Autocatalytic amplified detection of DNA based on a CdSe quantum dot/folic acid electrochemiluminescence energy transfer system†
Guifen
Jie
*,
Yingqiang
Qin
,
Qingmin
Meng
and
Jialin
Wang
*
Key Laboratory of Sensor Analysis of Tumor Marker, Ministry of Education, College of Chemistry and Molecular Engineering, Qingdao University of Science and Technology, Qingdao 266042, P.R. China. E-mail: guifenjie@126.com; wangjialin5518@sina.com
First published on 29th August 2014
Abstract
In this work, electrochemiluminescence energy transfer (ECL-ET) from CdSe quantum dots (QDs) to folic acid is reported for the first time and applied for amplified detection of DNA by using a novel DNAzyme-mediated autocatalytic system.
The development of DNA biosensors capable of amplified detection of DNA is being greatly motivated by its potential applications in molecular diagnostics, forensic investigations, genetics therapy, and biomedical development,1 and numerous electrical,2 optical,3 or microgravimetric4 amplified DNA sensors have been reported. Different catalysts, such as enzymes,5 catalytic nucleic acids (DNAzymes),6 or metal nanoparticles (NPs),7 were used for the amplified detection of DNA. Amplified DNA detection was accomplished by an autocatalytic and catabolic DNAzyme-mediated process.8 The easy synthetic preparation of DNAzymes and the reduced nonspecific adsorption of DNAzymes make catalytic nucleic acids attractive reporter units. The use of DNA-based machines has recently been suggested as a means to stimulate the autonomous replication of a catalytic reporter as the result of DNA sensing.9 A further approach for amplifying the DNA detection has included the use of the Zn2+-dependent ligation DNAzyme and the isothermal autonomous synthesis of the ligation DNAzyme units, as a result of the DNA recognition event.10
Electrochemiluminescence (ECL) as a promising tool has attracted much attention in sensing and detecting trace amounts of samples due to its high sensitivity, wide dynamic concentration response range, as well as its potential and spatial control.11,12 Recent research demonstrated that QDs are promising ECL tags for selective determination of macromolecules in real samples.13 The marriage of energy transfer with electrochemiluminescence has produced a new technology named electrochemiluminescence energy transfer (ECL-ET), which can realize effective and sensitive detection of biomolecules.14 To obtain optimal ECL-ET efficiency, a perfect energy overlapped donor/acceptor pair is of great importance. Notably, the tunable emission wavelength and broad absorption spectra of QDs render them to be an ideal active donor or acceptor for ECL-ET.14
Considerable work has been done in NC ECL resonance energy transfer (RET). Xu and coworkers first demonstrated that the ECL from manganese-doped CdS NCs could be quenched by proximal Au NPs, such an ECL energy transfer platform allowed DNA detection.14 Subsequently, QD-based ECL energy transfer assays for DNA detection,15 cytosensing,16 aptamers,17 and immunoassay have been further explored.18 However, most of the ECL-ET assays used gold nanoparticles as quenchers, which have limits in biolabeling due to the steric hindrance to the electrode. Folic acid as a micromolecule was reported to act as an effective quencher for fluorescence.19 Furthermore, DNAzymes as enzyme mimics have attracted considerable attention in ECL techniques due to their high catalytic activity and good stability.20 So far, QD-based ECL-ET assays using folic acid as the quencher combined with a cyclic amplification technique have not been reported. Herein, we present a novel strategy for DNAzyme-mediated autocatalytic amplified detection of DNA based on electrochemiluminescence energy transfer (ECL-ET) from CdSe quantum dots to folic acid.
Characterization of the CdSe QDs
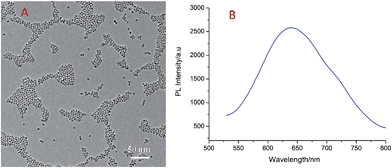
Fig. 1A shows the transmission electron microscopy (TEM) image of the CdSe QDs; the nanoparticles possessed uniform morphology and size and the average diameter was about 5–6 nm. Fig. 1B presents the photoluminescence (PL) spectra of the CdSe QDs; the PL emission peak of the QDs was at 650 nm and the PL intensity was high, indicating that CdSe QDs possess good luminescent properties and have promising optical applications.ECL behavior of the CdSe quantum dots
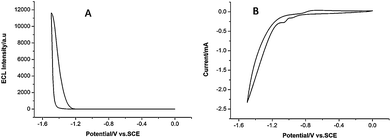
The ECL–potential curve and cyclic voltammogram (CV) of the CdSe quantum dots are shown in Fig. 2. A cathodic peak at −1.05 V is observed in the CV response, corresponding to the reduction of S2O82−. There is an ECL peak at −1.40 V in the ECL–potential curve, which results from the reaction of the CdSe QDs with S2O82−. The possible ECL mechanisms are as follows. The QDs are reduced to nanocrystalline species (QD−˙), while S2O82− is reduced to strong oxidant SO4−˙. Then SO4−˙ reacts with QD−˙ to form the excited state QD* that could emit light.| QD + e− → QD−˙ | (1) |
| S2O82− + e− → SO42− + SO4−˙ | (2) |
| QD−˙ + SO4−˙ → QD* + SO42− | (3) |
| QD* → QD + hν | (4) |
Mechanism of DNAzyme-mediated amplified detection of DNA based on ECL quenching of CdSe QDs by folic acid
The amplified ECL detection of DNA based on a Mg2+-dependent DNAzyme autocatalytic system was successfully designed.Scheme 1 shows the fabrication principle of the strategy. CdSe QDs were first assembled on the electrode by using PAMAM–CNT nanocomposites, showing a high ECL signal. The single-stranded hairpin loop of sequence 4 contains an adenine ribonucleotide and acts as the substrate for the Mg2+-dependent DNAzyme. The stem region of hairpin DNA4 contains the sequence of the target analyte DNA 1, and the quencher folic acid is tethered to the stem end. After the hairpin substrate with the quencher was linked to the CdSe QDs on the electrode, the QD ECL was effectively quenched. Upon interaction of analyte 1 with unit 3, the hairpin structure is opened, then units 2 and 3 were assembled into the active DNAzyme structure. The synergistic binding of the substrate 4 to the DNAzyme structure results in the cleavage of the hairpin substrate in the presence of Mg2+ ions, and the fragment with the quencher was released, then the QD ECL was enhanced. The quencher unit includes the base sequence of the analyte, which leads to the enhanced formation of the DNAzyme structure and the increased cleavage of unit 4, with the concomitant generation of a higher ECL signal for analyte sensing by the autocatalytic amplified technique.
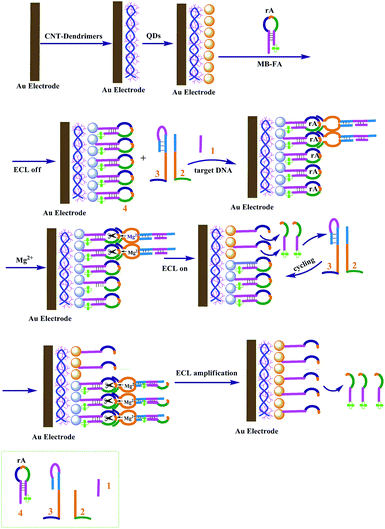 | ||
| Scheme 1 Schematic representation of the strategy of amplified ECL detection of DNA based on a Mg2+-dependent DNAzyme autocatalytic system. | ||
Preparation and characterization of the modified electrode for ECL detection
Fig. 3 shows the field-emission scanning electron microscopy (FE-SEM) images of the modified electrodes. The negatively charged multi-walled carbon nanotubes (CNTs) were first assembled on the electrode by using PAMAM dendrimers, then the CdSe QDs were covalently conjugated to the dendrimer NCs. It could be found that a thick film of PAMAM–CNTs nanocomposites was homogeneously formed on the electrode (Fig. 3A). By comparison, the well-dispersed CdSe QDs spread over the electrode more uniformly due to the smaller size of QDs (Fig. 3B).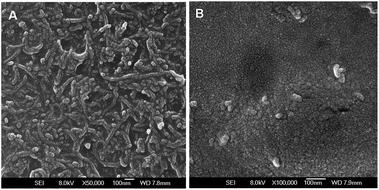 | ||
| Fig. 3 Representative SEM images of (A) PAMAM–CNTs; (B) PAMAM–CNT–QDs on the surface of the electrode. | ||
DNAzyme-mediated amplified detection of DNA based on ECL quenching of CdSe QDs by folic acid
The strategy feasibility of DNA detection based on ECL quenching of CdSe QDs by folic acid was investigated. As shown in Fig. S1,† a very low background ECL signal was observed on the bare electrode (curve a). When the PAMAM–CNT–CdSe composite QDs were assembled on the electrode, a high and stable ECL signal was obtained (curve b), indicating that PAMAM–CNT was an excellent immobilization matrix for CdSe composite QDs. Then folic acid as the ECL quencher was close to the QDs on the electrode; the ECL signal obviously decreased (curve c), demonstrating that folic acid efficiently quenched the QD ECL. In the presence of the target analyte together with the mixture containing units 2, 3 and Mg2+ ions, the ECL signal obviously enhanced (curve d), indicating that the DNAzyme-mediated amplification strategy based on ECL quenching of the QDs could be applied to the detection of analyte DNA.Fig. 4 shows the ECL signal changes upon analyzing different concentrations of target DNA. In the presence of target DNA by the DNAzyme-mediated autocatalytic amplification strategy, the ECL peak signal gradually increased with increasing concentrations of analyte DNA (curves a to f), indicating that the autocatalytic amplification strategy based on ECL quenching of CdSe QDs by folic acid enables the detection of the target DNA.
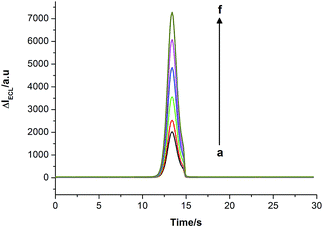 | ||
| Fig. 4 ECL signal changes for detection of different concentrations of target DNA. The concentrations of DNA (nmol L−1): (a) 0.05; (b) 0.1; (c) 1.0; (d) 10.0; (e) 100.0; (f) 1000.0. | ||
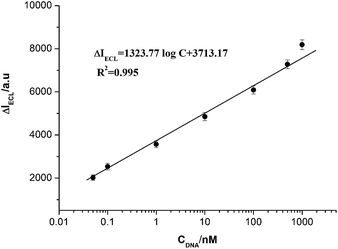
Fig. 5 displays the relationship between the ECL signal changes (ΔI) and the concentrations of analyte DNA (0.05 nmol L−1 to 1000 nmol L−1). ΔI was logarithmically related to the analyte concentrations in the range from 0.05 nmol L−1 to 1000 nmol L−1 (R = 0.995) with a detection limit of 0.05 nmol L−1 at 3σ, which is comparable to the previously reported detection of DNA. A series of five duplicate measurements of 1.0 nmol L−1 were used for estimating the precision, and the relative standard deviation (RSD) was 6.8%, showing good reproducibility of the ECL method.
The selectivity of the autocatalytic amplified detection of DNA based on ECL quenching of CdSe QDs was investigated by using unit 3 to hybridize with completely complementary tDNA, single-base mismatched DNA and noncomplementary DNA sequences, respectively (Fig. S2†). The ECL signal change for single-base mismatched sequences was significantly smaller than that of the completely complementary sequences, and the noncomplementary sequences showed no response. The results suggest that the autocatalytic amplified strategy has good selectivity for DNA detection.
In summary, we provide a novel autocatalytic amplification strategy for detection of DNA based on ECL energy transfer from CdSe QDs to folic acid. Several advantages of this method have been demonstrated: (1) ECL-ET from CdSe QDs to folic acid was reported for the first time and applied to amplified detection of DNA, which overcome steric hindrance in traditional quenchers such as gold nanoparticles; (2) the DNAzyme-mediated autocatalytic system was combined with QD ECL for DNA detection, which exhibits higher sensitivity and specificity, and rapid speed; (3) the present work is the first report on the ECL-ET from QDs to folic acid combined with a cyclic amplification technique for amplified detection of DNA, which opens up a promising new approach to QD-based ECL bioassays, and has extensive potential in analytical applications. In addition, the ECL-ET system also has the disadvantage of time-consumption in electrode modification, which should stimulate more promising technology in ECL bioassay of QDs.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (no. 21175078), and the Program for Changjiang Scholars and Innovative Research Team in University (PCSIRT).Notes and references
- (a) F. Wang, J. Elbaz, R. Orbach, N. Magen and I. Willner, J. Am. Chem. Soc., 2011, 133, 17149 CrossRef CAS PubMed; (b) M. Zhou and S. J. Dong, Acc. Chem. Res., 2011, 44, 1232 CrossRef CAS PubMed.
- W. Tan, K. Wang and T. J. Drake, Curr. Opin. Chem. Biol., 2004, 8, 547 CrossRef CAS PubMed.
- Y. Lu and J. Liu, Curr. Opin. Biotechnol., 2006, 17, 580 CrossRef CAS PubMed.
- F. Lucarelli, S. Tombelli, M. Minunni, G. Marrazza and M. Mascini, Anal. Chim. Acta, 2008, 609, 139 CrossRef CAS PubMed.
- G. Liu, Y. Wan, V. Gau, J. Zhang, L. Wang, S. Song and C. Fan, J. Am. Chem. Soc., 2008, 130, 6820 CrossRef CAS PubMed.
- (a) C. H. Lu, F. Wang and I. Willner, J. Am. Chem. Soc., 2012, 134, 10651 CrossRef CAS PubMed; (b) F. Wang, J. Elbaz, C. Teller and I. Willner, Angew. Chem., 2011, 123, 309 CrossRef.
- S. J. Kwon and A. J. Bard, J. Am. Chem. Soc., 2012, 134, 10777 CrossRef CAS PubMed.
- F. Wang, J. Elbaz, C. Teller and I. Willner, Angew. Chem., Int. Ed., 2011, 50, 295 CrossRef CAS PubMed.
- W. Xu, X. Xue, T. Li, H. Zeng and X. Liu, Angew. Chem., 2009, 121, 6981 CrossRef.
- F. Wang, J. Elbaz and I. Willner, J. Am. Chem. Soc., 2012, 134, 5504 CrossRef CAS PubMed.
- S. F. Liu, X. Zhang, Y. M. Yu and G. Z. Zou, Anal. Chem., 2014, 86, 2784 CrossRef CAS PubMed.
- G. F. Jie, L. Wang, J. X. Yuan and S. S. Zhang, Anal. Chem., 2011, 83, 3873 CrossRef CAS PubMed.
- G. Liang, S. Liu, G. Zou and X. Zhang, Anal. Chem., 2012, 84, 10645 CrossRef CAS PubMed.
- M.-S. Wu, H.-W. Shi, L.-J. He, J.-J. Xu and H.-Y. Chen, Anal. Chem., 2012, 84, 4207 CrossRef CAS PubMed.
- H. Zhou, Y.-Y. Zhang, J. Liu, J.-J. Xu and H.-Y. Chen, Chem. Commun., 2013, 49, 2246 RSC.
- G. F. Jie and J. X. Yuan, Anal. Chem., 2012, 84, 2811 CrossRef CAS PubMed.
- C.-Y. Tian, J.-J. Xu and H.-Y. Chen, Chem. Commun., 2012, 48, 8234 RSC.
- Y. Shan, J.-J. Xu and H.-Y. Chen, Chem. Commun., 2010, 46, 5079 RSC.
- S. Santra, C. Kaittanis, O. J. Santiesteban and J. M. Perez, J. Am. Chem. Soc., 2011, 133, 16680 CrossRef CAS PubMed.
- H. Zhou, Y. Y. Zhang, J. Liu, J. J. Xu and H. Y. Chen, Chem. Commun., 2013, 49, 2246 RSC.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental details, UV-vis, TEM and the optimization, selectivity and additional figures. See DOI: 10.1039/c4an01465k |
| This journal is © The Royal Society of Chemistry 2015 |