates, the gas displaces a liquid within the cartridge radially inward. Similar approaches for displacement pumping have been presented, employing an additional liquid that is introduced into a microfluidic cartridge. When the displacer liquid is pumped radially outward, it forces the sample liquid to move to a position situated closer to the center of rotation.44,45
Other approaches have exploited on-chip gas generation or expansion to displace and pump liquids. For this purpose, external heat sources have been used to heat up a gas volume entrapped in a microfluidic chamber, causing it to expand thermally. Thereby, water was transferred radially inward at constant spin frequencies between 5 and 20 Hz (Fig. 4a).46 The same principle was applied in reverse. A decrease in temperature was used for the thermal contraction of an entrapped gas volume. The resulting underpressure “pulled” the liquid into a chamber located at a radially inward position.47 Instead of thermal expansion, the on-chip electrolysis of water has been used to generate a gas volume that displaces liquids radially inward (Fig. 4c).48 All of the methods described so far require additional external or disk-integrated means for operation (Table 2).
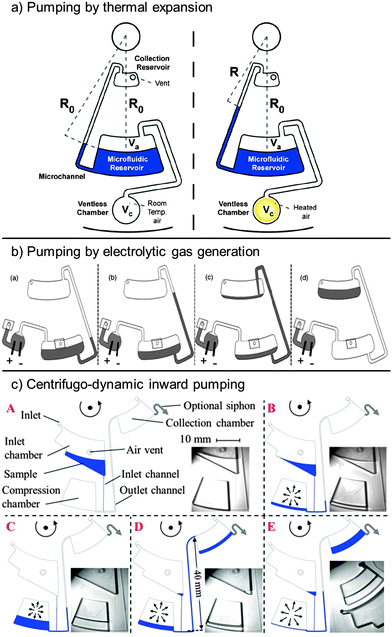 | ||
| Fig. 4 Liquid transport on centrifugal microfluidic platforms exploiting (a) gas-overpressure generated by heat46 (with kind permission from Springer Science and Business Media) and (b) electrolytic gas generation.48 (Reproduced with permission of the Electrochemical Society.) In (c), air compression at high centrifugation, followed by air expansion at a low spin frequency is used in combination with different hydraulic resistances of the inlet and outlet channels to pump liquids radially inward.49 (Reproduced with permission from The Royal Society of Chemistry.) | ||
| Ref. | Actuation principle | External means | Actuation pressures | Pumping ratea (μL s−1) | Pump efficiencya (%) |
|---|---|---|---|---|---|
| a Maximum values reported in the cited publication. | |||||
| Zehnle S. et al.49 | Centrifugo-dynamic | — | Δpc, Δppneu, Δpv | 18.2 | 91 |
| Kong M. C. R. et al.44 | Displacer liquid | — | Δpc, Δppneu | 0.6 | 60 |
| Noroozi Z. et al.48 | Electrolytic gas generation | Electrical connection | Δpc, Δppneu | 9.0 | 100 |
| Abi-Samra K. et al.46 | Thermal gas expansion | Radiation source | Δpc, Δppneu | 17.6 | 100 |
| Kong M. C. R. et al.12 | Pneumatic (external) | Pressurized gas | Δpc, Δppneu | 1.1 | 100 |
Recently, centrifugo-dynamic pumping has been presented, which does not require any external means but relies solely on the dynamics of deceleration from higher to lower spin frequencies.49 At high spin frequencies, a sample liquid is directed into a microfluidic dead-end chamber, where it entraps and compresses an air volume. The access channel to this dead end chamber branches into a narrow inlet channel, through which the liquid enters and into a wider outlet channel. The fast deceleration to a low spin frequency (6 Hz) leads to a fast expansion of the compressed air volume and, because of the lower flow resistance, most of the liquid is pumped from the dead-end chamber through the wider outlet channel to a radially more inward position (Fig. 4b).
Other methods for temporary liquid displacement to a radially inward position include capillary priming,50,51 pneumatic pumping,52 magneto-pneumatic pumping,53 and suction-enhanced siphon priming.54 These pumping techniques do not transfer liquids permanently to a position situated radially more inward. Instead, they can be used for enhanced fluid control. In combination with siphon valves for example, these pumping techniques are used to prime the siphon for subsequent transfer of liquid to a radially outward position.
2.3 Valving and switching
Valving is regarded as one of the most essential unit operations on the centrifugal microfluidic platform35 because it controls the flow of the fluid through the fluidic network. Typical requirements include rapid liquid passage at a distinctive point in the spatio-temporal domain, compatibility with a broad range of physicochemical liquid properties, and low dead-volumes.55 Valves can be grouped into active and passive valves, the latter referring to an actuation principle solely controlled by centrifugal forces.55 Obviously, passive actuation is advantageous to reduce the need for external means, which add to the complexity of the entire centrifugal microfluidic system.11 The initial state of a valve can be normally closed (NC) or normally open (NO). An overview of the implementations of valves in centrifugal microfluidics is given in Table 3. Embodiments of valves that feature more than one outlet and allow a liquid flow to be directed to a defined outlet are referred to as “switches”. The following sections discuss valves and switches, starting with passive ones.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) hydrophobic = counter pressure of hydrophobic capillary (eqn (7)), and Δppneu = pneumatic counter pressure (eqn (6))
hydrophobic = counter pressure of hydrophobic capillary (eqn (7)), and Δppneu = pneumatic counter pressure (eqn (6))
| Ref. | External means | Actuation principle | Mode | Vapor-tight |
|---|---|---|---|---|
| a Valving principle based on reduction of under pressure after defined opening of air vents. b Vapor-tightness has not been demonstrated, but valve is expected to be vapor-tight. | ||||
| Lai S. et al.59 | — | Δpc > Δpcap | NC | — |
| Duffy D. C. et al.58 | — | Δpc > Δpcap | NC | — |
| Gorkin R. et al.74 | — | Integrated film dissolves when brought into contact with liquid. Fluidic pathway is opened. | NC | ✓ |
| Mark D. et al.73 | — | Δpc > Δpcap + Δppneu | NC | — |
| Andersson P. et al.69 | — | Δpc > Δpcap hydrophobic | NC | — |
| Siegrist J. et al.81 | — | Δpcap > Δpc | NC | — |
| Gorkin R. et al.54 | — | Pressure drop at T-junction caused by auxiliary liquid pulls sample liquid over siphon crest. | NC | — |
| Hwang H. et al.79 | — | Integrated membrane valve opens above critical centrifugal pressure. Fluidic pathway is opened. | NC | ✓b |
| Gorkin R. et al.52 | — | Δppneu > Δpc | NC | — |
| LaCroix-Fralish A. et al.66 | — | Δpc > Δpcap | NC | — |
| Hoffmann J. et al.78 | — | Delamination of weakly bonded interface by exceeding critical centrifugal pressure. Fluidic pathway is opened. | NC | ✓ |
| Ukita Y. et al.57 | — | Time-dependent decrease of fill level opens connection to venting.a | NC | — |
| Zhang H. et al.65 | — | Δpc > Δpcap![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) hydrophobic hydrophobic |
NC | — |
| Kinahan D. J. et al.56 | — | Integrated film dissolves when brought into contact with liquid on paper strip. Air vent is opened.a | NC | ✓ |
| Kinahan D. J. et al.86 | — | First liquid dissolves a film to trigger valving of the a next liquid | NC | ✓b |
| Siegrist J. et al.76 | — | Δpc > Δppneu | NC | — |
| Abi-Samra K. et al.35 | Active: stationary halogen lamp | Integrated wax valves melted by infrared heating. Fluidic pathway is opened. | NC | ✓ |
| Park J. M. et al.87 | Active: mobile laser diode | Integrated ferrowax valves are melted by laser. Fluidic pathway is opened or closed. | NO/NC/reversible | ✓ |
| Amasia M. et al.90 | Active: thermo-electric module | Freezing of a liquid plug blocks fluidic pathway. | NO | ✓ |
| Garcia-Cordero J. L. et al.33 | Active: laser | Laser melts orifices in polymer separation layer. Fluidic pathway is opened. | NC | ✓ |
| Al-Faqheri W. et al.55 | Active: hot air gun | Integrated wax valves are melted by heat gun. Connection to venting is opened.a | NO/NC | ✓ |
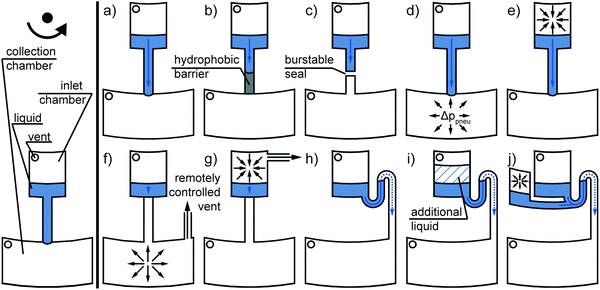 | ||
| Fig. 5 Passive valves solely actuated by centrifugal forces (eqn (1)): (a) capillary, (b) hydrophobic, (c) burstable seal, (d) centrifugo-pneumatic overpressure, (e) centrifugo-pneumatic under pressure, (f) remotely vented collection chamber (e.g., by wetting a dissolvable film56), (g) remotely vented inlet chamber (e.g., by a clepsydra structure57), (h) capillary siphon, (i) overflow siphon, and (j) pneumatic siphon valve. | ||
Early implementations of passive valves used the effect of liquid meniscus pinning at abrupt and sharp channel widenings. To pass the valve, the centrifugal pressure (eqn (4)) has to exceed the capillary counter pressure (eqn (7)). As the pinning effect of the fluid flow is solely based on the capillary counter pressure, these valves are referred to as “capillary valves” (Fig. 5a). Capillary valves have been demonstrated in complex fluidic networks, e.g., by Duffy et al.58 and Lai et al.59 Later, the flow sequencing of five different liquids using capillary valves with different burst pressures (as a result of defined channel cross sections at different radial positions) and parallel valving of up to 120 single 40 nL aliquots were successfully demonstrated by Madou et al.60 and Schwemmer et al.,61 respectively. Multiple studies have investigated the dependency of the burst pressure on the micro-channel dimensions, surface tension, and contact angle of the liquid using analytical modeling.62–65 In that context, deviations in the dimensions and low surface quality have been identified as critical parameters for burst pressure prediction and reproducibility.58,62,64 To circumvent stringent manufacturing requirements, the implementation of fused silica capillaries instead of monolithically integrated capillary valves was reported.66 Different burst pressures were realized by integrating fused silica capillaries with different inner diameters ranging from 12 to 100 μm. The concept of integrated fused silica capillaries was later adopted by Kong et al.44 and Kazarine et al.67
Geometric capillary valves become increasingly unstable for wetting liquids when the contact angles drop below 45°.68 To increase the reproducibility for liquids with low contact angles, local hydrophobic surface coatings have been applied. The valving principle is then based on stopping a liquid flow at the hydrophobic coating, and corresponding valves are referred to as “hydrophobic valves” (Fig. 5b). The flow continues when the centrifugal pressure (eqn (4)) overcomes the capillary pressure (eqn (7)). The demonstrated surface coatings include mostly fluorinated polymer solutions, which are applied by spraying69 or dispensing.70 An example of the highly parallel integration of 208 hydrophobic valves was given by Honda et al.71 Another approach demonstrated rapid surface modification for hydrophobic valves by means of a laser printer. Printed toner spots in a microchannel led to an increase in the contact angles from 51° to 111° (measured for DI-water). Depending on the density of the toner spots, a broad range of burst pressures, ranging from 158 ± 18 Pa to 573 ± 16 Pa, was realized.72
Another approach to circumvent the need for local surface coatings and high-precision manufacturing, termed “centrifugo-pneumatic valve” (Fig. 5d), was demonstrated by Mark et al. Here, the liquid flow is stopped by a combination of the capillary counter pressure (eqn (7)) at the interface of a channel to a dead-end chamber and the pneumatic counter pressure (eqn (6)) of the compressed air inside the dead-end chamber. Valving is triggered by the centrifugal pressure (eqn (4)) overcoming the counter pressures. After the breakthrough, the complete release of the liquid is ensured by the Rayleigh–Taylor instability of the liquid/air interface. Centrifugo-pneumatic valving makes it possible to handle highly wetting/low surface tension liquids with reported burst pressures of 1300 ± 400 Pa for ethanol and 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 ± 2800 Pa for water.73 The centrifugo-pneumatic valve was later combined by Gorkin et al. with an integrated water-dissolvable membrane. The membrane was applied to close an outlet of the dead-end chamber, which allowed centrifugo-pneumatic valving. After contact with the liquid, the membrane dissolved in as little as 10 seconds, which allowed for downstream fluidic post processing.74 Subsequently, microfluidic networks have been presented with multiple integrated dissolvable films to allow the auto-cascading of valving sequences.75 An inversion of the centrifugo-pneumatic valve, representing a centrifugo-pneumatic under pressure valve (Fig. 5e), was reported by Siegrist et al. The liquid is initially allocated in an unvented inlet chamber, and a retaining pneumatic under pressure (eqn (6)) in the enclosed air volume is generated when the liquid is forced radially outward through the centrifugo-pneumatic under pressure valve during rotation.76 Al-Faqheri et al. demonstrated that burst pressures in both centrifugo-pneumatic over- and under pressure valves can be controlled by blocking air vents with an auxiliary liquid.77
000 ± 2800 Pa for water.73 The centrifugo-pneumatic valve was later combined by Gorkin et al. with an integrated water-dissolvable membrane. The membrane was applied to close an outlet of the dead-end chamber, which allowed centrifugo-pneumatic valving. After contact with the liquid, the membrane dissolved in as little as 10 seconds, which allowed for downstream fluidic post processing.74 Subsequently, microfluidic networks have been presented with multiple integrated dissolvable films to allow the auto-cascading of valving sequences.75 An inversion of the centrifugo-pneumatic valve, representing a centrifugo-pneumatic under pressure valve (Fig. 5e), was reported by Siegrist et al. The liquid is initially allocated in an unvented inlet chamber, and a retaining pneumatic under pressure (eqn (6)) in the enclosed air volume is generated when the liquid is forced radially outward through the centrifugo-pneumatic under pressure valve during rotation.76 Al-Faqheri et al. demonstrated that burst pressures in both centrifugo-pneumatic over- and under pressure valves can be controlled by blocking air vents with an auxiliary liquid.77
To handle evaporating reagents, vapor-tight valves are required. Hoffmann et al. presented a valve that applied centrifugal pressure (eqn (4)) for the well-defined delamination of the sealing foil of a centrifugal microfluidic cartridge, thereby opening up the fluidic pathway. This valve is called a “burstable seal valve” (Fig. 5c). For centrifugal pressures of 2 bar, release times of 31 s were reported.78 In another approach, polydimethylsiloxane (PDMS) membranes were integrated into a microfluidic network to close the fluidic pathway by bonding the PDMS membrane to the thermoplastic cartridge. With increasing centrifugal pressure (eqn (4)), the membrane is deflected and opens up the fluidic pathway. Depending on the membrane thickness and spin speed, various flow rates were achieved.79
In contrast to passive valves that open with an increase in centrifugal pressure, “capillary siphon valves” (Fig. 5h) require a temporary state of low centrifugal pressure (eqn (4)) to trigger the burst event.80 This valving principle is based on the capillary priming of an S-shaped siphon channel and thus requires advancing contact angles <90°. The siphon channel connects an inlet reservoir and outlet reservoir and has to fulfill the following requirements: (a) the inlet of the siphon is located radially inward of the outlet and (b) the crest of the siphon is situated radially inward of the filling level of the inlet reservoir.3 The siphon channel is thus primed by capillary forces (eqn (7)) against the direction of the centrifugal forces at a low spin speed, while at higher spin speeds, the centrifugal forces dominate and prevent capillary priming.80 After priming the siphon, the inlet reservoir is emptied through the outlet at a sufficiently high centrifugal pressure. Siegrist et al. demonstrated flow sequencing based on serial siphon valving, i.e. the concatenation of multiple capillary siphons with integrated capillary valves. The integrated capillary valves prevent the premature priming of the capillary siphon and allow for the release of liquid after a defined number of rotate-and-halt cycles. However, this results in a minor dead-volume of liquid that does not reach the outlet. In this approach, plasma treatment has been recommended to render the surface hydrophilic for liquids with contact angles >90°.81 Because many of the materials used for centrifugal microfluidic cartridges exhibit hydrophobic properties and surface treatment adds to the complexity of cartridge fabrication, Godino et al. demonstrated the integration of paper-based siphons as a low-cost alternative.82 Alternatively, siphon valves can be primed by increasing the filling height inside the inlet chamber above the siphon crest by adding additional liquid. Such valves are referred to as “overflow siphon valves” (Fig. 5i).83
To circumvent the demand for hydrophilic coatings, siphon priming by the release of pneumatic energy (eqn (6)) from an enclosed and compressed air bubble was exploited in the so-called “pneumatic siphon valve” (Fig. 5j).52 Later, the cascading of pneumatically actuated siphons for sequential release was employed.84 Another approach demonstrated suction-enhanced siphon priming by creating an under pressure at the siphon outlet through an auxiliary liquid. However, in this approach, the siphoned reagent inevitably mixes with the auxiliary liquid.54
A small group of passive valves does not rely on centrifugal pressure but provides a time-dependent release of liquids. Recently, Schwemmer et al. introduced a microfluidic timer that could be used to trigger liquid actuation independent from the spinning speed: the timer employs temporary storage of pneumatic energy (eqn (6)), which is suddenly released after a pre-defined period of time. The timer is set by overfilling a first pneumatic chamber, which results in liquid flowing into a secondary pneumatic chamber through a narrow channel at high rotational frequencies. Upon decrease of centrifugal pressure (eqn (4)), emptying of the secondary chamber and channel results in a delay before the pneumatic energy is released.85 Kinahan et al. demonstrated the integration of a paper strip into a centrifugal microfluidic cartridge. This paper strip is “connected” to multiple integrated dissolvable films that sequentially open fluidic pathways as soon as the part of the paper strip in contact with the dissolvable film is wetted56 (Fig. 5f). Kinahan et al. also demonstrated event-triggered valving, where the completed valving of one liquid opens an air vent by dissolving a film to trigger the valving of a next liquid. By combination of a fluidic network with dissolvable films 10 sequential valving events at one rotational frequency were demonstrated in a single cartridge.86 Ukita et al. reported a microfluidic clepsydra structure connected to the venting of a loading structure for the sequential release of liquids. Over time, the liquid level in the clepsydra decreases and thereby sequentially opens the venting for the single loading structures57 (Fig. 5g).
Optofluidic valves actuated by a solid state laser were reported by Garcia-Cordero et al. Printed toner spots on a polymer separation layer, COP or polyethylene terephthalate (PET) film, were used to increase the light absorbance to melt orifices (30–280 μm in diameter) into the separation layer, thereby opening the fluidic pathway. When using 100 and 300 mW of laser power, the response time of the valve was reported to be 0.5 seconds. A fluidic network with 106 laser printed single addressable optofluidic valves has been presented. Contact between the liquid and valve had to be avoided during melting because the liquid could be contaminated by combustion products and absorb thermal energy.33
Instead of melting the cartridge substrate, paraffin wax valves have been integrated into centrifugal microfluidic cartridges. Stationary infrared sources were used to melt the wax under rotation, thereby opening the fluidic pathway. The sequential opening of valves has been demonstrated by using waxes with different melting temperatures. Response times of 25 seconds were reported for the simultaneous actuation of nine valves.35 Another approach used handheld heat guns instead of infrared lamps to melt wax valves.88 However, it has to be considered that the molten wax and heat input to the cartridge could have a negative effect on the reagents used.35 As an improvement to overcome these limitations, Al-Faqheri et al. relocated the wax valves away from the reagents, thereby preventing direct contact. Instead of opening the fluidic pathway, connections to the air vents were opened or closed by melting the valves55 (Fig. 6b).
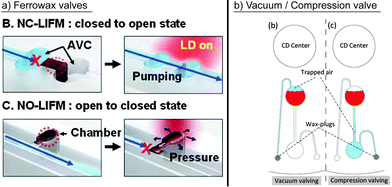 | ||
| Fig. 6 Prominent concepts for active valving. (a) Park et al. demonstrated laser irradiated ferrowax microvalves (LIFM) to open and close the fluidic pathways of normally closed LIFM (NC-LIFM) and normally opened LIFM (NO-LIFM), respectively, activated by a laser diode (LD).87 The layout includes assistant valve chambers (AVC). (Reproduced with permission from The Royal Society of Chemistry.) (b) Al-Faqheri et al. used wax plugs to open connections to the ventilation.55 (Reproduced under the Creative Commons Attribution License O.) | ||
Aiming at minimizing the energy input, single addressable, laser-irradiated ferrowax microvalves (LIFM) were introduced by Park et al.87 and later implemented for different applications.89 For efficient heating, iron oxide nanoparticles were mixed into the wax, which allowed valve actuation via low-power lasers (1.5 W) and a response time of only 0.5 seconds. The laser ensured that only the nanoparticles were heated and not the surrounding liquids. The LIFM were reported to be leak-free at a centrifugal pressure of up to 403.0 ± 7.6 kPa. Normally closed, normally open, and even reversible valve actuation has been demonstrated (Fig. 6a).
Amasia et al. demonstrated ice valving to avoid evaporation during PCR thermocycling. Liquid plugs were frozen in defined channel areas when the disk was at rest using thermoelectric modules. The response time for these ice valves was 30 seconds.90
An alternative to using thermal energy for active valving has been demonstrated by Swayne et al. A focused air stream opens a fluidic path for the liquid, which had previously been blocked by a gel. Postulated advantages of the valve are the small footprint and ease of fabrication.91
Other approaches for passive flow switching have been demonstrated, including that based on fluidic capacitance by Kim et al.93 and that based on the pneumatic counter pressure (eqn (6)) of an enclosed air volume by Mark et al. The latter exploits centrifugal pressures (eqn (4)), depending on the speed of rotation to direct liquids to either one of the outlets.95 Later, Müller et al. demonstrated passive unidirectional switching by closing the connection to the venting with the overflow volume of one of the assay reagents.96
2.4 Metering and aliquoting
Most microfluidically integrated applications require precise input volumes of liquids in order to obtain quantitatively reproducible results. Consequently, unit operations for the metering of liquid volumes are widely employed. Splitting an input liquid volume into multiple defined sub-volumes is referred to as aliquoting, which mostly involves multiple parallel metering steps. Aliquoting itself was subcategorized by Mark et al. into one-stage and two-stage aliquoting (Fig. 7b). The latter refers to a microfluidic aliquoting process in which single aliquots are transferred into fluidically separated chambers after metering.98 The embodiments of centrifugal microfluidic unit operations for metering and aliquoting are listed in Table 4. In the simplest case, a metering structure consists of a connection channel to an inlet, a metering chamber with a defined volume, and an overflow to a waste chamber for excess volume (Fig. 7a). The metering can be combined with valves at the radially outer end of the metering chamber to allow for further fluidic processing. The demonstrated valves include hydrophobic,69 capillary siphon,99 and centrifugo-pneumatic valves.73 The metering accuracy is mainly affected by the variation of the cavity size within the fabrication tolerances98 and the wicking effects at liquid interfaces due to capillary forces.100 Capillary forces (eqn (7)) can be counteracted by centrifugal forces (eqn (4)), which produces a high metering accuracy in centrifugal microfluidics even at nanoliter volumes.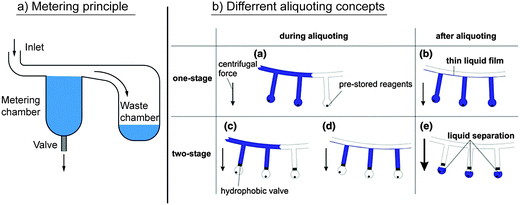 | ||
| Fig. 7 Centrifugal microfluidic unit operations for metering and aliquoting. (a) Basic principle of metering. A liquid fills a metering chamber with a defined volume. The excess is gated into a waste chamber. The metered volume can subsequently be transferred into the microfluidic network via suitable valves. (b) Different aliquoting concepts.98 (With kind permission from Springer Science and Business Media.) | ||
| Ref. | Integrated valve type | Aliquoted volume | CV (%) | Number of parallel aliquots |
|---|---|---|---|---|
| Schembri C. T. et al.80 | No valve | Not reported | <2 | 4 or 21 |
| Sundberg S. O. et al.101 | No valve | 33 nL | 16 | 1000 |
| Andersson P. et al.69 | Hydrophobic valve | 200 nL | 0.75 | 112 |
| Andersson P. et al.69 | Hydrophobic valve | 20 nL | 1.90 | 1 |
| Mark D. et al.98 | Centrifugo pneumatic valve | 6–10 μL | 2.2–3.6 | 8 or 16 |
| Steigert J. et al.99 | Capillary siphon | 500 nL | <5 | 1 |
| Schwemmer F. et al.61 | Capillary valve | 40 nL | 1–5.5 | 120 |
| Li G. et al.102 | Capillary valve | 31 nL | 2.80 | 24 |
| Hwang H. et al.30 | Ferrowax-based microvalves | 100 μL | Not reported | 5 |
In single-stage aliquoting, fluid volumes are metered directly into the receiving chamber. Thus, the aliquoting process simply involves the transport of the liquid from an inlet into multiple receiving chambers, while the excess is gated into an overflow. As mentioned by Mark et al., single stage aliquoting bears the problem of cross contamination between adjacent aliquots, because they might still be connected by a liquid film.98 To avoid cross contamination, Sundberg et al. used a mineral oil to fill the microfluidic channel and separate the aliquoted volumes after the aliquoting process.101
Two-stage aliquoting allows for full fluidic separation between adjacent aliquots, and therefore is usually applied when cross contamination is an issue,37 or when further fluidic processing of the individual aliquots is required. Two-stage aliquoting combines the parallel metering of one-step aliquoting with normally closed valves at the radial outer side of each metering finger. After metering, the single aliquots can pass the valve and be used for further fluidic processing.30,69
2.5 Mixing
The purpose of mixing in microfluidics is to reach a sufficiently high distribution and homogeneity of sample and reagent molecules such that chemical reactions are accelerated. Conventional mixing in macroscopic standard laboratory processes is mostly performed by stirring, shaking, or vortexing. However, on a centrifugal microfluidic platform, mixing becomes difficult because the cartridge is rigidly attached to a motor shaft, which rotates the cartridge with a relatively high moment of inertia. The artificial gravity generated by this rotation makes the centrifugal microfluidic platform particularly useful for the separation of phases with different mass densities, but not for mixing. Moreover, for liquid volumes ranging from several hundred nanoliters to a few milliliters, purely diffusive mixing is rather inefficient.103,104 Since mixing is nevertheless crucial for many biochemical assays, several methods have been researched to mix fluids on the centrifugal microfluidic platform.A concept for the batch-wise “shake-mode” mixing of liquids that relied on continuous changes in the spin speed of the centrifugal microfluidic cartridge was demonstrated by Grumann et al. The angular momentum caused by the acceleration or deceleration induced Euler forces (eqn (3)) and resulted in layer inversion of the liquids in the microfluidic chamber (Fig. 8a). As a measure of the mixing quality, the standard deviation of all the recorded pixel grayscale values of a mixture containing dyed and undyed liquids was determined using image processing. The mixing time was defined as the time required to reach a 1/e decay in the standard deviation. As a result, the mixing time in the reported embodiment could be reduced from 7 minutes for purely diffusive mixing down to 3 seconds for shake-mode mixing. It was found that the mixing quality depended on the acceleration and deceleration rates, as well as the azimuthal span of the rotation and radial position of the mixing chamber. Adding magnetic beads and pulling them through the mixing chamber further reduced the mixing time to 0.5 seconds. A deflection of the magnetic beads was induced by a set of external permanent magnets that attracted the beads radially in- and outward.103
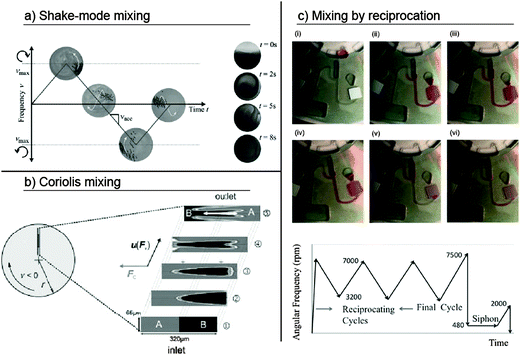 | ||
| Fig. 8 Different concepts for mixing of liquids employed in centrifugal microfluidics. (a) Shake-mode mixing at alternating spin frequencies.103 (Reproduced with permission from The Royal Society of Chemistry.) (b) Coriolis mixing exploiting Coriolis force induced transversal flow.108 (Preprinted with permission of John Wiley and Sons.) (c) Mixing by reciprocating the flow at alternating spin frequencies.106 (Reprinted with permission from AIP Publishing LLC.) | ||
Noroozi et al. presented another mixing concept that employs the interplay of centrifugal and pneumatic pressures (eqn (4) and (6)) to transport liquids between two chambers (Fig. 8c).105 This mixing-by-reciprocation concept was later used to maximize the incubation and hybridization efficiency for the centrifugal microfluidic integration of an immunoassay and showed a reduction in the processing time and reagent consumption by one order of magnitude.106 In this approach, mixing occurs due to micro-vortices and Taylor dispersions, which are both present in each mixing cycle. The use of the pneumatic counter-pressures of an entrapped air volume enables frequency oscillations at elevated spin speeds, thus making mixing by reciprocation easily combinable with pneumatic siphon valving.
Instead of pneumatic energy storage, Aeinehvand et al. recently integrated a latex membrane in a stack of PMMA layers and pressure sensitive adhesives. At the radial distal end of the mixing chamber, the latex membrane could freely expand out of the disk plane through a hole in the solid PMMA, thus forming a micro-balloon. The reciprocating flow of the reagents to be mixed was induced by oscillations of the spin frequency. At a high spin speed, the centrifugal pressure drove the reagents into the inflating micro-balloon, thereby stretching the latex membrane. At rest, the absence of the centrifugal pressure allowed the latex membrane to return to its initial flat shape. This version of mixing by reciprocation was proven to be suitable for low operating frequencies in the range of 0–14 Hz and chamber depths in the range of a few hundred micrometers. For such shallow chambers, mixing by reciprocating the flow was shown to be a good alternative to shake-mode mixing.107 This is because shake-mode mixing requires moderate aspect ratios in the range of one to provide sufficient advection.
Mixing based on Coriolis pseudo-forces (eqn (2)) was demonstrated by Haeberle et al. Here, two liquids were dispensed into two separate microfluidic inlets on the centrifugal microfluidic cartridge (Fig. 8b). These liquids merged within a Y-shaped channel, where they were mixed due to transversal convection as a result of the Coriolis forces acting perpendicular to the flow direction. After mixing, the product was spun from the cartridge into a receiving vessel, thereby allowing for continuous mixing.108 Coriolis mixing was later improved by the multilamination of flows via a split-and-recombine concept.109 In another work, Coriolis mixing was used to fold laminar flows and thereby shorten mixing times by two orders of magnitude.110 Further investigations on the mixing regimes of two fluids in a T-shaped microchannel showed Coriolis force-based mixing at intermediate spin speeds.111 The channel geometry, speed of rotation, and flow rates were identified as key impact parameters on the mixing quality.109,112 Recently, Coriolis mixers have been employed in serpentine configurations that also use the Dean effect in channel bends to improve the overall mixing efficiency.113,114 The independence from changes in the spin speed makes Coriolis mixing suitable for applications on a wide range of processing devices, e.g., standard laboratory centrifuges. A challenge for the integration of Coriolis mixing is that the flow rates of the fluids entering the mixing channels have to be accurately controlled.
Other approaches for mixing at a constant spin speed have recently been explored. Burger et al. used the disruption of continuous liquid flows to generate discrete droplets and create multiple alternating lamellae with two different liquids. In this way, the interface between the two liquid phases was significantly increased, and mixing by diffusion was supported. By generating droplets with 60 nL volumes, blood plasma and PBS were mixed and divided into single aliquots. The protein concentrations in all of the aliquots showed good agreement with the value expected for a perfect mixture.83
Liebeskind et al. used the catalytic decomposition of H2O2 to water and oxygen as an on-chip gas source to generate gas bubbles for mixing. The generated gas was pumped into a mixing chamber, where, due to the buoyancy force in the artificial gravitational field, the bubbles moved through the liquids to be mixed and caused perturbations. The mixer was used to perform the lysis and binding steps in the extraction of DNA from whole blood.115
Active mixing employing an external air stream was used by Kong et al. to stir liquids within a microfluidic chamber. The air stream was directed from outside the disk through an orifice into the microfluidic structures, which allowed mixing at constant and low spin frequencies. Within 11.2 seconds, a 30-fold increase in mixing quality was reported compared to diffusive mixing at a spin frequency of 7.5 Hz.116
2.6 Separation
The separation of different substances from each other is an essential unit operation in many (bio-) chemical processes. The target substances can be small molecules such as metabolites, macromolecules like nucleic acids and proteins, and larger elements such as cells or solid particles that have to be isolated from a surrounding medium. Typically, differences in the chemical or physical properties of these substances are exploited for the technical implementation. This review chapter is structured as follows. First, we review publications on physical separation techniques, including filtering and sedimentation, followed by a discussion of the implementations of chemical separation within centrifugal microfluidics.Filtering by cartridge-integrated geometric restrictions was demonstrated by Czugala et al. In this implementation, the height of a microfluidic channel was decreased step-wise from 1500 μm to 86 μm. Via these restrictions, up to 94% of the particles were filtered from a river-water sample.117 Instead of geometric restrictions, filter membranes have successfully been integrated into centrifugal microfluidic cartridges to remove bacteria from water samples18 or particulates from soil.118 Both publications report filtration efficiencies of 100% of the tested particulates. Also based on filter membranes, selective filtering of circulating tumor cells from a whole blood sample was demonstrated. Filtration efficiencies were reported to be up to 84%.119
Specific filtering by di-electrophoresis exploiting the electrical polarizability of molecules has been demonstrated by Martinez-Duarte and co-workers. Cartridge integrated carbon electrodes powered via electrical contacts with a slip-ring on the rotor shaft specifically filtered yeast cells from a mixture of yeast cells and latex particles.120 Boettcher and colleagues presented the manipulation of particles and cells using a rotating microfluidic di-electrophoresis chip. Two co-rotating batteries powered the chip, while a co-rotating generator provided the required alternating currents. Using the described di-electrophoretic setup, sedimenting cells and particles could be directed to a defined branch of a Y-shaped channel.121
Burger et al. presented an implementation for capturing beads during sedimentation using arrays of V-shaped structures. The implementation aimed at a sharp peak in bead-distribution, i.e., capturing exactly one bead per cup. The size and density of the V-cup structures, as well as the size of the beads, were identified as important parameters for the bead distribution and number of empty cups. Up to 99.7% single bead-occupancy per V-cup was reported with 5% of the cups remaining empty.122
Kirby et al. presented a concept for centrifugo-magnetophoretic particle separation. Magnetic particles sediment in a stagnant fluid due to centrifugal forces. Permanent-magnets integrated into the rotating cartridge cause a defined deflection of the magnetic particles perpendicular to the centrifugal forces while non-magnetic particles sediment in direction of the centrifugal force. Thereby, particles could be routed to one of three outlets depending on their size, density, and magnetic properties and on the spin speed.123 This concept was later employed by Glynn et al. for separating beads with captured CD4+ cells from whole blood.124
A unit operation for the sedimentation of solid particles from turbid samples and the subsequent transfer of clear supernatant was demonstrated by LaCroix-Fralish et al. Fused silica capillaries (<110 μm in diameter) were used as the connection between two microfluidic chambers. The liquid above the sedimented fraction of solid particles was decanted by placing one end of the capillary in the upstream chamber.66 In another implementation, saw-toothed obstacles in an inlet chamber were used to hold back sedimented particles from seawater samples. After sedimentation, a wax valve was opened to release the clear seawater into an aliquoting structure.30
Similar concepts have been employed for blood-plasma separation based on the sedimentation of the denser cellular blood content from the cell-free blood plasma. The implementations basically differ in the implemented unit operations for plasma transfer after sedimentation, which included centrifuge-pneumatic gating,125 centrifuge-pneumatic siphon valving,126 capillary siphon valving,99 decanting,127 or using an integrated Y-channel that allowed denser cell content to enter the radially outward branch of the Y-channel, while the plasma was transferred into the downstream microfluidics via the radial inward channel.128 Because blood-plasma separation is a discrete process chain in many laboratory workflows, it is discussed in detail with respect to the reported performance parameters in Section 3.3.1.
A common affinity mechanism for the separation of nucleic acids exploits the binding of DNA and RNA to silica surfaces under high chaotropic salt conditions.129 Implementations have been demonstrated using non-mobile cartridge-integrated silica membranes,24,96 glass bead columns,130 or silica sol–gel.131 Other separation principles involve the hybridization of nucleic acids to complementary strands that are immobilized to the cartridge surface.132–134 The affinity mechanism exploited for immunoassays and immunoseparation relies on the binding of antibodies to antigens. Antibodies (and in rare cases antigens) have been immobilized to a variety of non-mobile solid supports, including trapped antibody-coated polystyrene beads,71,135 glass beads,136 silica beads,137 PMMA disks,59 and nitrocellulose membranes,106 which are then passed by the sample and other liquid reagents.
Demonstrated implementations with mobile support include a simple approach for the separation of nucleic acids using magnetic silica beads as the mobile support. Depending on the azimuthal position of the centrifugal microfluidic cartridge with respect to an external magnet, the beads could be transported through multiple reagent-filled microfluidic chambers.138 Cho et al. used antibody-coated magnetic beads for pathogen capturing and immuno-magnetic separation from a whole blood sample. The beads were manipulated by a cartridge-integrated magnet and an external magnet on a linear gear. Thereby, the mixing of the beads or temporary immobilization of the beads in a dedicated location could be achieved while the surrounding media were exchanged.89 Another approach for immunomagnetic separation was demonstrated by Chen and co-workers, where antibody-labeled magnetic beads were used to capture target cells. After binding, the beads were trapped by a co-rotating magnet, while the cell sample was gated into a waste reservoir.139 Glynn et al. and Kirby et al. demonstrated centrifugo-magnetophoretic separation to separate magnetic from non-magnetic particles or cells. In this approach, co-rotating disk-integrated magnets were used to deflect sedimenting magnetic particles with attached target cells to designated reservoirs.123,124
Schaff and Sommer demonstrated the sedimentation of beads through a density media for an immunoassay. Antibody-labeled beads were used to capture antigen and detection antibodies from a sample layered on top of a density medium. After capture, the beads were separated from the sample by sedimentation through the density medium.88
2.7 Droplet handling
While droplet-based microfluidics is a very active field in pressure-driven microfluidics, so far little work on droplet handling has been performed in centrifugal microfluidics. The reported unit operations are limited to the generation of droplets140 or bubbles.141 In these publications, both the droplets and bubbles were generated in oil.With respect to applications, droplet generation in centrifugal microfluidics has been employed to create particles. Chitosan/alginate droplets142,143 were generated at a nozzle in air and dispensed into a cross-linking solution. Upon contact with the hardening solution, the droplets became solid, forming microparticles. The reported advantages compared to other microfluidic bead generation methods are low dead volumes, uniform droplets due to the pulse free propulsion, and possible parallelization by a straightforward and even distribution of hydrostatic pressure on an array of nozzles. In particular, the dispensing method using an air gap, which prevents contact between the nozzle and hardening solution and thus circumvents nozzle clogging, is reported to be a unique feature.
Dispensing through an air gap was later applied to form 3D multi-compartmental particles using a multi-barreled capillary as a nozzle.144 Up to six-compartment body compositions with custom designed geometries were reported in this work. These were produced on a tabletop centrifuge equipped with a swinging bucket rotor.
Within centrifugal microfluidics, besides particle generation, we see the potential for the automation of highly parallel applications such as emulsion-based nucleic acid amplification as sample preparation for sequencing or digital amplification, or the implementation of digital immunoassays. The advantages include artificial gravity-based pulse-free propulsion, and thus the ability to form well-defined highly parallel micro-droplets with minimal dead volume. For example, centrifugal step emulsification can be employed for absolute quantification of nucleic acids by digital droplet RPA.145 Furthermore, the integration of droplet-based operations, together with complex sample preparation such as nucleic acid purification, may enable sample-to-answer implementations of digital assays.
2.8 Detection
Although not a classical fluidic functionality, we consider detection to be a unit operation because it represents a basic building block for the assessment or quantification of the result of an assay. With respect to fluidics, detection usually requires maintaining the analyzed volume at a certain position or defined flow rate. The more relevant aspect of detection, however, is the general principle with which the quantification is assessed. Therefore, we categorize the unit operations used for detection into optical, electrochemical, and other detection principles.Kim et al. presented a centrifugal microfluidic cartridge with an integrated lateral flow strip. Gold nanoparticle-stained antibodies were bound to a DNA amplification product and created a visible line on the lateral flow strip.146 Another molecular biological application exploited a color change from purple to blue during isothermal DNA amplification.147 Riegger et al. presented a system for the visual detection of hematocrit. A disk-imprinted scale next to a dead-end channel allowed for the visual read-out of hematocrit after centrifugation by identifying the location of the interface between the sedimented red blood cells and the plasma.148
Grumann et al. employed the total internal reflection for absorbance measurements. A light beam directed onto the disk plane was deflected by a cartridge-integrated V-groove and gated through a microfluidic chamber in the azimuthal direction. A second V-groove deflected the light beam out of the disk plane to the detector. Thereby, the path length for the absorption measurement (and thus the sensitivity) was increased from 1 mm to 10 mm compared to direct light incidence (Fig. 9a).149 Czugala et al. used a paired emitter detector diode (PEDD) device for absorption detection. In the PEDD setup, two light emitting diodes were used. One diode served as the light source and was placed above the cartridge, while the second diode, operated in the reverse bias mode, served as the light detector for the transmitted light. An improved sensitivity and signal-to-noise ratio along with a low cost, small size, and low power consumption, were reported as the major advantages of the PEDD setup compared with the standard setup using an LED and a photodiode (Fig. 9b).117 LaCroix-Fralish et al. presented the spectrophotometric detection of a bioassay using a halogen light source, which emitted light in the ultraviolet and visible regime, and a Czerny–Turner type spectrometer with a photodiode array for the detection of the transmitted light. For the detection, the disk had to be removed from the spinning device and mounted in the path of the spectrometer.42
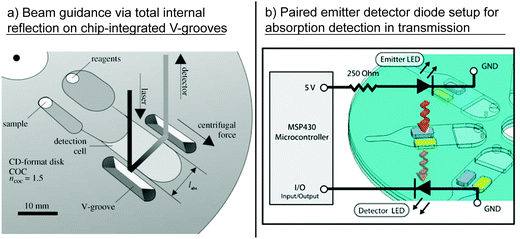 | ||
| Fig. 9 Different setups for optical detection. (a) Enhancement of sensitivity by on-chip beam guidance using chip-integrated V-grooves.149 (With kind permission from Springer Science and Business Media.) (b) Paired emitter detector diode (PEDD) setup as sensitive and cheap alternative to common LED–photodiode setups for absorption measurement in transmission.117 (Reproduced with permission from The Royal Society of Chemistry.) | ||
Detection via fluorescence measurement is frequently conducted for nucleic acid analysis and in some cases also for immunoassays, and typically provides a more sensitive and specific detection150 compared to other optical detection methods. Focke et al. presented a microfluidic cartridge with a line-up of reaction cavities close to the rim of the cartridge. Fluorescence signals from these reaction cavities were detected using a commercially available PCR thermocycler by exploiting the inbuilt fluorescence detection unit, i.e., an LED excitation source and a photo-multiplier for detection.38 The same concept was later adapted for other applications.26,37,39,41 Nwankire et al. presented a microfluidic cartridge with an integrated supercritical angle fluorescence chip that allowed the selective measurement of fluorescent signals generated in close proximity to the surface. The optical setup was completed by a laser for fluorescence excitation and a photomultiplier for detection.150 Various papers have reported the implementation of CCD cameras, especially for spatially resolved optical information. Riegger et al. demonstrated a detection concept for multiplexing via color-coding composed of an LED for excitation and a CCD camera for detection. In a first step, the camera acquired the spectral information of a layer of quantum dot beads for decoding the various bead types used and subsequently detected the fluorescence signals on the bead surfaces to quantify the bead-specific analyte reactions. The fluorescence on the bead surfaces was associated with the assay result, while the color of the beads corresponded to the assay target.151 Ukita et al. presented a stroboscopic fluorescence microscope for observation of fluorescent objects such as 6 μm particles on a spinning disk at a rotational frequency of up to 3000 rpm.152 The detection of multiple ions using a cartridge-integrated optode array was demonstrated by Watts et al. The detection principle was based on a change in the fluorescence signal due to the exchange of cations from the sample with the hydrogen in the optode membrane.153
Otsuka and colleagues developed a cartridge-integrated surface plasmon resonance sensor for the detection of protein adsorption to a gold surface. The adsorption of proteins influenced the resonance frequency of the surface plasmons, which resulted in a shift in the light intensity distribution with respect to the wavelength. The light intensity was measured using a CCD camera.154
Recently, multiple papers have been published on the use of standard optical CD and DVD pick-up heads for detection. One of the driving forces for their implementation is the cost benefit155,156 because they are already produced in large numbers for consumer electronics. Li and coworkers demonstrated the read-out of different binding assays using an unmodified CD read-out system by exploiting the error-signals in the detection because biomolecule/nanoparticle conjugates, bound to the surface of a CD, block the laser beam. The detected error-signal corresponded to a physical location or spot on the disk.157 A similar principle for the detection of immobilized immunoreaction products based on the error distribution as a function of the “playtime” was presented by Morais et al. using a standard DVD drive. In the same work, another detection concept was introduced, where signal changes from the DVD drive-integrated detection photodiode were acquired, as the reflection of the laser beam was attenuated when striking the immunoreaction product.155 Lange et al. used a modified CD pick-up head for the detection of silver grains on the CD surface, which were catalyzed by surface immobilized, gold-labeled antibodies. The silver grains caused a change in reflectivity.158 A DVD pickup head for the detection of binding events was employed by Bosco et al. Binding biomolecules to gold-coated cantilevers caused a deflection, a change in the resonant frequency and optical roughness, which was detected by the DVD laser.159
Steinert et al. promoted a system for protein structure analysis using X-ray crystallography as the detection principle. In this approach, X-rays from a beamline were transmitted to a cartridge-integrated crystallization chamber and produced characteristic diffraction patterns.163 Schwemmer and colleagues later proposed a platform for the small-angle X-ray (SAXS) scattering-based analysis of protein structures based on the scattering of X-rays transmitted to reaction chambers on a centrifugal microfluidic cartridge.61
2.9 Conclusion of unit operations and introduction of process chains
Traditionally, centrifugal microfluidics has mainly used the interplay of centrifugal forces and capillary forces to control the liquid flow.62,64,80 Both forces are present on centrifugal microfluidic platforms, because centrifugation is inherently available in rotating systems and capillary forces become dominant as dimensions shrink. Yet, the increasing demand on centrifugal microfluidic cartridges, namely for the integration of complex assays and high reliability/robustness, has led to an expansion of the means that are used to realize specific unit operations.One of these means is on-chip air compression or expansion by the processing liquid, which enables new principles for valving and pumping.49,52,74,98 Similar to centrifugation, this method is also intrinsically available, but compared to capillary action, it is less dependent on the surface tension and wetting properties, as well as the fabrication tolerances. Moreover, the pneumatic forces are usually orders of magnitude higher than the capillary forces, making pneumatic action particularly robust.
Another trend is the use of external radiation sources to selectively heat up areas of the cartridge or to perform optical measurements.35,46 The simple implementation of radiation sources and detectors into processing devices, as well as their non-contact characteristic and applicability in numerous unit operations, make them exceedingly promising. Furthermore, such unit operations are widely independent of the liquid properties. These advantages also apply to external magnets, which are mostly used in combination with magnetic beads.138,164 Another advantage of external active means is the extension of the degrees of freedom in cartridge operation, which allows some unit operations to become independent of the rotational speed.
The portfolio of unit operations that has been discussed in this review article so far includes sample and reagent supply, liquid transport, valving and switching, metering and aliquoting, mixing, separation, droplet generation, and detection. Combining these fluidic unit operations makes it possible to implement tasks with higher complexity such as blood plasma separation, cell lysis, nucleic acid purification, and nucleic acid amplification. Here, we introduce the term “process chain” in order to refer to these tasks with higher complexity. “Process chains” can usually be implemented by combining “unit operations,” and they are very useful to describe assay implementations on a higher hierarchical level. Complex applications such as genotyping assays in molecular diagnostics can be implemented to a great extent in a straightforward manner by simply concatenating several of the above-mentioned “process chains” such as “cell lysis,” “nucleic acid purification,” and “nucleic acid amplification.” Developers may re-use validated “process chains” from other assay implementations within the same microfluidic platform without the need to know the underlying fluidic unit operations in great detail, which reduces the costs and risks of implementing new assays. In that context, applying “process chains” in an assay implementation is very similar to applying “modules” and/or “subroutines” in programming. Introducing process chains is advantageous for all kinds of microfluidic platforms.
In the following sections, the most relevant applications and underlying process chains that have been published so far are presented and discussed.
3. Applications
The review of the applications in centrifugal microfluidics starts with a discussion of nucleic acid-based analysis, which can be subdivided into sample preparation, amplification and detection, and the implementation of sample-to-answer nucleic acid-based analysis. Here, the term process chain is used to categorize how the lysis of cells, purification of nucleic acids, and subsequent amplification and detection are implemented in centrifugal microfluidics. Subsequently, immunoassay-based analysis is reviewed by separately discussing the largest group of enzyme-linked immuno-sorbent assays (ELISA) and other implementations of immunoassays. Thereby, the implementations of process chains for blocking, immunocapture, and washing are discussed. A review of clinical chemistry applications follows, including a discussion of the implemented process chains for blood plasma separation as an example. Then, we discuss centrifugal microfluidic cell handling; the analysis of food, water, and soil; and the analysis of protein structures and functions. Finally, applications are reviewed that do not fit into the above-listed categories such as the generation of photonic crystals.3.1 Nucleic acid analysis
Bench top nucleic acid analysis is applied to a wide range of applications where information on the DNA or RNA level is required. Because of the multiplicity of processing steps within standard laboratory workflows, significant efforts have been put into automation by microfluidic integration, aiming at reducing the laboratory time as well as reagent and equipment costs.165 The automation and integration of all the required steps on one cartridge, which can potentially be processed in a portable processing device, will facilitate complex nucleic acid testing at the point-of-care because minimal resources and no special laboratory training will be required to perform the test.The standard laboratory workflow for a nucleic acid analysis can be roughly divided into two parts.166 (1) The first part is sample preparation with the aim to make nucleic acids accessible. Process chains include the lysis of eukaryotic or bacterial cells and nucleic acid purification or concentration for subsequent analysis. (2) The second part involves the post processing of nucleic acids with process chains such as nucleic acid amplification, e.g., mostly PCR and unit operations for the detection of the amplification result.
| Ref. | Lysis | Purif. | Lysis/purification method | Sample matrix and volume | Target | Performance parameters | Time | Notes |
|---|---|---|---|---|---|---|---|---|
| a ML: mechanical lysis; TL: thermal lysis; SPE: solid phase extraction; IMS: immunomagnetic separation. | ||||||||
| Kim J. et al.167 | ✓ | ML: beads in rimming flow | Culture media | CHO-K1 cells, E. coli and S. cerevisiae | Lysis efficiency 65% vs. conventional lysis protocol | 5–7 min | No connection to downstream fluidics demonstrated | |
| Kido H. et al.168 | ✓ | ML: magnetically assisted bead beating | 70 μL: (1) LB culture media (2) YPD culture media | (1) E. coli and (2) S. cerevisiae | Released DNA from (1) ≤40 μg mL−1; (2) ≤60 μg mL−1 | 30–480 s | Two processing stages required | |
| Siegrist J. et al.76 | ✓ | ML: magnetically assisted bead beating | 4 × 90–95 μL or 1 × 360–380 μL; (1) DI water; (2) clinical nasopharyngeal aspirate (NPA) | (1) B. subtilis spores (2) human metapneumo-, entero- or adeno-virus | (1) Equivalent lysis vs. reference; (2) correct identification of all viruses | <6 min | Lysis of spores demonstrated | |
| Hoffmann J. et al.24 | ✓ | SPE; silica matrix integrated into cartridge | 32 μL lysed whole blood | Human DNA | ≤77% vs. off-chip reference | Not stated | Liquid reagent prestorage in glass ampoules | |
| Müller M. et al.96 | ✓ | SPE; silica matrix integrated into cartridge | 32 μL lysed blood | Human DNA | 53 ± 8% vs. reference | 66 min | Commercially available reagents prestored | |
| Park B. H. et al.131 | ✓ | SPE; silica sol–gel integrated into cartridge | Virus lysate (5 μL) | RNA from influenza H1N1 virus | RNA capture yield 80% | 5 min | Small reagent volumes | |
| Jung J. H. et al.130 | ✓ | SPE; integrated glass bead column | 3.5 μL RNA sample (0.5 μL virus lysate, 1.25 μL EtOH, 1.75 μL 6M Gu-Hcl) | RNA from influenza H3N2 virus | RNA capture yield ∼81% | 440 s | Lysis process not included. Elution with RT-PCR cocktail demonstrated | |
| Strohmeier O. et al.138 | ✓ | SPE; magnetic silica beads | LB media (50 μL) | DNA from lysed Listeria innocua and lambda phage | Up to 68% ± 24% for L. innocua and 43% ± 10% for lambda phage vs. manual reference | 12.5 min | Novel handling concept for magnetic beads | |
| Wadle S. et al.170 | ✓ | ✓ | SPE; magnetic silica beads | 200 μL whole blood | Human DNA | Extracted DNA: 4.6 ± 0.7 ng μL−1 (disk) vs. 4.1 ± 0.4 ng μL−1 (reference) | Not stated | Commercially available extraction reagents |
| Strohmeier O. et al.171 | ✓ | ✓ | SPE; magnetic silica beads | 200 μL: culture media, blood plasma, whole blood | Human DNA, DNA from B. subtilis and E. coli and RNA from Rift Valley fever virus | Up to 98.5% for B. subtilis, 102.1% for E. coli and 34.2% for Rift Valley fever | ∼30 min | Measurement of PCR inhibitors included. Commercially available reagents |
| Cho Y. K. et al.89 | ✓ | ✓ | IMS with beads; TL by laser-induced heating | 100 μL whole blood | E. coli and Hepatitis B virus (HBV) | Comparable to bench top extractions | 12 min | Blood plasma separation included |
| Kloke A. et al.36 | ✓ | ✓ | SPE; silica matrix integrated into cartridge | 200 μL whole blood | Human DNA | Equal to manual reference | 50 min | Operated on standard laboratory centrifuge |
A process chain for mechanical lysis on a centrifugal microfluidic PDMS cartridge was first integrated by Kim et al. using the collision and friction of glass beads in a rimming flow. The rimming flow in a co-axially arranged microfluidic chamber was a result of alternating rotation, which depended on the bead density, solid volume fraction, acceleration rate, and angular velocity.167 Another centrifugal microfluidic cartridge for mechanical lysis was presented by the same group. Lysis was supported by the collision of glass beads, agitated by an oscillating magnetic disk in a radially arranged microfluidic chamber. The cell debris was centrifuged radially outward, while the supernatant was transferred to a collection port via a capillary siphon. To induce the oscillation of the ferromagnetic disk, integrated permanent magnets were rotated above the non-rotating microfluidic cartridge on a second spin stand, which consequently required the manual transfer of the cartridge between the different processing devices.168
An improved version of the aforementioned work was presented by Siegrist et al., in which the ferromagnetic disk in the microfluidic lysis chamber was actuated by the defined rotation of the centrifugal microfluidic polycarbonate cartridge over a set of external stationary magnets. In this approach, four lysis chambers were arranged isoradially, making it possible to process up to four samples in parallel. Centrifugo-pneumatic under-pressure valves were used to prevent sample flow into the clarification chamber during lysis. After centrifugation, the clear supernatant was transferred to a collection port via a capillary siphon. For the subsequent PCR analysis, heat inactivation of the inhibitors in the sample was required.76
For nucleic acid purification from lysed whole blood via a bind-wash-elute protocol, the so-called “Boom chemistry”,129 a centrifugal microfluidic cyclic olefin copolymer cartridge with on-board liquid reagent prestorage was presented by Hoffmann et al. (Fig. 10a). As the solid phase for DNA purification, silica membranes from commercially available QIAGEN spin columns were integrated into the cartridge. During processing, the pre-lysed sample and binding buffer mixture first passed through the silica membranes to capture the DNA. This was followed by a washing buffer. Finally, an elution buffer was supplied to elute the purified DNA from the membrane. An integrated Coriolis switch92,169 was used to separate the waste (lysed sample and washing buffers) and elution buffer containing the purified DNA.24 A similar system was presented by Müller et al., which was designed to be operated in a standard laboratory centrifuge.96 In this work, the Coriolis switch was replaced by a switch for unidirectional rotation because the centrifuge only supports one direction of rotation. Neither approach integrated lysis of the blood.
 | ||
| Fig. 10 Centrifugal microfluidic process chains for nucleic acid purification and extraction. (a) DNA purification from lysed whole blood via integrated silica matrix “d” with onboard liquid reagent prestorage “a.” An integrated Coriolis switch “e” is used to direct purified DNA and waste to different microfluidic chambers “f” and “g”,24 (reproduced with permission from The Royal Society of Chemistry.) (b) RNA purification from virus lysates via sol–gel matrix.131 (Reproduced with permission from The Royal Society of Chemistry), and (c) DNA extraction in LabTube via integrated silica matrix.36 (Reproduced with permission from The Royal Society of Chemistry.) | ||
A microscope slide-shaped microchip for RNA purification from low volumes (5 μL) of virus lysates via a bind-wash-elute chemistry was reported by Park et al. A sol–gel matrix in a microfluidically patterned PDMS layer was used as a solid phase for the separation of RNA from the lysate (Fig. 10b). A lysed sample premixed with ethanol for binding, washing buffer, and elution buffer were added to microfluidic reservoirs prior to rotation and sequentially released using the differences in the flow resistances of the connecting channels.131 In a later work, the sol–gel solid phase was replaced by a column of tetraethoxy orthosilicate (TEOS)-activated glass beads contained in a zig-zag-shaped microfluidic channel. Here, capillary valves between the washing buffer reservoirs and the zig-zag channel and a capillary siphon between the elution buffer reservoir and the zig-zag channel were exploited for the sequential release of the reagents to the glass bead bed.130 In both approaches, lysis of the virus samples was conducted off chip. Although all the reagents could be added to the chips at the beginning, the waste (washing buffer and lysate) had to be removed manually from the capture chamber during processing.
The purification of DNA from lysate samples with silica-coated magnetic beads was demonstrated using integrated-gas-phase transition magnetophoresis (GTM) on a microthermoformed foil cartridge. Bead transport was a result of the defined positioning of the foil cartridge in relation to an external stationary permanent magnet and did not require any human interaction. Initially, beads bound the DNA from the lysate in a first chamber. After binding, the beads were automatically transported through an air-gap into a second chamber containing washing buffer and finally into a third chamber with elution buffer.138 The modular concatenation of multiple chambers with different volumes was then applied for bead-based DNA extraction from whole blood, including lysis.170 In a later work, this process chain for nucleic acid extraction was extensively characterized for extractions from logarithmic dilutions of various target pathogens and sample matrices including Gram-positive Bacillus subtilis, Gram-negative Escherichia coli, Rift Valley fever RNA viruses from blood plasma and human genomic DNA from whole blood.171
Recently, the LabTube was introduced as a versatile centrifugal microfluidic platform for bind-wash-elute-based DNA extraction from blood and other samples.36 Microfluidic and micromechanical elements are integrated in a centrifuge tube with the outer dimensions of a 50 mL centrifuge tube, as depicted in Fig. 10c. An integrated centrifugally actuated ball-pen mechanism enables reagent release and liquid routing. Unit operations for mixing and separation-based extraction are also integrated. Using LabTube, extractions of genomic DNA from whole blood were demonstrated with yields and purities equal to manual reference runs. Sample addition, the transfer of LabTube into the centrifuge, and the withdrawal of a standard reaction tube containing the eluate remained as the only manual steps.
A highly comprehensive approach for pathogen specific DNA extraction on a centrifugal microfluidic polycarbonate cartridge was presented by Cho et al.89 In this work, target pathogens were separated from a sample by immunomagnetic separation using antibody-coated magnetic beads subsequent to disk-integrated blood plasma separation. Pathogens were thermally lysed by heating the beads with a laser. Multiple integrated ferrowax microvalves (LIFM) could be opened or closed by laser irradiation, thereby defining the fluidic routing.
Detection can be achieved using fluorescently labeled probes, by intercalating fluorescent dyes, after PCR, e.g., by the detection of the PCR product via gel- or capillary electrophoresis, or by hybridization to immobilized DNA capture probes (DNA microarrays). Although the application of centrifugal microfluidics for automating process chains like nucleic acid amplification has advantages (i.e., a reduced risk of cross contamination because of the closed systems, homogeneous temperature distribution, and recondensation of vapor), the interfaces required for thermocycling and optical readout remain technically challenging. In this context, the review of the amplification and detection methods is structured as follows. First, centrifugal microfluidic systems that only integrate the amplification process chain are reviewed. Then, systems with additionally integrated unit operations for detection are reviewed. These systems are compared by the degree of multiplexing (i.e., the ability to simultaneously detect different target nucleic acids), sensitivity, and time to result (Table 6). At the end of the section, we review centrifugal microfluidic systems that were exploited for processing microarrays.
| Ref. | Amplification | Target | Degree of geometric multiplexing | Sensitivity | Time (cycles) | Detection technology | Heating technology |
|---|---|---|---|---|---|---|---|
| PCR: polymerase chain reaction; RT-PCR: reverse transcriptase polymerase chain reaction; RPA: recombinase polymerase amplification; S. aureus: Staphylococcus aureus.a Time for heating and cooling not included. | |||||||
| Focke M. et al.38 | PCR | Resistance genes in S. aureus | 7 + 1 internal control | <10 DNA copies per well | 110 min (50 cycles) | FAM-labeled hydrolysis probes; real-time fluorescence detection | Air mediated in commercially available PCR thermocycler |
| Lutz S. et al.26 | RPA | mecA gene in S. aureus | Monoplex | <10 DNA copies per well | <15 min | Real-time fluorescence detection | Air mediated in commercially available PCR thermocycler |
| Focke M. et al.37 | Multiplex-preamplification, nested PCR | Resistance genes in S.aureus | Up to 4 | Down to 7 copies per sample | 17 min (10 cycles) pre-amp, 52 min (50 cycles) main amplificationa | FAM-labeled hydrolysis probes; real time fluorescence detection | Air mediated in commercially available PCR thermocycler |
| Strohmeier O. et al.39 | Allele-specific PCR | KRAS point mutations on tumor cell DNA | 8 | Not stated | Not stated | FAM-labeled hydrolysis probes; real-time fluorescence detection | Air mediated in commercially available PCR thermocycler |
| Strohmeier O. et al.41 | PCR | DNA from 6 different food borne pathogens | 6 | 0.1 pg DNA per well for Salmonella and Listeria | ∼2 h (50 cycles) | FAM-labeled hydrolysis probes; real-time fluorescence detection | Air mediated in commercially available PCR thermocycler |
| Sundberg S. O. et al.101 | Digital PCR | 300 base pair plasmid DNA | Monoplex | Amplification in 58 out of 1000 wells (DNA concentr.: 6 × 100 copies μL−1) | ∼25 min (45 cycles); additional 25 min for loading, fluorescent imaging and image analysis | Intercalating dye; “accumulated” real-time fluorescence detection of hundreds of wells for melting curve analysis; post PCR image acquisition with CCD camera for digital well analysis | Air mediated |
| Furutani S. et al.173 | PCR | invA gene in Salmonella enterica | Monoplex | PCR on isolated single cells | 95 °C/2 min for thermal lysis; denat. 95 °C/5 s, anneal. 55 °C/10 s; elongate 72 °C/10 s optimum 40 cycles | FAM-labeled hydrolysis probes; post-PCR fluorescence detection | Contact |
| Amasia M. et al.90 | PCR | Bacillus anthracis; Bacillus cereus | Monoplex | Not stated | 53 min (35 cycles) | Off-chip (analysis of PCR products by gel electrophoresis) | Contact; with thermoelectric modules |
| Jung J. H et al.172 | RT-PCR | Influenza A subtypes: H3N2, H5N1, and H1N1 | Monoplex and duplex | ∼2 RNA copies (demonstrated for H3N2) | 25.5 min | Off-chip (microcapillary electrophoresis) | Contact; serially on thermal blocks |
A centrifugal microfluidic cartridge for PCR-based amplification has been presented where contact heating and cooling using three thermoelectric modules was employed for thermocycling (1 module) and in parallel for freezing sub-volumes of the PCR buffer in the channel network (2 modules) to ice valves. These ice valves were integrated to block the connection channel between the PCR chamber and venting hole and thus prevent cross contamination through the vent because stationary thermocycling was conducted, without rotating the disk.90 Jung et al. developed a PDMS/glass cartridge for the reverse transcriptase PCR detection of viral RNA. The microfluidic cartridge was serially rotated over three temperature blocks at different temperatures for denaturation, annealing, and extension.172 In both approaches, the detection of the generated PCR product had to be conducted off-disk using gel electrophoresis90 or microcapillary electrophoresis.172
Further applications have been demonstrated using centrifugal forces to force a bacterial sample through 24 zig-zag shaped channels integrated into a centrifugal microfluidic PDMS cartridge. Single bacterial cells from the sample were distributed into multiple 1.5 nL microchambers connected to each zig-zag channel. For the thermal lysis of the cells and PCR-based amplification, the cartridge was placed on a custom-made thermocycling system for contact heating. After PCR, the fluorescence intensity was measured by placing the cartridge into an image analyzer.173
Digital PCR on centrifugal microfluidic cartridges was presented by Sundberg et al. By spinning the disk, a PCR mixture that included plasmid DNA was forced through a spiral channel and aliquoted into one thousand 33 nL amplification wells (Fig. 11a). Afterward, the PCR mixture aliquots in the wells were separated by forcing mineral oil through the spiral channel. An air-mediated temperature setting for thermocycling allowed PCR cycle times of 33 seconds.101 The proposed digital PCR platform has been commercialized and distributed by Espira Inc.174
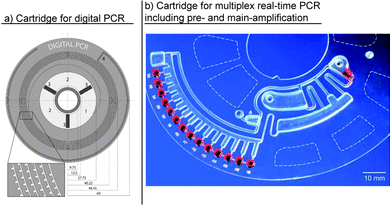 | ||
| Fig. 11 Centrifugal microfluidic cartridges for nucleic acid amplification: (a) cartridge for digital PCR using unit operation for one-step aliquoting to 1000 1 nL amplification wells.101 (Reprinted with permission from the American Chemical Society.) and (b) Cartridge for pre amplification and subsequent multiplex real-time PCR-based main amplification, including integrated two-stage aliquoting into fourteen 10 μL amplification wells.37 (Reproduced with permission from The Royal Society of Chemistry.) | ||
Centrifugal microfluidic cartridges have been exploited for the real-time PCR-based genotyping of methicillin-resistant Staphylococcus aureus (MRSA).38 Cartridges were fabricated from thin polymer foils using microthermoforming175 to allow fast, air-mediated, heat transfer (Fig. 11b). An integrated unit operation for two-step aliquoting made it possible to divide and fluidically separate an initial volume of PCR mastermix into eight aliquots of 10 μL each. The aliquots were then transferred into a separate amplification chamber harboring a set of dryly prestored primers and probes. Thereby, “geometric” multiplexing was achieved. Up to four separate DNA samples could be analyzed per cartridge.38 To increase the sensitivity, an advanced version of the aforementioned cartridge was presented by the same group, which included pre-amplification prior to aliquoting and a downstream nested PCR. A translation of the integrated functionality into a schematic description highlighting the implemented process chains and unit operations is depicted in Fig. 12. As an advantage, the integration of the pre- and main amplification into the same cartridge circumvented the risk of cross contamination by sample handling after pre-amplification.37
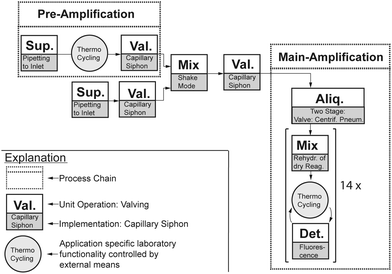 | ||
| Fig. 12 Schematic interpretation of integrated functionality of Focke et al.37 Dashed boxes represent process chains, and solid boxes depict unit operations and the demonstrated implementation (Sup.: sample or reagent supply; Val.: valving; Mix: mixing; Aliq.: aliquoting; Det.: detection). Circles illustrate application specific laboratory functionalities that are controlled by external means. | ||
A similar cartridge has been used for isothermal real-time amplification by recombinase polymerase amplification (RPA). In this work, a lyophilized polymerase pellet and liquid rehydration buffer were prestored on the cartridge. Thus, only a template DNA addition was required. The rehydration of the lyophilized polymerase pellet was achieved by integrated shake mode mixing before the RPA mastermix was transferred into an aliquoting structure via a capillary siphon valve. Up to six samples could be analyzed per cartridge.26 For multiplex point mutation detection, an allele-specific PCR has been integrated into centrifugal microfluidic foil disk-segments to allow the independent processing of up to four samples per run. The automation comprises the aliquoting of a PCR mastermix into multiple fluidically separated amplification chambers with dryly prestored primers and probes, followed by an allele-specific PCR.39 In another approach, Strohmeier et al. presented a cartridge for the detection of six common food borne pathogens. This cartridge included amplification chambers for integrated positive and negative controls and demonstrated the capability for quantitative real-time PCR by the parallel amplification of integrated DNA standards.41 As an advantage, all the cartridges and disk segments could be processed in a modified, commercially available centrifugal real-time PCR thermocycler for fluidic processing, amplification, and fluorescence detection, and did not require additional equipment. Recently, Czilwik et al. presented a passive microfluidic vapor diffusion barrier to reduce pressure increase during thermocycling. The application of this unit operation was demonstrated for PCR amplification and subsequent transport of the amplification product for further analysis.176
Recently, Focus Diagnostics and 3M introduced the integrated cycler, a real time PCR cycler, to the market. Up to 96 pre-extracted nucleic acid samples can be pipetted to a universal single-use disk. Each of the 96 radially inward inlet wells is directly connected to one of 96 amplification wells located at the outer rim of the cartridge. Contact heating is employed for thermocycling. Up to four fluorescence channels are available in the instrument for real-time detection. In 2012, Focus Diagnostics' Flu Test for use in combination with the 3M integrated cycler was approved by the FDA.177 A list of the relevant patents for the disposable disk and device can be found on the website.178
In addition to the integration of process chains like those for nucleic acid amplification and detection, in the past, multiple centrifugal microfluidic cartridges have been presented for automating microarray processing.
Peng et al. presented a glass disk that was first attached to a PDMS disk with 96 radial channels. Using centrifugal forces, DNA probes were then pumped through the channels for “printing” radially DNA probe lines on the glass disk. The first PDMS disk was then peeled off and replaced by a second PDMS disk with 96 spiral channels that orthogonally intersected the 96 probe lines. Finally, DNA samples were forced through the spiral channels and hybridized to the probe lines. Successful hybridization was detected using a fluorescence scanner.132 This centrifugal microfluidic cartridge for DNA hybridization with slightly increased channel dimensions was later used by the same group for the detection of PCR products from the fungal pathogens Botrytis cinerea and Didymella bryoniae. The presented system was capable of detecting 3 ng PCR products after hybridization for 2 h at 45 °C.179 By improving the flow control and channel design and adding an additional fluorescent dye, the detection of less than 0.2 ng of PCR products derived from three different fungal pathogens (Didymella bryoniae, Botrytis cinerea, and Botrytis squamosa) within 3 min at 23 °C180 was presented.
Peytavi et al. developed a slide-shaped PDMS chip with integrated microfluidic channels for the discrimination of the single nucleotide polymorphisms of four clinically relevant Staphylococcus species. The serial release of samples (PCR products with incorporated Cy-labeled dUTPs), washing buffer, and rinsing buffer into the array chamber was controlled by the spin speed and integrated capillary valves. Afterward, the slide was dried during rotation at a high spin speed. For readout, the glass slide was transferred into an array scanner. A 10-fold increase in the hybridization signal was reported for the microfluidic flow-through approach compared to passive systems that solely rely on the diffusion of an analyte to the capture probe.134 A similar microfluidic chip was later used for the hybridization of 25-mer DNA samples. Enzyme-labeled fluorescence technology was used to generate the signal for detection. A threefold increase in fluorescence intensity compared to passive assays was reported for similar hybridization times.133
Hoehl et al. presented a LabTube36 with an integrated process chain for solid-phase-based DNA purification from lysates of a verotoxin produced by Escherichia coli spiked in water, milk, and apple juice samples, combined with the subsequent isothermal LAMP amplification. In this work, a battery-driven heating system was integrated for the direct amplification in the tube. The positive LAMP amplification resulted in a visible color change for the LAMP reaction. A reduction in the manual labor time from 45 to 1 minute was reported, requiring only a single pipetting step to load the LabTube with the pre-lysed bacterial sample.181
Kim et al. presented a centrifugal microfluidic cartridge for the detection of Salmonella from PBS and milk samples that included process chains for laser-induced thermal lysis89 and isothermal amplification via RPA. For sequential fluid control, several ferrowax valves89 were integrated. Read-out of the result was possible via visual detection on an integrated lateral flow strip. Detection limits of 10 CFU mL−1 and 102 CFU mL−1 were reported for the PBS and milk samples, respectively, with a time to result of 30 minutes. Not included in the microfluidically automated process was the process chain for immunomagnetic sample enrichment from the 1 mL milk and PBS samples. After capturing the pathogens, the capture beads were magnetically collected, washed twice, and resuspended in 5 μL of distilled water, which was then loaded onto the cartridge.146
Strohmeier et al. presented a centrifugal microfluidic polymer foil cartridge for the sample-to-answer analysis of bacterial targets from a blood plasma sample. The following process chains were combined on the cartridge in sequential order: chemical lysis, magnetic bead-based DNA purification, and isothermal amplification via RPA with real-time fluorescence detection relying on unit operations such as capillary siphons, gas-phase transition magnetophoresis for DNA separation,138 and aliquoting.98 The disk could be processed in a portable device, and successful sample-to-answer detection was demonstrated for 6 × 104 genome equivalents of Bacillus anthracis and 6 × 106 genome equivalents of Francisella tularensis spiked into blood plasma samples. A total processing time of 45 minutes was reported.43 An updated version of the aforementioned work demonstrated real-time PCR-based detection of Staphylococcus warneri, Streptococcus agalactiae, Escherichia coli and Haemophilus influenzae from a 200 μL serum sample. Limits of detection were reported to be 3, 150, 5 and 18 colony forming units, respectively. In addition to the above-mentioned process chains, a stickpack for prestorage and on-demand release of rehydration buffer and a process chain for pre-amplification prior to target specific PCR was integrated to increase the sensitivity.29,182 Pre-amplification required further unit operations for metering the eluate and pumping49 the pre-amplified solution toward the center of the cartridge. Processing was conducted in a portable PCR device.182
Jung et al. presented a centrifugal microfluidic cartridge for the purification of viral RNA from H3N2 influenza combined with the subsequent amplification and detection. No process chain for sample lysis was included. RNA separation from the lysate and purification were conducted using a microglass bead solid phase, while an RT-PCR cocktail was used to elute the purified RNA from the bead bed. The sample, washing buffers, and RT-PCR mix were sequentially released from their inlet chambers by differences in the flow resistance values of the respective channels or by capillary siphons.183
3M recently commercialized the “direct amplification disc”184 for the sample-to-answer analysis of influenza virus A/B and respiratory syncytial virus (RSV). The “direct amplification disc” can be operated in the 3M integrated cycler. The disk allows the real-time amplification of up to eight unprocessed clinical samples by making use of direct amplification chemistries185 that can perform nucleic acid extraction and amplification in one protocol. For processing, a 50 μL patient sample and 50 μL reaction mix have to be pipetted to the direct amplification disc prior to processing. The microfluidic layout has not been published, although several patents might disclose the functionalities of single unit operations such as metering186 and valving.187 Up to four fluorescence channels are available for detection.
The Canadian company GenePOC Inc. is approaching the market with a centrifugal microfluidic disk segment with sample-to-answer capability, which includes process chains for mechanical lysis and subsequent amplification and detection. Up to eight disk segments can be processed in parallel, allowing the independent analyses of up to eight samples with volumes of 100–200 μL in parallel. According to the corresponding patent application,188 the system features mechanical lysis using glass beads that are actuated by an additional magnetizable element in the microfluidic chamber similar to the system presented by Kido et al.168 Afterwards, a portion of the lysate is diluted with a dilution buffer, heated up, and aliquoted into three separate amplification chambers that contain specific PCR reagents. By using four different dyes, up to 12 targets should be detectable from one sample in less than 1 hour with less than 1 minute of hands-on time.189
Although showing full sample-to-answer capability, neither commercial system has an integrated process chain for nucleic acid purification after lysis. On the one hand, this makes microfluidic integration easy because of the reduced complexity. On the other hand, the approach might only be suitable for certain sample materials with low amounts of inhibitors and sufficient pathogen-loads because no DNA/RNA concentration step is included.
To circumvent the need for additional equipment, the processing of centrifugal microfluidic cartridges for sample preparation36 or amplification and detection26,37–39,41,181 in commercially available equipment has been demonstrated. These microfluidic chips, which extend the functionality of an existing processing device, have been called “microfluidic apps”.191 Other cartridges could be processed in small and portable devices, making them suitable for single sample testing and application at the point-of-care.43,170,182 In addition to single sample and point-of-care testing, first applications have been demonstrated for highly parallel applications such as digital PCR.101
The application of centrifugal microfluidics for automation of nucleic acid analysis provides unique advantages for assay automation as multiple standard laboratory process chains already exploit centrifugal forces when conducted manually. The advantages include the possibility to perform density based separations during sample preparation such as the separation of blood plasma from whole blood or the concentration of bacterial pathogens by sedimentation. Furthermore, nucleic acid extraction on the bench commonly uses so called “spin columes” where the sample and liquid reagents are serially forced through solid phase membranes by centrifugation. With respect to PCR based nucleic acid amplification, centrifugal microfluidic cartridges may benefit from the straight forward approach to remove bubbles (due to buoyancy in the centrifugal gravity field) at elevated temperatures.
3.2 Immunoassays
Immunoassays (IA) are widely established in (bio-) medical diagnostics, biological and biochemical studies, drug development, environmental analyses, and food safety.59,156,192 Immunoassays are based on the highly specific affinity of antibodies (Ab) to antigens (Ag), allowing for the detection of bioanalytes that provide appropriate binding sites (epitopes). Either the antigen or antibody can be the target bioanalyte. In heterogeneous immunoassays, the capture antibody is immobilized either on macroscopic solid supports or on microscopic beads suspended in the solution. The analyte is present in the liquid phase. After a certain incubation period, the bound analyte is measured directly on the surface using a suitable transducer or biosensor system, or using a secondary antibody in solution conjugated with a suitable tracer. In the latter case, an active bound/free separation step, e.g., by washing, is required. Alternatively, homogeneous immunoassays do not require a bound/free separation step. In this case, a signal is generated by the binding of the appropriate tracer or tracer combination to the analyte.A wide variety of immunoassay formats are in place, and two main categories can be considered. An immunometric assay employs an antibody labeled with a tracer, which is advantageous if the target analyte exposes multiple binding sites or epitopes. In this case, for example, the primary or capture antibody binds the analyte to the solid phase, and the secondary labeled antibody builds up a sandwich-type structure with the analyte. After the bound/free separation, the tracer bound via the sandwich to the solid phase can be quantified. Competitive assay formats are often used for small analytes, which expose only one binding site or epitope. In this case, an analyte analogon conjugated with a tracer competes with the analyte in the sample. The analyte analogon is applied in a defined, limited concentration to enable balanced competition with the analyte for the binding antibody.
The integration and automation of immunoassays on centrifugal microfluidic platforms are especially regarded as attractive because conventional assay protocols are labor intensive and consist of a large number of manual processing steps.59 As most of the steps are similar for a broad variety of assays, platform-based automation offers unique advantages to reduce costs and ensure consistent results.60,135,193 Yet, the most commonly employed platform for immunoassays are microtiter plates having, for example, 96 wells in a well-defined pitch,194 where liquid handling can be automated by pipetting robots. In contrast, the microfluidic automation of immunoassays offers some unique advantages such as reduced reaction times due to reduced diffusion distances, as well as reductions in the reagent and sample volumes.59,156
As the accuracy of diagnostic findings can be enhanced by simultaneous analyses of multiple biomarkers, the degree of multiplexing of one sample within an IA automation is an additional important characteristic.194 Similar to nucleic acid analysis, multiplexing is typically achieved by differentiation in the spatiotemporal or spectroscopic domain.194 In this context, we propose an evaluation of centrifugal microfluidic cartridges for immunoassays based on the following criteria: the analytical sensitivity (limit of detection, LOD) and reproducibility/precision (coefficient of variation, CV) achieved for the specific analysis. Further, if the performance criteria for a specific analyte can be met, the time to result and degree of automation, integration, parallelization, and multiplexing should be evaluated. Table 7 summarizes important key characteristics of the reviewed systems. The review section is split into two subchapters, centrifugal microfluidic systems for ELISA followed by a section on other immunoassay formats.
| Ref. | Assay format/solid phase/detection | Sample matrix | Multiplexing | Parallelization | Reagent pre-loading/storage | Total time [min] | Target analyte/LOD |
|---|---|---|---|---|---|---|---|
| LOD = limit of detection, ELISA = enzyme-linked immunosorbent assay, IgG = immunoglobulin G, FIA = fluorescence based immunoassays, CLIA = chemiluminescent IA, Ag = antigen, PBS = phosphate buffered saline, BSA = bovine serum albumin, lf = label-free, Ab = antibody, IgA = immunoglobulin A, HRP = horseradish peroxidase, MB = muscle-brain type, ELISPOT = enzyme-linked ImmunoSpot assay, AuNP = gold nanoparticle, SAW = surface acoustic wave, SPR = surface plasmon resonance, SAF = supercritical angle fluorescence.a Reagents are automatically dispensed by a robotic loading system.b Off-chip sample and detection Ab incubation requires 90 min.c Essential assay steps take place off-chip. | |||||||
| Lai et al.59 | ELISA/channel/florescence | Cell culture | 1 | Up to 24 | Yes | >60 | rat IgG/31 nM |
| Honda et al.71 | FIA/beads/florescence | PBS with BSA | 1 | 104 | Yesa | 50 | α-Fetoprotein/0.15; interleukin 6/1.25; carcinoembryonic Ag/1.31 pmol L−1 |
| Inganäs et al.15 | FIA/beads/florescence | Whole blood | 1 | 104 | Yesa | 50 | Human interleukin 2; human interleukin 1β; myoglobin/all subpicomolar |
| Cho et al.199 | lf IA/cantilever/resonance frequency | Buffer solution | 1 | 5 | Yes | N/A | Prostate specific Ag/picomolar |
| Riegger et al.151 | FIA/beads/florescence | Serum | 15 | 4 | No | N/A | Tetanus Ab/158; hepatitis A Ab/215 mIU mL−1 |
| Riegger et al.195 | ELISA/beads/chemiluminescence | Plasma | 1 | 8 | No | N/A | Myoglobin/12.2 ng mL−1 |
| Nagai et al.136 | ELISA/beads/fluorescence | Mixture of secretory IgA and HRP-labeled anti-IgA antibodies | 1 | 18 | Yesc | 30b | Secretory IgA/6.4 nM |
| B. S. Lee et al.135 | ELISA/beads/absorbance | Whole blood | 1 | 3 | Yes | 30–50 | Hepatitis B Ag/0.51 ng mL−1; anti-hepatitis B Ab/8.6 mIU mL−1 |
| Koh et al.198 | FIA/beads/florescence | Serum | N/A | N/A | Yesc | <20 | Shiga-like toxin 1/0.8; ricin/1; anthrax/1.9 pM |
| B. S. Lee et al.137 | ELISA/beads/absorbance | Whole blood | 1 | 1 | Yes | 22 | Creatine-kinase MB/0.92 ng mL−1 |
| Noroozi et al.106 | ELISPOT/membrane/colorimetric | Serum | 25 | 8 | No | N/A | Burkholderia Ag/N/A |
| Schaff and Sommer88 | FIA/beads/florescence | Plasma/whole blood | >15 | 20 | Yes | 15 | Interleukin 6/63; C-reactive protein/92 pmol L−1 |
| Park et al.194 | ELISA/beads/absorbance | Whole blood*/saliva** | 3 | 2 | Yes | 20 | High sens. C-reactive protein/0.27*, 0.30**; cardiac troponin I/0.27*, 0.51**; N-terminal pro-B type natriuretic peptide/0.32*, 0.24** ng mL−1 |
| Burger et al.122 | FIA/beads/florescence | PBS with BSA | 3 | 4 | No | N/A | Mouse anti-ERα IgG; human IgG; rabbit anti-fd bacteriophage IgG/N/A |
| W. Lee et al.162 | AuNP IA/SAW sensor/mass enhancement | Plasma/whole blood | 1 | N/A | Yes | 20 | Cardiac troponin I/6.7 pg mL−1 |
| Kim et al.34 | ELISA/beads/electrochemical | PBS | 1 | 3 | Yes | <20 | C-reactive protein/4.9 pg mL−1 |
| Nwankire et al.150 | FIA/SAF chip/fluorescence | Bioprocess sample | 1 | N/A | No | <30 | Human IgG/N/A |
| Welte et al.196 | CLIA/chamber/chemiluminescence | Standard solution | 1 | 24 | No | 45 | Estradiol/60 pg mL−1 |
| Otsuka et al.154 | lf IA/SPR sensor/optical | Buffer solution | 1 | 8 | Yes | N/A | Human IgA/N/A |
The majority of the steps in the laboratory workflow for a typical heterogeneous sandwich ELISA can be automated by utilizing the immunocapture process chain: (1) the immobilization of the primary/capture Ab or Ag on a solid phase, (2) binding of the target Ag or Ab in the sample to the primary Ab or Ag on the solid phase, and (3) binding of the enzyme-labeled secondary/detection Ab to the target Ag or Ab. The blocking process chain is thereby applied between the first and second steps to prevent unspecific binding, whereas all the steps are separated by multiple washing process chains to rinse away the unbound material. The remaining steps for signal generation and detection involve unit operations for (4) supplying the substrate solution for the enzymatic reaction, (5) the eventual termination of the enzymatic reaction by supplying a stopping solution, and (6) the quantification of the enzymatic reaction product. An early centrifugal microfluidic cartridge for ELISA-based immunoassays was reported by Lai et al. Integrated capillary valves allow for the sequential release of pre-loaded reagents into a microchannel with immobilized primary antibodies. Each liquid solution displaces the aforementioned into a waste chamber. A singleplex analysis of rat IgG from a hybridoma culture proved advantageous with respect to reagent consumption and assay time.59 Later, a similar system was used for the detection of cytokine interferon-gamma.192
A later approach for direct ELISA was presented by Riegger et al. Up to eight separate immunoassays could be processed per cartridge in parallel for the detection of the relevant biomarkers for acute myocardial infarction. High-speed chemiluminescence detection with a photo-multiplier was performed under rotation in less than 1 second.195
An increase in parallelization to 18 immunoassays per cartridge was presented by Nagai et al. A single bead served as the solid phase for the competitive, indirect ELISA targeting a mental stress biomarker. Prior to the on-cartridge automation, time-consuming off-chip steps had to be performed.136 An injection-molded cartridge featuring 24 parallel immunoassays was reported by Welte et al. A multiplicity of unit operations, including capillary siphon and hydrophobic valves were integrated to route the reagents. All the reagents had to be loaded step-by-step during the protocol.196
A totally integrated ELISA for detecting the antigens and antibodies of the hepatitis B virus was presented by B. S. Lee et al. An integrated process chain for blood-plasma separation allowed the use of a whole-blood sample. The routing of the sample and reagents was controlled by integrated active laser irradiated ferrowax microvalves. Shake-mode mixing was implemented to mix beads (solid phase) with the plasma, detection probe, washing buffers, or tetramethyl benzidine (TMB) solution. The parallelization of three separate immunoassays allowed tests to be performed for the antigen and antibody of the hepatitis B virus, HBsAg and anti-HBs, and a control, in parallel on a single cartridge. The assay time increased by 2/3 compared to processing a single IA. All the required assay components were pre-loaded onto the disk.135 Later, an advanced version of the aforementioned injection-molded cartridge, combining the demonstrated IA principle and a biochemical analysis applying a lipid test panel (see Section 3.3) was presented. These tests were performed simultaneously from one whole-blood sample, aiming at the detection of CK-MB (muscle and brain fraction of creatine kinase) as a biomarker for recent heart attacks.137
The combination of a high degree of integration with multiplexing ability was reported by Park et al. The cartridge featured two ELISAs in parallel (Fig. 14a), each capable of testing a sample for three targets or controls, respectively. Reagents were pre-loaded onto the cartridge prior to the test. An analysis of cardiovascular disease biomarkers in whole saliva or blood was performed. The reaction chambers were first flushed with common liquids simultaneously. Later, the fluidic pathways were isolated from each other by active laser-actuated microvalves for individual substrate and stop solution supply, as well as for detection.194 A schematic representation of the integrated application highlighting the implemented process chains and unit operations is depicted in Fig. 13.
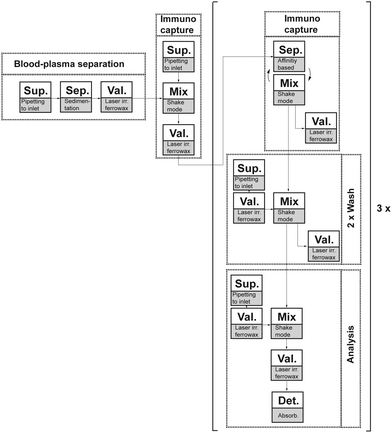 | ||
| Fig. 13 Schematic representation of integrated process chains (dashed boxes: blood-plasma separation, immunocapture, washing, and analysis) and corresponding sequence of unit operations (solid boxes: Sup.: supply of reagents or sample; Sep.: separation; Val.: valving; Det.: detection).194 | ||
Recently, new readout concepts were the subject of intensified research. A cartridge featuring flow-enhanced electrochemical detection under rotation was shown by Kim et al. This measuring method featured an adjustable sensitivity (LOD values of 21.3, 4.9, and 84.5 pg mL−1 for stagnant, flow, and reference, respectively) due to its demonstrated dependency on the flow rate. Flow control was realized by integrated active ferrowax microvalves. The target biomarkers for cardiovascular disease (CVD) were indirectly detected by measuring an electroactive substrate catalyzed by an enzyme conjugated with the detection Ab. Liquid reagents were pre-stored on the cartridge prior to sealing.34
Multiplexed FIA for centrifugal microfluidics applying colored beads as the solid phase was shown earlier by Riegger et al. Here, the beads were color-encoded with dyes or quantum dots with theoretical degrees of multiplexing of fifteen and five, respectively. Prior to fluorescence readout of the detection Ab, dye and quantum dot beads were identified with >90% and >80% reliabilities, respectively. The detection was realized within 15 seconds using a color CCD-camera and software algorithm.151 Noroozi et al. demonstrated a cartridge with decreased assay time due to enhanced Ag–Ab interaction employing micro-mixing by flow reciprocation. Multiplexing was achieved by spotting an array of antigens on the surface of the detection chamber.106 In both setups, reagents had to be loaded step-by-step onto the cartridge. Later, the combination of color-coded multiplexing with beads, captured in V-shaped cups, was presented by Burger et al., where reagents had to be introduced to the cartridge step-by-step.122
A cartridge replacing the conventional washing steps by the centrifugation of beads through a density medium was presented by Schaff and Sommer. Sedimentation allowed the multiplexing of two inflammation biomarkers (interleukin 6 (IL-6)/C-reactive protein (CRP)) inside a single channel by separating beads of different sizes and densities. A theoretical multiplexing degree of >15 was reported. A whole-blood sample (IL-6) could be processed without the need of plasma separation. Wax valves employing phase change paraffin were integrated into the cartridge for fluidic routing.88 The presented work was extended by Koh et al., who showed the detection of three high priority potential bioterrorism agents (Fig. 14b).198
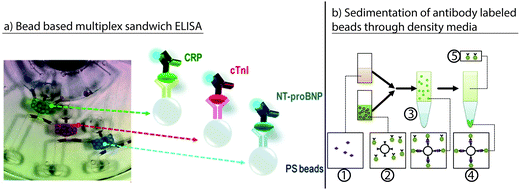 | ||
| Fig. 14 Various implementations of immunoassays on the centrifugal microfluidic platform. (a) Bead-based multiplex sandwich ELISA.194 Depicted are three reaction cavities with differently labeled solid phases and individual substrate solutions (green, red, blue). Shadows were caused by the image acquisition. (Reprinted with permission from the American Chemical Society.) (b) Immunoassay based on the sedimentation of antibody-labeled beads through a density medium according to:198 (1) sample with analyte and (2) detector suspension with beads and labeled antibodies are mixed, forming a layer on (3) a density medium for incubation. Upon rotation, (4) a pellet is formed in the density medium with (5) the sample with unbound label remaining above. (Reprinted with permission from The Chemical and Biological Microsystems Society.) | ||
An early demonstration of label-free IA on a centrifugal cartridge was presented by Cho et al.199 Resonant frequency changes in electromechanical cantilever sensors were used for the IA readout. The cantilever required drying via centrifugation prior to readout. Reagents were pre-loaded prior to testing. Later, a cartridge applying a surface plasmon resonance (SPR) sensor for label-free detection was reported by Otsuka et al. The SPR allowed for the real-time measurement of biomolecular interactions.154 In this work, the serial fluid transport of all the required reagents was realized, similar to Lai et al.,59 by the integration of cascades of capillary valves.
A cartridge applying an injection-molded COC surface-confined supercritical angle fluorescence (SAF)-chip in a hybrid assembly for readout was demonstrated by Nwankire et al. The readout concept allowed simple and cost-efficient hardware components. Hybrid assembly via the stacking of different layers enabled “3D fluidic flow.” Serial capillary siphon valving allowed the sequential release of pre-loaded reagents. All the reagents had to be adjusted for siphon-priming using Tween® 20.150
A rectangular injection-molded cartridge, which could be inserted into a centrifugal processing device, was demonstrated by W. Lee et al. The cartridge incorporated a dual-type architecture with two surface acoustic wave (SAW) immunosensors for readout. The liquid flow was controlled by active laser-irradiated ferrowax microvalves, allowing for the preloading of reagents and their release on demand. The sensitivity of the sensor was increased by mass enhancement using gold staining with gold nanoparticle conjugates, along with the detection of Ab targeting biomarkers for acute myocardial infarction. A comparison with a standard laboratory instrument was conducted with 44 patient samples, yielding a correlation coefficient of 0.998.162
Conversely, the required handling steps for cartridges featuring a high degree of parallelization may be conventionally automated off-chip by robotic dispensing, as demonstrated in the Gyrolab Workstation™.197 The corresponding systems must thus be operated at (already automated) laboratories, with the benefit of bringing the aforementioned improvements in centrifugal microfluidics to them.
Independent of the operational site, centrifugal microfluidic systems feature mature process chains for the automation of immunoassays. Unique unit operations that are available solely on centrifugal microfluidic platforms, are the density difference based separation of plasma from blood cells as sample preparation and the excellent performance of bound/free separation by scalable volume forces. The latter enabled the miniaturization of immunoassays to the nanoliter volume while maintaining sufficient sensitivity and specificity, as demonstrated by the Gyrolab Bioaffy LabCD series.200
Future research is expected to further improve automation of immunoassays with respect to point-of-care applications. An emphasis could lie on read-out concepts to increase the parallelization, sensitivity, and multiplexing, or to improve specificity of label-free detection. Another emphasis could lie on the reduction of turnaround times.
3.3 Clinical chemistry
If clinical chemistry parameters can be measured at the point-of-care, patients can be diagnosed faster, and treatment can start immediately. A reduced turnaround time for laboratory tests offers the opportunity to better monitor a patient's health, reduce unnecessary treatments, and reduce hospital costs.201 Examples of parameters that especially benefit from short turnaround times are glucose and electrolytes (e.g., sodium or potassium).201 Centrifugal microfluidics makes it possible to analyze such parameters in a portable device directly from whole blood, by combining centrifuge-based plasma separation with subsequent automated assays.80This has made blood-based clinical chemistry analyzers the most commercially successful field of centrifugal microfluidics. Among the centrifugal microfluidic systems available are the Piccolo Xpress (Abaxis), and the Cobas b 101 (Roche). With a total of 1.5 million cartridges sold in 2011, the Abaxis Piccolo Xpress is currently the most-used system.7
By nature, most commercial systems do not reveal the detailed fluidics. Nonetheless, to discuss blood separation methods as a preparation step for clinical chemistry, this section starts with a review of the blood separation techniques presented in scientific journals. Subsequently, we highlight the major advances in both commercially available and scientific applications of clinical chemistry on centrifugal microfluidic platforms.
| Ref. | Separation principle | Sample volume [μL] | Duration [s] | Yielda [%] | Purity | Maximum hematocrit (HCT) [%] |
|---|---|---|---|---|---|---|
| a Yield is defined as the portion of plasma volume extracted from the total plasma volume. | ||||||
| Burger R. et al.125 | Centrifugo-pneumatic gating | 5 | 120 | 80 | 20 cells μL−1 | N/A |
| Zehnle S. et al.126 | Centrifugo-pneumatic valving | 40 | 43 | 88 | 99.8% | 60 |
| Amasia M. et al.203 | Capillary siphon | 2000 | 320 | 77 | >99.99% | 49 |
| Zhang J. et al.202 | Multi-force separation | 0.5 | 1–2 | 22 | 99% | 6 |
| Haeberle S. et al.127 | Separation by decanting | 5 | 20 | N/A | >99.89% | N/A |
Continuous plasma separation has been demonstrated employing a quasi-isoradial channel, in which the blood cells sediment at the outer perimeter and eventually slide into a waste chamber.127 During this process, the blood plasma also flows into the waste chamber, but remains at a radially inner position due to its lower density. As the waste chamber becomes full, the purified plasma decants into a collection chamber and is available for further downstream processing. The process of cell sedimentation can be amplified by the Coriolis force and the inertial force that pushes the cells toward the outer rims of bent channels.128,202
In batch plasma separation, for the decantation of supernatant plasma after cell sedimentation, a siphon is used in combination with a sedimentation chamber, where the cells are concentrated by centrifugation. Dynamics of cell sedimentation are described by the equilibrium of centrifugal force and drag force (eqn (1)vs.eqn (10)). The inlet position of the siphon is chosen such that it is located radially inward of the shock interface, i.e., the interface between the concentrated cells and purified plasma. Subsequent siphon priming can be accomplished either by capillary action at a greatly reduced spin speed203 or by pneumatic action.84,126 The latter does not require any surface treatment because the pneumatic action is independent of the surface properties. In addition, it enables plasma extraction at a relatively high spin speed, which allows the cell resuspension by Euler forces to be suppressed. Apart from resuspension, the diffusion of cells back into the purified plasma should also be minimized, which can be achieved by creating a small interface between the two chamber compartments for cells and purified plasma.204
An alternative method for batchwise plasma separation without siphon valving has been presented for bead-based immunoassay135 and ELISA.194 After loading the blood sample into the microfluidic disk and the sedimentation of cells by centrifugation, valving of the supernatant plasma was performed by opening a ferro-wax valve. The normally closed valve opened upon laser irradiation with response times of less than 1 s when the disk was at rest.
Nwankire et al. presented a system for point-of-care liver function screening. The analyzer consisted of a small portable disk player and centrifugal microfluidic cartridge. The cartridge included automated blood plasma separation from finger-prick samples. After separation, the purified blood plasma was aliquoted into five reaction chambers via centrifugo-pneumatic aliquoting based on dissolvable films. The reactions were quantified via colorimetric measurements. A translation of the integrated functionality into a schematic description highlighting the combination of process chains and unit operations is depicted in Fig. 15. The authors successfully tested the system in a centralized lab in Nigeria, with a time to result for the complete assay panel of 20 min.206
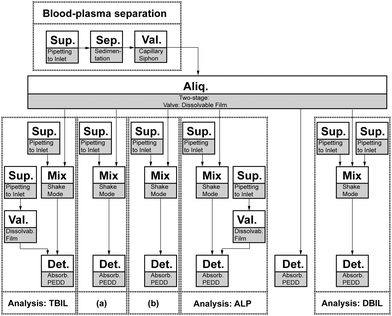 | ||
| Fig. 15 Schematic representation of microfluidic process, including implemented process chains (dashed boxes; TBIL: total bilirubin; (a) albumin; (b) γ-glutamyltransferase; ALP: alkaline phosphatase; DBIL: direct bilirubin) and unit operations (solid boxes; Sup.: supply of reagents or sample; Sep.: separation; Aliq.: aliquoting; Val.: valve; Det.: detection; Mix.: mixing).206 | ||
Lin et al. demonstrated a centrifugal disk for blood coagulation. The disk detects both, partial thromboplastin time and activated partial thromboplastin time. After aliquoting of blood, the blood plasma is separated.207 The separated plasma aliquots are then combined with either a first reagent for quantification of partial thromboplastin time or with a first and second reagent for quantification of the activated partial thromboplastin time.208 Both parameters were quantified via colorimetric measurements in a microfluidic disk analyser.209
Typically, clinical chemistry testing involves absorbance-based measurements such as those applied to determine the concentrations of glucose149 and alcohol100,203 in whole blood.
Recently, an electrochemical lab-on-a-CD system for whole blood analysis was introduced.161 This system incorporates nanoporous electrodes coated with an enzyme layer that triggers the production of H2O2 in the presence of a specific analyte. By applying a potential, the concentration of H2O2 can then be detected electrochemically. The system performance was comparable to colorimetric methods for the tested analytes (glucose, lactate, and uric acid) and could easily be extended to other enzymatic reactions producing H2O2.
Most of the centrifugal microfluidics systems for clinical chemistry reported so far have focused on blood samples. However, a notable exception is a recently presented cartridge featuring an assay for the determination of N-acetyl-β-D-glucosaminidase activity from urine.210 From 15 μL of artificial urine, 330 nL was metered using two-stage metering with capillary valves and mixed with 5 μL of a substrate solution. After 20 min of enzyme reaction, the incubated mixture was transferred via a second capillary valve to the read-out cavity, where it was mixed with a stop solution, and readout was performed using fluorescence detection.
The Abaxis Piccolo Xpress offers a range of cartridges with different lyophilized reagents for a wide variety of whole-blood and blood-plasma tests, including a lipid panel and an electrolyte panel for veterinary and medical diagnostics. All the cartridges are based on the same microfluidic operations, making it a perfect example of a platform-based approach.8 Blood plasma is separated from 100 μL of the patient's blood. At the same time, a pre-stored diluent is released from a central container. A defined volume of diluent and blood plasma are then combined via capillary siphons and mixed using shake-mode mixing. The mixture is subsequently aliquoted into 21 test cavities via one-stage aliquoting. Up to 12 test reactions can be monitored on one cartridge using nine different wavelengths. For online quality control, multiple cuvettes are used to ensure that the sample is introduced and the diluent is released properly.80,211
The Samsung LABGEO A20A system is based on a previously reported combined immunoassay (see Section 3.2) and biochemical analysis of whole blood.137,212 The system reported by B. S. Lee et al. uses up to 350 μL of a patient's blood for both the immunoassay and biochemical analysis. Plasma separation, valving, incubation, washing, mixing, and aliquoting are controlled on the disk using ferrowax valves. In contrast to earlier published methods, the system generates two different dilutions of blood plasma. According to the authors, this allows for the integration of a wider range of assays. Read-out is done by the absorbance at 10 different wavelengths.137 The total reported analysis time for all the liquid operations was 22 min.
The Roche Cobas b 101 currently offers disks for HbA1c and a complete lipid profile. The required blood volumes are 2 μL for the HbA1c test and 19 μL for the lipid profile. The analysis time for each disk is about 6 minutes. A unique feature of the disks is a sideways lid within the disk plane. This lid covers the inlet, which can be used to aspirate a patient's blood directly from a finger stick onto the disk, thereby eliminating the need for pipettes or capillaries.
3.4 Cell handling, separation, and analysis
In the last few years, a growing interest in cell handling on centrifugal microfluidic platforms could be observed.213 Starting from cell suspensions with concentrations generally in the range of 10–103 cells per microliter, researchers have developed methods to isolate, count, and separate different cell types. To date, these methods can be categorized into three different types: geometric, magnetophoretic, and dielectrophoretic approaches.Geometric cell isolation employs centrifugation to pump a suspension of cells along micro-cavities in a centrifugal disk. These cavities are arranged to capture and trap mammalian cells or bacteria, where they can be used to perform an assay.173,214,215 Cell isolation enables studies and analyses of single cells in a defined environment. As an example, the cytotoxicity of paraformaldehyde has been tested using isolated HEK293 cells, and apoptosis tests have successfully been performed with isolated Jurkat cells after UV exposure.214 In order to test the applicability of such isolation methods, cell isolation has been combined with cell viability tests based on cell staining and fluorescence microscopy. In this way, the isolation performance can also be determined by testing the cell occupancy of the cavities on-disk. After cell isolation, single cell PCR makes it possible to determine the cell type, as demonstrated with Salmonella enterica. The bacteria were lysed thermally within the disk, and a specific Salmonella gene was amplified. In this work, the disk consisted of a micro-structured silicon wafer bonded to glass.173 Burger et al. extended their V-cup array for geometrical cell capture under stopped flow (cf. Section 2.6.1) by an optical setup comprising optical tweezers and a fluorescence microscope. In that, cells from different cell lines could be discriminated by fluorescence imaging. As a preparative step for single cell assaying, a single target cell of the HL-60 line could be selected and moved to a defined location within the PDMS disk using the optical tweezers.216
While geometrical cell isolation aims at all cell types within a certain size range, magnetophoresis can be employed to extract specific cells that are tagged to magnetic beads. In this process chain, magnets are used on-disk or off-disk to attract magnetically labeled target cells (positive selection) or non-target cells (negative selection). The magnetically labeled cells can be either deflected or immobilized using the interplay of centrifugal and magnetic forces, and can thus be separated from the non-labeled cells. In positive selection approaches, rare MCF-7 cancer cells have been separated from background Jurkat cells217 or whole blood218 using on-disk magnets. In a negative selection approach, non-target cells labeled with magnetic beads were separated from target MCF-7 cells with rarities down to 10−6. While the labeled non-target cells were kept at a radially inner position, the target cells were centrifuged and concentrated radially outward.164
A further cell-handling possibility was shown using electrically contacting centrifugal microfluidic cartridges.120,219 These made it possible to combine centrifugation with dielectrophoresis. In a hybrid setup, platinum coated glass slides that formed a microfluidic channel were mounted onto a centrifugal disk, together with two 9 V batteries for the power supply and a signal generator. At a spin frequency of 25 Hz, U-937 lymphocytes were separated from erythrocytes in diluted human whole blood.219
Apart from the isolation and purification of cells, the cell count is a central parameter to obtain quantitative diagnostic results. In particular, the hematocrit is a significant indicator for the physiological condition of a patient. With the use of a single dead-end channel in a microfluidic disk, cell sedimentation has been demonstrated in a standard CD drive. After processing, the hematocrit was determined visually from a scale bar on the disk.148
A similar method has been employed to assess the count of bovine somatic cells in milk, as well as the fat content of milk.220 For a case where discrimination between different cell types is not required, a standard CD drive was used to run a modified data CD that incorporated a microfluidic PDMS layer. Once a cell suspension was injected into the microfluidic layer, the CD was run to check the data error rate arising from defects (or biological cells) on the CD. It was shown that the error rate was proportional to the concentration of cells.221
The increasing demand for mobile diagnostic platforms also includes the ability to isolate, count, and discriminate between different white blood cells (WBCs). The first publications in this field had the goal of centrifugation using gradient density media. Such methods take advantage of the fact that different cells have different mass densities. Blood constituents are concentrated by centrifuging the blood, together with one or more gradient density media (DGM) with densities ranging between those of the blood constituents. In this way, concentrated layers of the desired species can be formed, made visible, and quantified by specific fluorescent labeling, and even isolated by siphon valving the different layers.222,223 Park et al. presented a way to use anti-EpCAM to selectively bind rare circulating tumor cells (CTCs) to magnetic beads which were centrifuged and collected separately from a 5 mL blood sample. The high density of the magnetic beads made it possible to centrifuge the bead-bound CTCs through a density gradient medium (DGM) that had a lower density than the beads, but a higher density than the blood sample. In this process chain, the fluidic routing was realized using laser-triggered ferro-wax valves. The procedure included an incubation time of 1 hour to bind the CTCs (100 HCC827 lung cancer cells per 5 mL) to the beads, while a recovery rate of over 95%, cell viability of around 90%, and purity of approximately 12 remaining leukocytes per milliliter could be achieved.224 The implemented sequence of process chains and unit operations for this work is depicted in Fig. 16. Recently, Lee et al. isolated CTCs from whole blood samples circumventing the need for functionalized beads. Instead, a thin membrane with a pore size of 8 μm was implemented in a leak-proof fashion in the centrifugal disk. In this way, more than 50% of MCF-7 cells could be captured from whole blood samples with different concentrations of spiked MCF-7 cells. While red blood cells could be discarded completely, the number of captured white blood cells could be reduced by a factor of 20, compared to the ScreenCell system that was used for reference.119
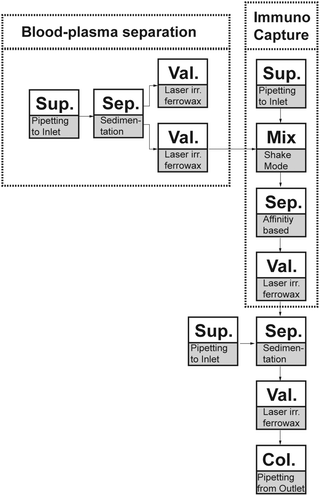 | ||
| Fig. 16 Schematic representation of implemented sequence of process chains (dashed boxes) and unit operations for separation of CTC by immunocapture (solid boxes; Sup.: supply of reagents or samples; Sep.: separation; Val.: valving; Col.: collection of product).224 | ||
3.5 Water, food, and soil analyses
Currently, complex environmental and food quality analyses mostly depend on manual sample collection and analyses with standard laboratory procedures such as autosamplers.225 However, in many cases, these methods are too labor- and cost-intensive for continuous sampling at point-of-care. A possible solution would be a portable bio-sensor, capable of sampling environmental or food samples directly on-site with minimal sample preparation. For this purpose, centrifugal microfluidics is a promising approach. In the following, we describe the available centrifugal microfluidic cartridges for water, food, and soil analyses.Spa and pool water is one of the largest markets for on-site water analysis.226 One commercially available system is the LaMotte Water Spin for pH and ion sensing. Water is inserted into the cartridge via a syringe and split into 10 receiving cavities, containing pre-stored reagents, using one-stage aliquoting. Two different test panels with up to ten different parameters are available for the system: a chlorine disk and biguanide disk.227 These disks are processed, and reactions are read out on a portable instrument using spectrophotometry. According to LaMotte, the system achieves “[…] greater precision than current water labs without time consuming procedures or sacrificing accuracy by using test strip scanners”.14
Other fields for water analysis are waste, river, lake, and sea water. Czugala et al. introduced a cartridge used for turbidity measurement and colorimetric pH analysis. The turbidity is measured from particles at a filter structure integrated directly after the sample inlet. Different pH levels can be measured via the absorbance of prestored ion-gels. Up to seven samples can be processed on one disk (Fig. 17a). The capability of the system was first demonstrated using water samples from the Tolka River (Dublin, Ireland).117
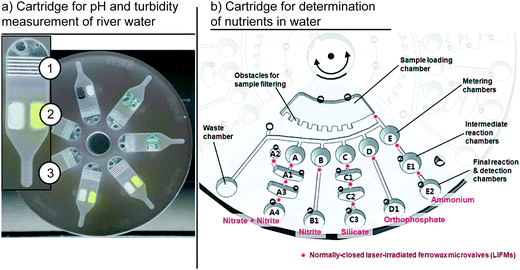 | ||
| Fig. 17 Embodiments of centrifugal microfluidic cartridges for water analysis. (a) Cartridge for turbidity and pH measurement reported to Czugala et al. This cartridge includes a filter region for the removal of solid contaminants larger than 86 μm (1), along with a sensing area (2) and sedimentation region for solid contaminants smaller than 86 μm.117 (Reproduced with permission from The Royal Society of Chemistry.) (b) Cartridge for measurement of nutrients in water.30 Five different reactions can be performed in parallel using a single sample. (Reproduced with permission of the American Chemical Society.) | ||
Hwang et al. showed a disk for the colorimetric detection of nutrients in water. The disk was loaded with up to four samples (Fig. 17b). After the on-disk filtration of particulates, each sample was aliquoted, and the concentrations of five different targets, ammonium, nitrite, nitrate, silicate, and orthophosphate, could be measured in parallel. The integration of the high number of independent tests per sample was made possible via the use of ferrowax-based microvalves for both liquid routing and reagent pre-storage. The first demonstrations of the cartridge were performed using seawater from Chunsu Bay, Korea.30 The integrated process, highlighting the implemented process chains and unit operations, is shown in a schematic representation in Fig. 18.
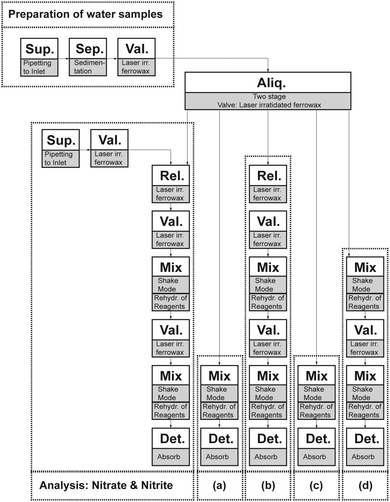 | ||
| Fig. 18 Schematic of integrated functionality reported by Hwang et al.30 The dashed boxes represent the process chains: (a) analysis of nitrite, (b) analysis of silicate, (c) analysis of orthophosphate, and (d) analysis of ammonium. The solid boxes depict the unit operation and demonstrated implementation (Sup.: sample or reagent supply; Val.: valving; Mix: mixing; Aliq.: aliquoting; Det.: detection; Sep.: separation; Rel.: reagent release). | ||
Watts et al. employed four specific ion sensing optodes for the detection of potassium, sodium, calcium, and chloride from aquarium water samples. The presented cartridge incorporated six liquids that were sequentially released using capillary valves of different dimensions. First, a three-point calibration was performed by washing the optodes with three specifically designed calibration solutions. Subsequently, three replicates of the sample solution were measured. The results of the first test using aquarium water samples were in agreement with those of standard laboratory methods, but did not yet reach the same sensitivity.153
LaCroix-Fralish et al. presented a minimalistic single-step centrifugal microfluidic disk for the determination of nitrite and hexavalent chromium in natural water and wastewater. The disk consisted of 24 chambers loaded with dry reagents. In each cavity, an individual sample could be loaded, mixed, and measured using spectrophotometric detection.42 The platform was later extended to two-step reactions using a single capillary valve between two chambers. This cartridge was then used for simultaneous nitrate and nitrite analyses of up to twelve samples each.228 To further extend the dynamic range of the system, Kong et al. included a serial dilution step in the cartridge. After the first measurement in the first cavity, the sample is pumped inward using an external pneumatic source. Part of the sample is metered and mixed with a diluent in a second measurement cavity. The system can be used for the simultaneous determination of aqueous sulfide in up to three samples. The included three-fold dilution allowed for an increase in the dynamic range from 0.4–2.0 mg L−1 to 0.4–6.0 mg L−1.229
To detect trace metals and organic contaminants in drinking water, the pre-concentration of the contaminants is often required.230 Lafleur et al. proposed a cartridge for on-site pre-concentration using solid-phase extraction. This cartridge consisted of an inlet, a silica gel column, and an overflow reservoir. The capability of the cartridge was demonstrated for the quantification of trace metals via inductively coupled plasma mass spectrometry231 and for organic contaminations via fluorescent excitation using an external LED.232 The system could be used for the easy handling of sample material at the point of interest and the later analysis of the cartridge in a laboratory environment.232
A cartridge for the liquid–solid extraction of pyrene, an organic pollutant from soil was presented by Duford et al.233 In this cartridge, three cavities are radially connected via capillary valves. In the first cavity, soil samples are mixed by an inserted magnet and external magnetic fields. The extraction is then transferred to the second chamber, where solid particulates are filtered out via sedimentation. Subsequently, the liquid is transferred to the third chamber, where the target analyte can be quantified via UV-absorbance. The same cartridge concept was later used for the inhibition-based determination of pesticide residues of carbofuran in both soil and vegetable samples.232
A major risk to the integrity of foodstuff and the food supply chain are bio-terroristic attacks. One potential candidate for such attacks is botulinum neurotoxin. A large number of individuals could be affected if this neurotoxin was used to contaminate the environment or food chain. Currently, botulinum neurotoxin is mainly tested in mouse models, which takes several days. Alternative in vitro tests such as ELISA are not sensitive to a wide range of toxin forms and types. Thus, Van Oordt et al. developed a centrifugal microfluidic cartridge for the bioluminescence-based detection of botulinum neurotoxin in water, milk, and other food samples. First, the cartridge is filled with a sample and luciferase-coated bead mixture. The luciferase is linked to the beads via a peptide linker, which is cleaved specifically by enzymatically active botulinum toxin. After the incubation of the beads in the sample, the sample is separated by a siphon structure and combined with a luciferin substrate. The concentration of active botulinum toxin is determined by the intensity of the bioluminescence signal as a result of the luciferase reporter assay.234
Garcia-Cordero et al. developed a centrifugal microfluidic cytometer for milk quality analysis. A milk sample (150 μL) is pipetted into the cartridge. Under artificial gravity during centrifugation, denser cells are pelleted in a dead-end funnel structure. The less-dense fat rises to the top, forming a cream band. By reading out the cell pellets via a microscope, the system can determine cell numbers between 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 to 3
000 to 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 to diagnose bovine mastitis. The fat content of the milk is measured from the cream band in order to additionally estimate the health and nutritional status of the cow.220
000 to diagnose bovine mastitis. The fat content of the milk is measured from the cream band in order to additionally estimate the health and nutritional status of the cow.220
3.6 Analysis of protein structure and function
Proteins are one of the essential building blocks of life. Consequently, an analysis of the structure and function of protein is important for a variety of applications, from basic research to pharmaceutical studies. In the following, we present a selection of the contributions to protein structure analysis using a centrifugal microfluidic platform.Protein structures analyzed by X-ray crystallography still constitute the majority of proteins in the Protein Data Bank. Protein crystallography could benefit significantly from the reduced volumes and increased parallelization offered by microfluidics, because of the large number of different screening conditions needed for generating high-quality protein crystals and the limited amount of purified protein solutions available.102,235
A centrifugal microfluidic cartridge for protein crystallization was presented by Li et al. It automated the metering of 24 different precipitants and the two-stage aliquoting of the protein solution into the respective mixing wells. All the aliquoting and metering was controlled via the capillary filling of inverted V-shaped structures, with the valving controlled by capillary valves. The cartridge was used to demonstrate the on-disk crystallization and analysis of cyan fluorescent protein and lysozyme.102
Steinert et al. presented a cartridge for the protein crystallization screening of up to 100 different precipitants on one disk via free interface diffusion. The disks could be filled with protein volumes down to 1 nL using PipeJet dispensers.163 Protein crystals of lysozyme, proteinase K, insulin, and catalase were successfully grown and could be measured on-chip at a synchrotron beamline.163
3.7 Other applications of centrifugal microfluidics
Apart from the studies covered in the previous chapters, there are numerous creative solutions that do not fit into the previously discussed categories, but deserve to be covered in this review.Gubala et al. introduced a simple cartridge to study biomolecule adsorption in microfluidic channels. A 40 μL sample was introduced on one side of the chip. It was then transported through a microfluidic channel by spinning on a standard spin coater. Part of the volume was extracted, and the concentration of the Cy5 tagged biomolecules was quantified via a fluorescence measurement. The amount of molecules adsorbed could be calculated from the difference in the concentrations before and after processing.236
Bruchet et al. investigated the use of a centrifugal microfluidic platform for the analysis of nuclear spent fuels. In a typical setting, nuclear spent fuels are dissolved in nitric acid and analyzed in a specially shielded hot cell. The authors showed a 1000-fold reduction in the required volume using centrifugal microfluidics, which allowed the analysis to be performed in a glove box. In a first proof of concept, Bruchet et al. showed that a centrifugal microfluidic cartridge with an integrated monolithic anion exchange stationary phase was capable of extracting europium at a yield of ∼97%.237
S.-K. Lee et al. presented a cartridge for the generation of photonic crystals. The cartridge was used to centrifuge suspensions of monodisperse silica or polystyrene latex spheres into dead-end channels, where the nanoparticles formed closely packed columns with predefined shapes. By subsequently spinning different bead solutions, the authors were able to fabricate hybrid colloidal crystals.238
Glass et al. reported on a miniaturized centrifugal microfluidic cartridge for potential use in handheld devices (miniLOAD). The 10 mm disk could be rotated by acoustic actuation, eliminating the need for moving parts. The authors presented valving and mixing as the first simple unit operations on this platform.239
4. Embodiments of centrifugal microfluidic platforms
Many different embodiments (platforms) employing centrifugal microfluidics for a wide range of applications have been demonstrated in the quite short history of the field. Table 9 lists the systems that are either currently commercially available or are in a pre-commercial state. Additionally, we also want to give a brief overview of the history and mention companies that discontinued their developments, but still might be considered, e.g., for patent search.| Ref. | Provider (developer) | Identifier cartridge/name of system | Applications | Commercialization status |
|---|---|---|---|---|
| 13 | Abaxis | Piccolo Xpress | Blood parameter analysis | Commercially available |
| 240 | Samsung | LABGEO IB10 | Immunoassays | Commercially available |
| 241 | Focus Diagnostics (3M) | Universal Disc & Direct Amplification Disk/Integrated Cycler | Nucleic acid analysis | Commercially available |
| 242 | Roche (Panasonic) | Cobas 101b | Blood parameter analysis (HbA1c and lipid panel) | Commercially available |
| 243 | Capital Bio | RTisochip | Nucleic acid analysis (respiratory tract infections) | Commercially available |
| 197 | Gyros AB | Gyrolab Bioaffy CD | Immunoassays | Commercially available |
| 14 | LaMotte | Water Link Spin Lab | Water analysis | Commercially available |
| 244 | Skyla | VB 1 Veterinary Clinical Chemistry Analyzer | Blood chemistry testing for veterinary applications | Commercially available |
| 245 | Biosurfit | Spinit | Immunoassays/blood parameter analysis | Commercially available |
| 246 | Radisens Diagnostics | Unknown | Immunoassay, clinical chemistry, and hematology assays | Precom (planned 2015) |
| 247 | GenePOC-Diagnostics | Unknown | Nucleic acid | Precom (planned 2016) |
| 248 | Spin Chip Diagnostics | Unknown | Blood analysis | Development |
| 174 | Espira Inc. | Unknown | Nucleic acid analysis | Development |
| 36 | Hahn-Schickard | LabTube | Various applications | Development |
| 249 | Sandia National Labs | Spin DX | Various applications | Development |
The history of centrifugal microfluidics dates back to the 1960s, to Oak Ridge National Laboratories' (ORNL) centrifugal analyzer for clinical chemistry.11 At that time, the possibility of increasing the throughput for enzymatic assays compared to conventional flow-through systems led to the first commercialized centrifugal analyzer systems only a few years after the presentation of the original idea, the Electro-Nucleonics Inc. GEMSAEC system, in 1970.250 Centrifugal analyzers exploited centrifugal forces to pump liquid from one point to another, but did not make use of unit operations, e.g., valving to control the fluidic process.251 Following these early days, multiple companies developed and/or commercialized centrifugal analyzers (Centri Union Carbide's “CentrifiChem”, American Instruments' “Rotochem”, Instrumentation Laboratories Inc.'s “Multistat”, and Roche's “Cobas Bio”11). For a more detailed overview of the history, we refer the reader to “Landmark Papers in Clinical Chemistry”252 and Gorkin et al.11
The field gained momentum again with the introduction of the Abaxis Piccolo XPress for the panel analysis of different blood parameters in 1995, a still successful product (Table 9). Besides the success story of the Piccolo XPress, many well-known companies in the field of centrifugal microfluidics discontinued their development for different reasons. The US start-up Gamera developed a “LabCD” system for drug development assays. Gamera was acquired by Tecan in 2000, and Tecan discontinued the development program for “LabCD” in 2005, giving difficulties in the development and delays in the commercialization as the reasons (Tecan press release, July 14, 2005). Spin-X, which used a proprietary virtual laser valve technology for “on-the-fly” valve generation and generic cartridges, discontinued their developments in 2011. Other embodiments of centrifugal microfluidics that have generated IPs include “BCD” by Burstein Technologies; “BioCD” by Quadraspec, which later became Perfinity Biosciences Inc.; Advanced Array Technologies, which later (from 2002 on) became Eppendorf Array Technologies, and Lingvitae.
Furthermore, it is worth naming prominent research groups from academia that made great contributions to progress in the field. Based on the number of publications, the most prominent groups are UC Irvine (Prof. Marc Madou), UNIST (Prof. Yoon-Kyoung Cho), the joint group at IMTEK and Hahn-Schickard (Prof. Roland Zengerle), and BDI (Prof. Jens Ducrée), while many other groups are entering the field and moving forward the state of the art of centrifugal microfluidics at a high pace.
5. General conclusions and outlook
This review aimed to provide a comprehensive description of centrifugal microfluidics, together with its various embodiments (platforms). It also aimed to provide an up-to-date overview of the available set of unit operations (providing basic fluidic functionalities) and how they can be concatenated for the automation of complex laboratory workflows. Additionally, we outlined how recent advances in unit operation development might significantly contribute to the development of centrifugal microfluidics as an enabling technology in the future. We introduced the category “process chain” as an assembly of unit operations representing workflows on a higher level of integration. Process chains can be used as stand-alone solutions for the automation of a particular laboratory process step, or multiple process chains can be combined to realize more complex (bio-medical) applications. Vice versa, we demonstrated how some of the recently published applications using centrifugal microfluidics for automation are already based on the provided set of unit operations.When aiming at the automation of laboratory workflows, the suitability of using centrifugal microfluidics for the desired application must first be evaluated. The decision about the suitability depends (1) on rather general aspects like the overall feasibility of miniaturization, integration, and parallelization, but also (2) on assay-specific details like the available volumes and required assay sensitivity, specificity, yield/efficiency, and reproducibility. The manufacturing technologies for cartridges, which typically need to be disposable, the hybrid integration, and the need for surface treatments will have large influences on the price-per-part and need to be cross checked with the requirements and reimbursement. Equally important are the specifications of the processing device and required auxiliary means. Finally, all the involved processing steps must cope with the application-specific regulations and certifications. The platform approach, with its well-defined unit operations (e.g., known max/min volume, tolerances, and reproducibility) and process chains (e.g., known yield, sensitivity, and specificity) of prior knowledge and art, plays a key role in a cost- and time-efficient layout and design.
The above outlined features are valid for all microfluidic platforms. Nonetheless, we conclude that the specific advantages of centrifugal microfluidics are evident. The single propulsion mechanism of the rotating frame enables the standardization of unit operations with minimum waste of sample and reagent volumes. Volume forces can be adjusted by rotation which enables the efficient removal of any disturbing bubbles and the separation of residual volumes from channels, chambers and sensor matrixes. For sample preparation, the density based separation is inherently available, for example for blood plasma separation. Sample supply is particularly simple: the sample is applied to an inlet cavity and transported further by centrifugation. Hence, the known cross-contamination from systems that need to be connected by a pump is avoided.
Until today, high throughput analysis systems based on centrifugal microfluidics have been realized for clinical chemistry and immunoassays. Gyros, for example, demonstrated the generation of 112 immunoassay data points per cartridge in less than one hour.197 Different Gyrolab CDs comprise the same or very similar centrifugal microfluidic operations such as hydrophobic patch valves, overflow metering and the integration of same sized affinity columns, supporting the idea of using validated unit operations and process chains for efficient product development. For nucleic acid analysis, however, a remaining challenge is the limited number of individual samples that are processed in a given timeframe and a high-throughput nucleic acid analysis system for centrifugal microfluidics has not yet been presented, but might be addressed in future work.
Lately, the storage of pneumatic energy for liquid routing has enabled the monolithic integration of increasingly complex assays, which is a clear trend in centrifugal microfluidics. In this context, the overall system integration, including all aspects of the automation of laboratory workflows, still requires research. For immunoassays and clinical chemistry applications for example, Roche (cobas 101b) and Abaxis (Picolo Xpress) presented fully integrated concepts for the automated pre-storage and release of reagents. For nucleic acid applications however, the cost-efficient mass production of the disposables, including the onboard long-term storage and automated release of reagents, is still a major problem to be solved. Special care must be taken in relation to the properties of the different polymers used. The vapor permeability of the substrate material may cause liquid loss during storage, and the undesired adsorption of target molecules may occur during processing.
These are just a few examples where further research and development is needed. As a consequence, we foresee major research activity in the field of overall system integration, manufacturing, packaging, and parallelization.
Another approach, aiming at a lower market entry barrier, is the concept of using microfluidics as an “App”,191i.e., using already existing laboratory instruments for processing, and thus minimizing the need for high initial investments for processing devices. Microfluidic Apps have successfully been demonstrated for sample preparation in nucleic acid analysis36,181 and for the automated generation of dilution series.253 Both Apps are operated on standard laboratory centrifuges. Other examples have demonstrated multiplexed PCR on different targets on a centrifugal microfluidic cartridge that can be operated in a commercially available PCR thermocycler.254
References
- H. Becker, Lab Chip, 2010, 10, 271 RSC.
- N. Blow, Nat. Methods, 2009, 6, 683–686 CrossRef CAS.
- J. Ducrée, S. Haeberle, S. Lutz, S. Pausch, F. von Stetten and R. Zengerle, J. Micromech. Microeng., 2007, 17, S103 CrossRef.
- S. Haeberle and R. Zengerle, Lab Chip, 2007, 7, 1094 RSC.
- Cepheid - GeneXpert, Cepheid - GeneXpert, available at: http://www.cepheid.com/us/cepheid-solutions/systems/genexpert-systems/genexpert-iv, accessed 29 October 2014.
- Abbott Point of Care - i-STAT system, i-STAT® System - Point-of-Care Testing - Handheld Blood Analyzer', available at: http://www.abbottpointofcare.com/, accessed 29 October 2014.
- Yole Dévelopement SA: POC 2014 Point of Care Testing: Applications of Microfluidic Technologies, 2014.
- D. Mark, S. Haeberle, G. Roth, F. von Stetten and R. Zengerle, Chem. Soc. Rev., 2010, 39, 1153 RSC.
- M. L. Sin, J. Gao, J. C. Liao and P. K. Wong, J. Biol. Eng., 2011, 5, 6 CrossRef PubMed.
- M. Madou, J. Zoval, G. Jia, H. Kido, J. Kim and N. Kim, Annu. Rev. Biomed. Eng., 2006, 8, 601–628 CrossRef CAS PubMed.
- R. Gorkin, J. Park, J. Siegrist, M. Amasia, B. S. Lee, J.-M. Park, J. Kim, H. Kim, M. Madou and Y.-K. Cho, Lab Chip, 2010, 10, 1758 RSC.
- M. C. R. Kong and E. D. Salin, Anal. Chem., 2010, 82, 8039–8041 CrossRef CAS PubMed.
- Abaxis, Abaxis: Piccolo Xpress, available at: http://www.piccoloxpress.com/, accessed 8 May 2014.
- LaMotte, LaMotte: WaterLink Spin Lab, available at: http://www.lamotte.com/en/pool-spa/labs/3576.html, accessed 8 May 2014.
- M. Inganäs, H. Dérand, A. Eckersten, G. Ekstrand, A.-K. Honerud, G. Jesson, G. Thorsén, T. Söderman and P. Andersson, Clin. Chem., 2005, 51, 1985–1987 Search PubMed.
- S. Haeberle, T. Brenner, H.-P. Schlosser, R. Zengerle and J. Ducrée, Chem. Eng. Technol., 2005, 28, 613–616 CrossRef CAS PubMed.
- A. P. Bouchard, D. A. Duford and E. D. Salin, Anal. Chem., 2010, 82, 8386–8389 CrossRef CAS PubMed.
- M. Karle, J. Wöhrle, F. von Stetten, R. Zengerle, D. Mark, Proceedings of Transducers, 2013, pp. 1235–1238.
- M. Rombach, S. Lutz, D. Mark, G. Roth, R. Zengerle, C. Dumschat, A. Witt, S. Hensel, S. Frenzel, F. Aßmann, F. Gehring, T. Reiner, H. Drechsel, P. Szallies and F. von Stetten, Proc. of μTAS, 2012, pp. 782–784.
- Roche, cobas b 101 POC System, available at: http://https://www.roche-diagnostics.ch/de/ProductsRDS/Seiten/cobas-b-101.aspx.
- M. Hitzbleck and E. Delamarche, Chem. Soc. Rev., 2013, 42, 8494–8516 RSC.
- J. Hoffmann, S. Hin, F. von Stetten, R. Zengerle and G. Roth, RSC Adv., 2012, 2, 3885 RSC.
- S. K. Vashist, E. Lam, S. Hrapovic, K. B. Male and J. H. T. Luong, Chem. Rev., 2014, 114, 11083–11130 CrossRef CAS PubMed.
- J. Hoffmann, D. Mark, S. Lutz, R. Zengerle and F. von Stetten, Lab Chip, 2010, 10, 1480 RSC.
- Abaxis, Piccolo Xpress, available at: http://www.piccoloxpress.com/products/piccolo/overview/.
- S. Lutz, P. Weber, M. Focke, B. Faltin, J. Hoffmann, C. Müller, D. Mark, G. Roth, P. Munday, N. Armes, O. Piepenburg, R. Zengerle and F. von Stetten, Lab Chip, 2010, 10, 887 RSC.
- T. van Oordt, Y. Barb, R. Zengerle and F. von Stetten, J. Appl. Polym. Sci., 2014, 131, 40291 CrossRef PubMed.
- T. van Oordt, Y. Barb, J. Smetana, R. Zengerle and F. von Stetten, Lab Chip, 2013, 13, 2888–2892 RSC.
- G. Czilwik, T. Messinger, O. Strohmeier, F. von Stetten, R. Zengerle, P. Saarinen, J. Niittymäki, K. McAllister, O. Sheils, J. Drexler and D. Mark, Proc. of μTAS, 2014, pp. 2528–2529.
- H. Hwang, Y. Kim, J. Cho, J.-y. Lee, M.-S. Choi and Y.-K. Cho, Anal. Chem., 2013, 85, 2954–2960 CrossRef CAS PubMed.
- T. Kawai, N. Naruishi, H. Nagai, Y. Tanaka, Y. Hagihara and Y. Yoshida, Anal. Chem., 2013, 85, 6587–6592 CrossRef CAS PubMed.
- J. L. Garcia-Cordero, F. Benito-Lopez, D. Diamond, J. Ducrée and A. J. Ricco, Proc. of IEEE MEMS, 2009, pp. 439–442.
- J. L. Garcia-Cordero, D. Kurzbuch, F. Benito-Lopez, D. Diamond, L. P. Lee and A. J. Ricco, Lab Chip, 2010, 10, 2680 RSC.
- T.-H. Kim, K. Abi-Samra, V. Sunkara, D.-K. Park, M. Amasia, N. Kim, J. Kim, H. Kim, M. Madou and Y.-K. Cho, Lab Chip, 2013, 13, 3747 RSC.
- K. Abi-Samra, R. Hanson, M. Madou and R. A. Gorkin III, Lab Chip, 2011, 11, 723 RSC.
- A. Kloke, A. R. Fiebach, S. Zhang, L. Drechsel, S. Niekrawietz, M. M. Hoehl, R. Kneusel, K. Panthel, J. Steigert, F. von Stetten, R. Zengerle and N. Paust, Lab Chip, 2014, 14, 1527 RSC.
- M. Focke, F. Stumpf, G. Roth, R. Zengerle and F. von Stetten, Lab Chip, 2010, 10, 3210 RSC.
- M. Focke, F. Stumpf, B. Faltin, P. Reith, D. Bamarni, S. Wadle, C. Müller, H. Reinecke, J. Schrenzel, P. Francois, D. Mark, G. Roth, R. Zengerle and F. von Stetten, Lab Chip, 2010, 10, 2519 RSC.
- O. Strohmeier, S. Laßmann, B. Riedel, D. Mark, G. Roth, M. Werner, R. Zengerle and F. von Stetten, Microchim. Acta, 2014, 181, 1681–1688 CrossRef CAS.
- M. Rombach, D. Kosse, B. Faltin, S. Wadle, G. Roth, R. Zengerle and F. von Stetten, BioTechniques, 2014, 57, 151–155 CAS.
- O. Strohmeier, N. Marquart, D. Mark, G. Roth, R. Zengerle and F. von Stetten, Anal. Methods, 2014, 6, 2038 RSC.
- A. LaCroix-Fralish, J. Clare, C. D. Skinner and E. D. Salin, Talanta, 2009, 80, 670–675 CrossRef CAS PubMed.
- O. Strohmeier, B. Kanat, D. Bär, P. Patel, J. Drexler, M. Weidmann, T. van Oordt, G. Roth, D. Mark, R. Zengerle and F. von Stetten, Proc. of μTAS, 2012, pp. 779–881.
- M. C. R. Kong, A. P. Bouchard and E. D. Salin, Micromachines, 2012, 3, 1–9 CrossRef PubMed.
- S. Soroori, L. Kulinsky, H. Kido and M. Madou, Microfluid. Nanofluid., 2014, 16, 1117–1129 CrossRef CAS PubMed.
- K. Abi-Samra, L. Clime, L. Kong, R. Gorkin, T.-H. Kim, Y.-K. Cho and M. Madou, Microfluid. Nanofluid., 2011, 11, 643–652 CrossRef CAS.
- T. H. G. Thio, F. Ibrahim, W. Al-Faqheri, J. Moebius, N. S. Khalid, N. Soin, M. K. B. A. Kahar and M. Madou, Lab Chip, 2013, 13, 3199 RSC.
- Z. Noroozi, H. Kido and M. J. Madou, J. Electrochem. Soc., 2011, 158, P130 CrossRef CAS PubMed.
- S. Zehnle, F. Schwemmer, G. Roth, F. von Stetten, R. Zengerle and N. Paust, Lab Chip, 2012, 12, 5142 RSC.
- J. L. Garcia-Cordero, L. Basabe-Desmonts, J. Ducrée and A. J. Ricco, Microfluid. Nanofluid., 2010, 9, 695–703 CrossRef.
- C. Li, X. Dong, J. Qin and B. Lin, Anal. Chim. Acta, 2009, 640, 93–99 CrossRef CAS PubMed.
- R. Gorkin, L. Clime, M. Madou and H. Kido, Microfluid. Nanofluid., 2010, 9, 541–549 CrossRef.
- S. Haeberle, N. Schmitt, R. Zengerle and J. Ducrée, Sens. Actuators, A, 2007, 135, 28–33 CrossRef CAS PubMed.
- R. Gorkin, S. Soroori, W. Southard, L. Clime, T. Veres, H. Kido, L. Kulinsky and M. Madou, Microfluid. Nanofluid., 2012, 12, 345–354 CrossRef.
- W. Al-Faqheri, F. Ibrahim, T. H. G. Thio, J. Moebius, K. Joseph, H. Arof and M. Madou, PLoS One, 2013, 8, e58523 CAS.
- D. J. Kinahan, S. M. Kearney, O. P. Faneuil, M. T. Glynn, N. Dimov and J. Ducrée, RSC Adv., 2015, 5, 1818–1826 RSC.
- Y. Ukita, M. Ishizawa, Y. Takamura and Y. Utsumi, Proc. of μTAS, 2012, pp. 1465–1467.
- D. C. Duffy, H. L. Gillis, J. Lin, N. F. Sheppard and G. J. Kellogg, Anal. Chem., 1999, 71, 4669–4678 CrossRef CAS.
- S. Lai, S. Wang, J. Luo, L. J. Lee, S.-T. Yang and M. J. Madou, Anal. Chem., 2004, 76, 1832–1837 CrossRef CAS PubMed.
- M. J. Madou, L. J. Lee, S. Daunert, S. Lai and C.-H. Shih, Biomed. Microdevices, 2001, 3, 245–254 CrossRef CAS.
- F. Schwemmer, S. Zehnle, N. Paust, C. Blanchet, M. Rössle, F. von Stetten, R. Zengerle and D. Mark, Proc. of μTAS, 2012, pp. 1450–1452.
- H. Cho, H.-Y. Kim, J. Y. Kang and T. S. Kim, J. Colloid Interface Sci., 2007, 306, 379–385 CrossRef CAS PubMed.
- M. Liu, J. Zhang, Y. Liu, W. M. Lau and J. Yang, Chem. Eng. Technol., 2008, 31, 1328–1335 CrossRef CAS PubMed.
- J. M. Chen, P.-C. Huang and M.-G. Lin, Microfluid. Nanofluid., 2008, 4, 427–437 CrossRef.
- H. Zhang, H. H. Tran, B. H. Chung and N. Y. Lee, Analyst, 2013, 138, 1750 RSC.
- A. LaCroix-Fralish, E. J. Templeton, E. D. Salin and C. D. Skinner, Lab Chip, 2009, 9, 3151 RSC.
- A. Kazarine, M. C. R. Kong, E. J. Templeton and E. D. Salin, Anal. Chem., 2012, 84, 6939–6943 CrossRef CAS PubMed.
- M. Focke, R. Feuerstein, F. Stumpf, D. Mark, T. Metz, R. Zengerle and F. von Stetten, Proc. of μTAS, 2009, pp. 1397–1399.
- P. Andersson, G. Jesson, G. Kylberg, G. Ekstrand and G. Thorsén, Anal. Chem., 2007, 79, 4022–4030 CrossRef CAS PubMed.
- L. Riegger, M. M. Mielnik, A. Gulliksen, D. Mark, J. Steigert, S. Lutz, M. Clad, R. Zengerle, P. Koltay and J. Hoffmann, J. Micromech. Microeng., 2010, 20, 045021 CrossRef.
- N. Honda, U. Lindberg, P. Andersson, S. Hoffmann and H. Takei, Clin. Chem., 2005, 51, 1955–1961 CAS.
- Y. Ouyang, S. Wang, J. Li, P. S. Riehl, M. Begley and J. P. Landers, Lab Chip, 2013, 13, 1762 RSC.
- D. Mark, T. Metz, S. Haeberle, S. Lutz, J. Ducrée, R. Zengerle and F. von Stetten, Lab Chip, 2009, 9, 3599 RSC.
- R. Gorkin III, C. E. Nwankire, J. Gaughran, X. Zhang, G. G. Donohoe, M. Rook, R. O'Kennedy and J. Ducrée, Lab Chip, 2012, 12, 2894 RSC.
- D. J. Kinahan, S. M. Kearney and J. Ducrée, Proceedings of Transducers, 2013, pp. 2189–2192.
- J. Siegrist, R. Gorkin, M. Bastien, G. Stewart, R. Peytavi, H. Kido, M. Bergeron and M. Madou, Lab Chip, 2010, 10, 363 RSC.
- W. Al-Faqheri, F. Ibrahim, T. H. G. Thio, N. Bahari, H. Arof, H. A. Rothan, R. Yusof and M. Madou, Sensors, 2015, 15, 4658–4676 CrossRef CAS PubMed.
- J. Hoffmann, D. Mark, R. Zengerle and F. von Stetten, Proceedings of Transducers, 2009, pp. 1991–1994.
- H. Hwang, H.-H. Kim and Y.-K. Cho, Lab Chip, 2011, 11, 1434 RSC.
- C. T. Schembri, T. L. Burd, A. R. Kopf-Sill, L. R. Shea and B. Braynin, J. Autom. Chem., 1995, 17, 99–104 CrossRef CAS PubMed.
- J. Siegrist, R. Gorkin, L. Clime, E. Roy, R. Peytavi, H. Kido, M. Bergeron, T. Veres and M. Madou, Microfluid. Nanofluid., 2010, 9, 55–63 CrossRef.
- N. Godino, E. Vereshchagina, R. Gorkin and J. Ducrée, Microfluid. Nanofluid., 2013, 16, 895–905 CrossRef.
- R. Burger, N. Reis, J. G. Fonseca and J. Ducree, Proc. of IEEE MEMS, 2009, pp. 443–446.
- N. Godino, R. Gorkin III, A. V. Linares, R. Burger and J. Ducrée, Lab Chip, 2013, 13, 685 RSC.
- F. Schwemmer, S. Zehnle, D. Mark, F. von Stetten, R. Zengerle and N. Paust, Lab Chip, 2015, 15, 1545–1553 RSC.
- D. Kinahan, S. M. Kearney, N. Dimov, M. T. Glynn and J. Ducree, Lab Chip, 2014, 14, 2249–2258 RSC.
- J.-M. Park, Y.-K. Cho, B.-S. Lee, J.-G. Lee and C. Ko, Lab Chip, 2007, 7, 557 RSC.
- U. Y. Schaff and G. J. Sommer, Clin. Chem., 2011, 57, 753–761 CAS.
- Y.-K. Cho, J.-G. Lee, J.-M. Park, B.-S. Lee, Y. Lee and C. Ko, Lab Chip, 2007, 7, 565 RSC.
- M. Amasia, M. Cozzens and M. J. Madou, Sens. Actuators, B, 2012, 161, 1191–1197 CrossRef CAS PubMed.
- L. Swayne, A. Kazarine, E. J. Templeton and E. D. Salin, Talanta, 2015, 134, 443–447 CrossRef CAS PubMed.
- T. Brenner, T. Glatzel, R. Zengerle and J. Ducree, Lab Chip, 2005, 5, 146 RSC.
- J. Kim, H. Kido, R. H. Rangel and M. J. Madou, Sens. Actuators, B, 2008, 128, 613–621 CrossRef CAS PubMed.
- T. T. Thuy, M. Inganäs, G. Ekstrand and G. Thorsén, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2010, 878, 2803–2810 CrossRef CAS PubMed.
- D. Mark, M. Rombach, S. Lutz and R. Zengerle, Proc. of μTAS, 2009, pp. 110–112.
- M. Müller, D. Mark, M. Rombach, G. Roth, J. Hoffmann, R. Zengerle and F. von Stetten, Proc. of μTAS, 2010, pp. 405–407.
- M. C. R. Kong and E. D. Salin, Anal. Chem., 2011, 83, 1148–1151 CrossRef CAS PubMed.
- D. Mark, P. Weber, S. Lutz, M. Focke, R. Zengerle and F. Stetten, Microfluid. Nanofluid., 2011, 10, 1279–1288 CrossRef CAS.
- J. Steigert, T. Brenner, M. Grumann, L. Riegger, S. Lutz, R. Zengerle and J. Ducrée, Biomed. Microdevices, 2007, 9, 675–679 CrossRef CAS PubMed.
- J. Steigert, M. Grumann, T. Brenner, L. Riegger, J. Harter, R. Zengerle and J. Ducree, Lab Chip, 2006, 6, 1040 RSC.
- S. O. Sundberg, C. T. Wittwer, C. Gao and B. K. Gale, Anal. Chem., 2010, 82, 1546–1550 CrossRef CAS PubMed.
- G. Li, Q. Chen, J. Li, X. Hu and J. Zhao, Anal. Chem., 2010, 82, 4362–4369 CrossRef CAS PubMed.
- M. Grumann, A. Geipel, L. Riegger, R. Zengerle and J. Ducrée, Lab Chip, 2005, 5, 560 RSC.
- Y. Ren and W. W.-F. Leung, Int. J. Heat Mass Transfer, 2013, 60, 95–104 CrossRef PubMed.
- Z. Noroozi, H. Kido, M. Micic, H. Pan, C. Bartolome, M. Princevac, J. Zoval and M. Madou, Rev. Sci. Instrum., 2009, 80, 075102 CrossRef PubMed.
- Z. Noroozi, H. Kido, R. Peytavi, R. Nakajima-Sasaki, A. Jasinskas, M. Micic, P. L. Felgner and M. J. Madou, Rev. Sci. Instrum., 2011, 82, 064303 CrossRef PubMed.
- M. M. Aeinehvand, F. Ibrahim, S. W. Harun, W. Al-Faqheri, T. H. G. Thio, A. Kazemzadeh and M. Madou, Lab Chip, 2014, 14, 988–997 RSC.
- S. Haeberle, T. Brenner, H.-P. Schlosser, R. Zengerle and J. Ducrée, Chem. Eng. Technol., 2005, 28, 613–616 CrossRef CAS PubMed.
- J. Ducrée, T. Brenner, S. Haeberle, T. Glatzel and R. Zengerle, Microfluid. Nanofluid., 2006, 2, 78–84 CrossRef.
- J. Ducrée, S. Haeberle, T. Brenner, T. Glatzel and R. Zengerle, Microfluid. Nanofluid., 2006, 2, 97–105 CrossRef.
- D. Chakraborty, M. Madou and S. Chakraborty, Lab Chip, 2011, 11, 2823 RSC.
- Y. Ukita and Y. Takamura, Microfluid. Nanofluid., 2013, 15, 829–837 CrossRef CAS.
- J.-N. Kuo and L.-R. Jiang, Microsyst. Technol., 2014, 20, 91–99 CrossRef CAS PubMed.
- M. La, S. J. Park, H. W. Kim, J. J. Park, K. T. Ahn, S. M. Ryew and D. S. Kim, Microfluid. Nanofluid., 2013, 15, 87–98 CrossRef CAS.
- J. Liebeskind, A. Kloke, A. R. Fiebach, F. von Stetten, R. Zengerle and N. Paust, Proc. of μTAS, 2013, pp. 967–969.
- M. C. R. Kong and E. D. Salin, Microfluid. Nanofluid., 2012, 13, 519–525 CrossRef CAS.
- M. Czugala, R. Gorkin III, T. Phelan, J. Gaughran, V. F. Curto, J. Ducrée, D. Diamond and F. Benito-Lopez, Lab Chip, 2012, 12, 5069 RSC.
- E. J. Templeton and E. D. Salin, Microfluid. Nanofluid., 2014, 17, 245–251 CrossRef.
- A. Lee, J. Park, M. Lim, V. Sunkara, S. Y. Kim, G. H. Kim, M.-H. Kim and Y.-K. Cho, Anal. Chem., 2014, 86, 11349–11356 CrossRef CAS PubMed.
- R. Martinez-Duarte, R. A. Gorkin III, K. Abi-Samra and M. J. Madou, Lab Chip, 2010, 10, 1030 RSC.
- M. Boettcher, M. S. Jaeger, L. Riegger, J. Ducrée, R. Zengerle and C. DUSCHL, Biophys. Rev. Lett., 2006, 1, 443–451 CrossRef CAS.
- R. Burger, P. Reith, G. Kijanka, V. Akujobi, P. Abgrall and J. Ducrée, Lab Chip, 2012, 12, 1289 RSC.
- D. Kirby, J. Siegrist, G. Kijanka, L. Zavattoni, O. Sheils, J. O'Leary, R. Burger and J. Ducrée, Microfluid. Nanofluid., 2012, 13, 899–908 CrossRef CAS.
- M. Glynn, D. Kirby, D. Chung, D. J. Kinahan, G. Kijanka and J. Ducrée, J. Lab. Autom., 2013, 19, 285–296 CrossRef PubMed.
- R. Burger, N. Reis, J. G. da Fonseca and J. Ducrée, J. Micromech. Microeng., 2013, 23, 035035 CrossRef.
- S. Zehnle, M. Rombach, F. von Stetten, R. Zengerle and N. Paust, Proc. of μTAS, 2012, pp. 869–871.
- S. Haeberle, T. Brenner, R. Zengerle and J. Ducree, Lab Chip, 2006, 6, 776 RSC.
- B.-S. Li and J.-N. Kuo, NEMS, 2013, 462–465 Search PubMed.
- R. Boom, et al. , J. Clin. Microbiol., 1990, 495–503 CAS.
- J. H. Jung, B. H. Park, Y. K. Choi and T. Seo, Lab Chip, 2013, 13, 3383–3388 RSC.
- B. H. Park, J. H. Jung, H. Zhang, N. Y. Lee and T. S. Seo, Lab Chip, 2012, 12, 3875 RSC.
- X. Y. Peng, P. C. Li, H.-Z. Yu, M. Parameswaran and W. L. Chou, Sens. Actuators, B, 2007, 128, 64–69 CrossRef CAS PubMed.
- G. Jia, K.-S. Ma, J. Kim, J. V. Zoval, R. Peytavi, M. G. Bergeron and M. J. Madou, Sens. Actuators, B, 2006, 114, 173–181 CrossRef CAS PubMed.
- R. Peytavi, F. R. Raymond, D. Gagne, F. J. Picard, G. Jia, J. Zoval, M. Madou, K. Boissinot, M. Boissinot, L. Bissonnette, M. Ouellette and M. G. Bergeron, Clin. Chem., 2005, 51, 1836–1844 CAS.
- B. S. Lee, J.-N. Lee, J.-M. Park, J.-G. Lee, S. Kim, Y.-K. Cho and C. Ko, Lab Chip, 2009, 9, 1548 RSC.
- H. Nagai, Y. Narita, M. Ohtaki, K. Saito and S.-I. Wakida, Anal. Sci., 2007, 23, 975–979 CrossRef CAS.
- B. S. Lee, Y. U. Lee, H.-S. Kim, T.-H. Kim, J. Park, J.-G. Lee, J. Kim, H. Kim, W. G. Lee and Y.-K. Cho, Lab Chip, 2011, 11, 70 RSC.
- O. Strohmeier, A. Emperle, G. Roth, D. Mark, R. Zengerle and F. von Stetten, Lab Chip, 2013, 13, 146–155 RSC.
- K.-C. Chen, T.-P. Lee, Y.-C. Pan, C.-L. Chiang, C.-L. Chen, Y.-H. Yang, B.-L. Chiang, H. Lee and A. M. Wo, Clin. Chem., 2011, 57, 586–592 CAS.
- S. Haeberle, R. Zengerle and J. Ducrée, Microfluid. Nanofluid., 2007, 3, 65–75 CrossRef.
- D. Chakraborty and S. Chakraborty, Appl. Phys. Lett., 2010, 97, 234103 CrossRef PubMed.
- D. Mark, S. Haeberle, R. Zengerle, J. Ducree and G. T. Vladisavljević, J. Colloid Interface Sci., 2009, 336, 634–641 CrossRef CAS PubMed.
- S. Haeberle, L. Naegele, R. Burger, F. von Stetten, R. Zengerle and J. Ducrée, J. Microencapsul., 2008, 25, 267–274 CrossRef CAS PubMed.
- K. Maeda, H. Onoe, M. Takinoue and S. Takeuchi, Adv. Mater., 2012, 24, 1340–1346 CrossRef CAS PubMed.
- F. Schuler, F. Schwemmer, M. Trotter, S. Wadle, R. Zengerle, F. von Stetten and N. Paust, Lab Chip, 2015 10.1039/C5LC00291E.
- T.-H. Kim, J. Park, C.-J. Kim and Y.-K. Cho, Anal. Chem., 2014, 86, 3841–3848 CrossRef CAS PubMed.
- M. M. Hoehl, E. S. Bocholt, A. Kloke, N. Paust, F. von Stetten, R. Zengerle, J. Steigert and A. H. Slocum, Analyst, 2014, 16, 375–385 CAS.
- L. Riegger, M. Grumann, J. Steigert, S. Lutz, C. P. Steinert, C. Mueller, J. Viertel, O. Prucker, J. Rühe, R. Zengerle and J. Ducrée, Biomed. Microdevices, 2007, 9, 795–799 CrossRef CAS PubMed.
- M. Grumann, J. Steigert, L. Riegger, I. Moser, B. Enderle, K. Riebeseel, G. Urban, R. Zengerle and J. Ducrée, Biomed. Microdevices, 2006, 8, 209–214 CrossRef CAS PubMed.
- C. E. Nwankire, G. G. Donohoe, X. Zhang, J. Siegrist, M. Somers, D. Kurzbuch, R. Monaghan, M. Kitsara, R. Burger, S. Hearty, J. Murrell, C. Martin, M. Rook, L. Barrett, S. Daniels, C. McDonagh, R. O'Kennedy and J. Ducrée, Anal. Chim. Acta, 2013, 781, 54–62 CrossRef CAS PubMed.
- L. Riegger, M. Grumann, T. Nann, J. Riegler, O. Ehlert, W. Bessler, K. Mittenbuehler, G. Urban, L. Pastewka, T. Brenner, R. Zengerle and J. Ducrée, Sens. Actuators, A, 2006, 126, 455–462 CrossRef CAS PubMed.
- Y. Ukita and Y. Takamura, Microfluid. Nanofluid., 2015, 18, 245–252 CrossRef CAS.
- A. S. Watts, A. A. Urbas, E. Moschou, V. G. Gavalas, J. V. Zoval, M. Madou and L. G. Bachas, Anal. Chem., 2007, 79, 8046–8054 CrossRef CAS PubMed.
- K. Otsuka, A. Hemmi, T. Usui, A. Moto, T. Tobita, N. Soh, K. Nakano, H. Zeng, K. Uchiyama, T. Imato and H. Nakajima, J. Sep. Sci., 2011, 34, 2913–2919 CrossRef PubMed.
- S. Morais, L. A. Tortajada-Genaro, T. Arnandis-Chover, R. Puchades and A. Maquieira, Anal. Chem., 2009, 81, 5646–5654 CrossRef CAS PubMed.
- R. Burger, M. Kitsara, J. Gaughran, C. Nwankire and J. Ducrée, Future Med., 2014, 72–92 Search PubMed.
- Y. Li, L. M. L. Ou and H.-Z. Yu, Anal. Chem., 2008, 80, 8216–8223 CrossRef CAS PubMed.
- S. A. Lange, G. Roth, S. Wittemann, T. Lacoste, A. Vetter, J. Grässle, S. Kopta, M. Kolleck, B. Breitinger, M. Wick, J. K. H. Hörber, S. Dübel and A. Bernard, Angew. Chem., Int. Ed., 2006, 45, 270–273 CrossRef CAS PubMed.
- F. G. Bosco, E.-T. Hwu, C.-H. Chen, S. Keller, M. Bache, M. H. Jakobsen, I.-S. Hwang and A. Boisen, Lab Chip, 2011, 11, 2411–2416 RSC.
- K. Abi-Samra, T.-H. Kim, D.-K. Park, N. Kim, J. Kim, H. Kim, Y.-K. Cho and M. Madou, Lab Chip, 2013, 13, 3253–3260 RSC.
- T. Li, Y. Fan, Y. Cheng and J. Yang, Lab Chip, 2013, 13, 2634 RSC.
- W. Lee, J. Jung, Y. K. Hahn, S. K. Kim, Y. Lee, J. Lee, T.-H. Lee, J.-Y. Park, H. Seo, J. N. Lee, J. H. Oh, Y.-S. Choi and S. S. Lee, Analyst, 2013, 138, 2558–2566 RSC.
- C. P. Steinert, J. Mueller-Dieckmann, M. Weiss, M. Roessle, R. Zengerle and P. Koltay, Proc. of MEMS, 2007, pp. 561–564.
- C.-L. Chen, K.-C. Chen, Y.-C. Pan, T.-P. Lee, L.-C. Hsiung, C.-M. Lin, C.-Y. Chen, C.-H. Lin, B.-L. Chiang and A. M. Wo, Lab Chip, 2011, 11, 474 RSC.
- J. Kim, M. Johnson, P. Hill and B. K. Gale, Integr. Biol., 2009, 1, 574 RSC.
- P.-A. Auroux, Y. Koc, A. deMello, A. Manz and P. J. R. Day, Lab Chip, 2004, 4, 534 RSC.
- J. Kim, S. Hee Jang, G. Jia, J. V. Zoval, N. A. Da Silva and M. J. Madou, Lab Chip, 2004, 4, 516 RSC.
- H. Kido, M. Micic, D. Smith, J. Zoval, J. Norton and M. Madou, Colloids Surf., B, 2007, 58, 44–51 CrossRef CAS PubMed.
- T. Brenner, T. Glatzel, R. Zengerle and J. Ducree, Proc. of μTAS, 2003, pp. 903–906.
- S. Wadle, O. Strohmeier, M. Rombach, D. Mark, R. Zengerle and F. von Stetten, Proc. of μTAS, 2012, pp. 1381–1383.
- O. Strohmeier, S. Keil, B. Kanat, P. Patel, M. Niedrig, M. Weidmann, F. Hufert, J. Drexler, R. Zengerle and F. von Stetten, RSC Adv., 2015, 5, 32144–32150 RSC.
- J. H. Jung, S. J. Choi, B. H. Park, Y. K. Choi and T. S. Seo, Lab Chip, 2012, 12, 1598 RSC.
- S. Furutani, H. Nagai, Y. Takamura and I. Kubo, Anal. Bioanal. Chem., 2010, 398, 2997–3004 CrossRef CAS PubMed.
- Espira Inc., Espira Inc: Digital PCR, available at: http://www.espirainc.com/digital-pcr.html, accessed 8 May 2014.
- M. Focke, D. Kosse, D. Al-Bamerni, S. Lutz, C. Müller, H. Reinecke, R. Zengerle and F. von Stetten, J. Micromech. Microeng., 2011, 21, 115002 CrossRef.
- G. Czilwik, I. Schwarz, M. Keller, S. Wadle, S. Zehnle, F. von Stetten, D. Mark, R. Zengerle and N. Paust, Lab Chip, 2015, 15, 1084–1091 RSC.
- GenomeWeb, http://www.genomeweb.com/pcrsample-prep/fda-clears-focus-diagnostics-flu-test-3m-integrated-cycler, available at: http://www.genomeweb.com/pcrsample-prep/fda-clears-focus-diagnostics-flu-test-3m-integrated-cycler, accessed 21 October 2013.
- Focus Diagnostics, http://www.mikrogen.de/uploads/tx_oemikrogentables/dokumente/PI-UM-MOL1101-DE.pdf, available at: http://www.mikrogen.de/uploads/tx_oemikrogentables/dokumente/PI-UM-MOL1101-DE.pdf, accessed 21 October 2013.
- L. Wang, P. C. Li, H.-Z. Yu and A. M. Parameswaran, Anal. Chim. Acta, 2008, 610, 97–104 CrossRef CAS PubMed.
- L. Wang and P. C. Li, Anal. Biochem., 2010, 400, 282–288 CrossRef CAS PubMed.
- M. M. Hoehl, M. Weißert, A. Dannenberg, T. Nesch, N. Paust, F. Stetten, R. Zengerle, A. H. Slocum and J. Steigert, Biomed. Microdevices, 2014, 16, 375–385 CAS.
- G. Czilwik, O. Strohmeier, I. Schwarz, N. Paust, S. Zehnle, F. von Stetten, R. Zengerle and D. Mark, Proc. of μTAS, 2013, pp. 1607–1609.
- J. H. Jung, B. H. Park, S. J. Choi and T. S. Seo, Proc. of μTAS, 2012, pp. 1966–1968.
- The 3M™ Integrated Cycler Direct Amplification Disc - International, available at: http://https://www.focusdx.com/3m-integrated-cycler/dad-intl, accessed 15 July 2014.
- M. Exner, L. Jacky, Y.-P. Chen, H. Mai, J. Chen, M. M. Tabb, M. Aye and E. Eleazar, US20130022963A, 2013.
- P. D. Ludowise, D. A. Whitman and F. D. Smith, US20120291538A1, 2012.
- P. D. Ludowise and J. D. Smith, US2012/0291565A1, 2010.
- R. Peytavi and S. Chapdelaine, WP2012/120463A1, 2012.
- L. Bissonnette, S. Chapdelaine, R. Peytavi, A. Huletsky, G. Stewart, M. Boissinot, P. Allibert and M. G. Bergeron, ed. G. J. Kost and C. M. Corbin, Global Point of Care - Strategies for Disasters, Emergencies, and Public Health Resilience, AACC Press, Washington, DC, 2015, pp. 235–247 Search PubMed.
- P. J. Asiello and A. J. Baeumner, Lab Chip, 2011, 11, 1420 RSC.
- D. Mark, F. von Stetten and R. Zengerle, Lab Chip, 2012, 12, 2464 RSC.
- H. He, Y. Yuan, W. Wang, N.-R. Chiou, A. J. Epstein and L. J. Lee, Biomicrofluidics, 2009, 3, 22401 CrossRef PubMed.
- R. D. Johnson, I. H. A. Badr, G. Barrett, S. Lai, Y. Lu, M. J. Madou and L. G. Bachas, Anal. Chem., 2001, 73, 3940–3946 CrossRef CAS.
- J. Park, V. Sunkara, T.-H. Kim, H. Hwang and Y.-K. Cho, Anal. Chem., 2012, 84, 2133–2140 CrossRef CAS PubMed.
- L. Riegger, J. Steigert, M. Grumann, S. Lutz, G. Olofsson, M. Kayyami, W. Bessler, K. Mittenbuehler, R. Zengerle and J. Ducree, Proc. of μTAS, 2006, pp. 819–821.
- G. Welte, S. Lutz, B. Cleven, H. Brahms, C. Gärtner, G. Roth, D. Mark, R. Zengerle and F. von Stetten, Proc. of μTAS, 2010, pp. 818–820.
- Gyros AB, Gyros: Gyrolab, available at: http://www.gyros.com/, accessed 8 May 2014.
- C. Y. Koh, U. Y. Schaff, A. K. Singh and G. J. Sommer, μTAS, 2011.
- H. Cho, J. Kang, S. Kwak, K. Hwang, J. Min, J. Lee, D. Yoon and T. Kim, Proc. of IEEE MEMS, 2005, pp. 698–701.
- Gyrolab Bioaffy system, Gyrolab CDs, available at: http://www.gyros.com/products/gyrolab-cds/, accessed 7 May 2015.
- G. P. Zaloga, Chest, 1990, 97, 185S CrossRef CAS.
- J. Zhang, Q. Guo, M. Liu and J. Yang, J. Micromech. Microeng., 2008, 18, 125025 CrossRef.
- M. Amasia and M. Madou, Bioanalysis, 2010, 2, 1701–1710 CrossRef CAS PubMed.
- T. Li, L. Zhang, K. M. Leung and J. Yang, J. Micromech. Microeng., 2010, 20, 105024 CrossRef.
- C. A. Burtis, J. C. Mailen, W. F. Johnson, C. D. Scott, T. O. Tiffany and N. G. Anderson, Clin. Chem., 1972, 18, 753–761 CAS.
- C. E. Nwankire, M. Czugala, R. Burger, K. J. Fraser, T. M. O'Connell, T. Glennon, B. E. Onwuliri, I. E. Nduaguibe, D. Diamond and J. Ducrée, Biosens. Bioelectron., 2014, 56, 352–358 CrossRef CAS PubMed.
- C.-H. Lin, C.-H. Shih and C.-H. Lu, J. Nanosci. Nanotechnol., 2013, 13, 2206–2212 CrossRef CAS PubMed.
- C.-H. Lin, C.-Y. Liu, C.-H. Shih and C.-H. Lu, Biomicrofluidics, 2014, 8, 052105 CrossRef PubMed.
- C.-H. Lin, K.-W. Lin, D. Yen, C.-H. Shih, C.-H. Lu, J.-M. Wang and C.-Y. Lin, J. Nanosci. Nanotechnol., 2015, 15, 1401–1407 CrossRef CAS PubMed.
- Y. Tanaka, S. Okuda, A. Sawai and S. Suzuki, Anal. Sci., 2012, 28, 33–38 CrossRef CAS.
- T. L. Burd, Clin. Chem., 1992, 38, 1665–1670 Search PubMed.
- J. Suk-Anake and C. Promptmas, Clin. Lab., 2012, 58, 1313–1318 CAS.
- R. Burger, D. Kirby, M. Glynn, C. Nwankire, M. O'Sullivan, J. Siegrist, D. Kinahan, G. Aguirre, G. Kijanka, R. A. Gorkin and J. Ducrée, Curr. Opin. Chem. Biol., 2012, 16, 409–414 CrossRef CAS PubMed.
- S.-W. Lee, J. Y. Kang, I.-H. Lee, S.-S. Ryu, S.-M. Kwak, K.-S. Shin, C. Kim, H.-I. Jung and T.-S. Kim, Sens. Actuators, A, 2008, 143, 64–69 CrossRef CAS PubMed.
- H. Chen, X. Li, L. Wang and P. C. Li, Talanta, 2010, 81, 1203–1208 CrossRef CAS PubMed.
- R. Burger, D. Kurzbuch, R. Gorkin, G. Kijanka, M. Glynn, C. McDonagh and J. Ducrée, Lab Chip, 2015, 15, 378–381 RSC.
- K.-C. Chen, Y.-C. Pan, C.-L. Chen, C.-H. Lin, C.-S. Huang and A. M. Wo, Anal. Biochem., 2012, 429, 116–123 CrossRef CAS PubMed.
- D. Kirby, G. Kijanka, J. Siegrist, J. Burger, O. Sheils, J. O'Leary and J. Ducree, Proc. of μTAS, 2012, pp. 1126–1128.
- M. Boettcher, M. S. Jaeger, L. Riegger, J. Ducree, R. Zengerle and C. Duschl, Biophys. Rev. Lett., 2006, 1, 443–451 CrossRef CAS.
- J. L. Garcia-Cordero, L. M. Barrett, R. O'Kennedy and A. J. Ricco, Biomed. Microdevices, 2010, 12, 1051–1059 CrossRef CAS PubMed.
- S. M. Imaad, N. Lord, G. Kulsharova and G. L. Liu, Lab Chip, 2011, 11, 1448 RSC.
- U. Schaff, A. Tentori and G. Sommer, Proc. of μTAS, 2010, pp. 103–105.
- D. J. Kinahan, M. T. Glynn, S. M. Kearney and J. Ducrée, Proc. of μTAS, 2012, pp. 1363–1365.
- J.-M. Park, M. S. Kim, H.-S. Moon, C. E. Yoo, D. Park, Y. J. Kim, K.-Y. Han, J.-Y. Lee, J. H. Oh, S. S. Kim, W.-Y. Park, W.-Y. Lee and N. Huh, Anal. Chem., 2014, 86, 3735–3742 CrossRef CAS PubMed.
- S. Rodriguez-Mozaz, M. J. Lopez de Alda and D. Barceló, Anal. Bioanal. Chem., 2006, 386, 1025–1041 CrossRef CAS PubMed.
- Global Water Intelligence, 2009.
- LaMotte, WaterLink Spin Lab, available at: http://www.lamotte.com/en/pool-spa/labs/3576.html.
- Y. Xi, E. J. Templeton and E. D. Salin, Talanta, 2010, 82, 1612–1615 CrossRef CAS PubMed.
- M. C. R. Kong and E. D. Salin, Anal. Chem., 2012, 84, 10038–10043 CrossRef CAS PubMed.
- J. P. Lafleur and E. D. Salin, J. Anal. At. Spectrom., 2009, 24, 1511 RSC.
- J. P. Lafleur, A. A. Rackov, S. McAuley and E. D. Salin, Talanta, 2010, 81, 722–726 CrossRef CAS PubMed.
- D. A. Duford, Y. Xi and E. D. Salin, Anal. Chem., 2013, 85, 7834–7841 CrossRef CAS PubMed.
- Y. Xi, D. A. Duford and E. D. Salin, Talanta, 2010, 82, 1072–1076 CrossRef CAS PubMed.
- T. van Oordt, G. B. Stevens, S. K. Vashist, R. Zengerle and F. von Stetten, RSC Adv., 2013, 3, 22046 RSC.
- L. Li and R. F. Ismagilov, Annu. Rev. Biophys., 2010, 39, 139–158 CrossRef CAS PubMed.
- V. Gubala, J. Siegrist, R. Monaghan, B. O'Reilly, R. P. Gandhiraman, S. Daniels, D. E. Williams and J. Ducrée, Anal. Chim. Acta, 2013, 760, 75–82 CrossRef CAS PubMed.
- A. Bruchet, V. Taniga, S. Descroix, L. Malaquin, F. Goutelard and C. Mariet, Talanta, 2013, 116, 488–494 CrossRef CAS PubMed.
- S.-K. Lee, G.-R. Yi and S.-M. Yang, Lab Chip, 2006, 6, 1171 RSC.
- N. R. Glass, R. J. Shilton, P. P. Y. Chan, J. R. Friend and L. Y. Yeo, Small, 2012, 8, 1881–1888 CrossRef CAS PubMed.
- Samsung, Samsung: LABGEO IB10, available at: http://www.samsungmedison.de/labgeo_ib10.aspx, accessed 19 September 2013.
- Focus Diagnostics, 3M INTEGRATED CYCLER, available at: http://www.focusdx.com/3m-integrated-cycler, accessed 8 May 2014.
- Roche, Roche: COBAS b 101, available at: http://www.cobas.com/home/product/cobas-b-101-poc-system.html, accessed 8 May 2014.
- Capitalbio, Capitalbio: RTisochip, available at: http://www.bioon.com.cn/product/show_product.asp?id=284704, accessed 8 May 2014.
- Skyla, Skyla: VB 1: Veterinary Clinical Chemistry Analyzer, available at: http://www.skyla.com/veterinarydetail.php?id=3, accessed 16 September 2014.
- Biosurfit, Biosurfit: SpinIt, available at: http://www.biosurfit.com/, accessed 8 May 2014.
- Radisens Diagnostics, Radisens Diagnostics, available at: http://www.radisens.com/, accessed 8 May 2014.
- Gene POC, Gene POC, available at: http://www.genepoc-diagnostics.com/Home.shtml, accessed 8 May 2014.
- SpinChip Diagnostics AS, SpinChip Diagnostics AS, available at: http://www.spinchip.no/, accessed 8 May 2014.
- J. Euske, Sandia National Laboratories: Licensing/Technology Transfer SpinDx™: Point-of-Care Diagnostics Using Centrifugal Microfluidics, available at: http://https://ip.sandia.gov/technology.do/techID=82, accessed 8 May 2014.
- N. G. Anderson, Clin. Chim. Acta, 1969, 25, 321–330 CrossRef CAS.
- M. J. Felton, Anal. Chem., 2003, 75, 302A CAS.
- R. M. Rocco, Clin. Chem., 2006, 52, 1977 Search PubMed.
- O. Strohmeier, M. Rombach, D. Mark, R. Zengerle, G. Roth and F. von Stetten, Proceedings of Transducers, 2011, pp. 2952–2955.
- M. Focke, O. Strohmeier, P. Reith, G. Roth, D. Mark, R. Zengerle and F. von Stetten, Proc. of μTAS, 2011, pp. 659–661.
Footnote |
| † Authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2015 |