Open Access Article
Open Access ArticleInteraction of silver atomic quantum clusters with living organisms: bactericidal effect of Ag3 clusters mediated by disruption of topoisomerase–DNA complexes†
J.
Neissa‡
a,
C.
Pérez-Arnaiz‡
 b,
V.
Porto
a,
N.
Busto
b,
V.
Porto
a,
N.
Busto
 b,
E.
Borrajo
a,
J. M.
Leal
b,
E.
Borrajo
a,
J. M.
Leal
 b,
M. A.
López-Quintela
b,
M. A.
López-Quintela
 c,
B.
García
c,
B.
García
 *b and
F.
Dominguez
*a
*b and
F.
Dominguez
*a
aDepartment of Physiology and Centro de Investigaciones en Medicina Molecular y Enfermedades Crónicas (CIMUS), University of Santiago de Compostela, E-15782 Santiago de Compostela, Spain. E-mail: fernando.dominguez@usc.es
bDepartment of Chemistry, University of Burgos, E-9001 Burgos, Spain. E-mail: begar@ubu.es
cDepartment of Physical Chemistry, Fac. Chemistry and Nanomag Laboratory, IIT. University of Santiago de Compostela, E-15782 Santiago de Compostela, Spain
First published on 14th July 2015
Abstract
Essential processes for living cells such as transcription and replication depend on the formation of specific protein–DNA recognition complexes. Proper formation of such complexes requires suitable fitting between the protein surface and the DNA surface. By adopting doxorubicin (DOX) as a model probe, we report here that Ag3 atomic quantum clusters (Ag-AQCs) inhibit the intercalation of DOX into DNA and have considerable influence on the interaction of DNA-binding proteins such as topoisomerase IV, Escherichia coli DNA gyrase and the restriction enzyme HindIII. Ag-AQCs at nanomolar concentrations inhibit enzyme activity. The inhibitory effect of Ag-AQCs is dose-dependent and occurs by intercalation into DNA. All these effects, not observed in the presence of Ag+ ions, can explain the powerful bactericidal activity of Ag-AQCs, extending the knowledge of silver bactericidal properties. Lastly, we highlight the interest of the interaction of Ag clusters with living organisms, an area that should be further explored due to the potential consequences that it might have, both beneficial and harmful.
Introduction
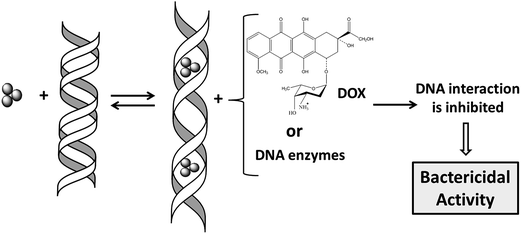
The bactericidal properties of silver have been known since ancient times; however, nowadays the underlying mechanisms are only poorly understood. Whether silver nanoparticles (NPs),1 silver ions (Ag+)2 or both are responsible for the cytotoxicity of silver is open to discussion,2–4 largely because of their relative bioavailability.5 Cells can reduce Ag+ to the metallic form, which constitutes a well-known biosynthetic method to prepare NPs.6 On the other hand, NPs taken up into eukaryotic cells dissolve quickly and chemical species of silver change over time from Ag0 to Ag–O– to Ag–S– forms.7 NPs are unstable in the presence of excess of S-containing chemicals such as glutathione, present in cells at millimolar levels,8 which can etch NPs to produce Ag clusters.9 At the same time, in the course of the formation of NPs by reduction of Ag+, stable Ag clusters can also be formed.10 Therefore, the possibility that Ag clusters are present in bacteria exposed to silver should not be excluded.Earlier, we have reported that Ag3 clusters (denoted here as Ag-AQCs) interact with DNA through intercalation.11 The effect exerted by Ag-AQCs on the DNA conformation is stronger than by classical intercalators such as acridines or ethidium bromide, mainly for two reasons: (a) Ag atoms have larger radii than C atoms and the unwinding and subsequent lengthening of the double helix induced by Ag-AQCs is in consequence larger than that occasioned by polycyclic aromatic ligands;11 and (b) the dissociation rate constant for the Ag-AQCs/DNA complex is several orders of magnitude lower than those with classical intercalators,11 thus prolonging the residence time in the intercalated position. In a topologically closed DNA domain, changes in the secondary structure are immediately reflected by a change in the overall shape, helping to disrupt the protein binding. This outcome raises naturally the question of whether Ag-AQCs can inhibit the binding of DNA-ligands. To shed some light on this issue, we firstly address the effect of Ag-AQCs, incubated in DNA, on the DOX–DNA binding. Likewise, for comparison purposes, a parallel study replacing Ag-AQCs by Ag+ ions was carried out. DOX is a renowned member of the anthracycline family, a very effective type of anticancer drugs currently in use.12–14 Recently, we have studied the interaction of DOX with calf thymus DNA (ct-DNA), reporting that this system is able to form a strong intercalated-groove DOX–DNA complex. The formation of this complex is categorized kinetically as a two-step mechanism in which the fast step is the groove binding of the amino sugar moiety and the slow step is the intercalation of the anthracycline group.15 We hypothesize that the distortion of the DNA groove as a consequence of the intercalation of Ag-AQC reported earlier11 is responsible for the inhibition of the binding of DOX to DNA.
We then study the effect of the Ag-AQCs incubated in DNA on major groove binding proteins such as topoisomerase IV, E. coli DNA gyrase and the restriction enzyme HindIII. Lastly, the effects of Ag-AQCs in living organisms were assessed by evaluation of their antibacterial activity. A schematic diagram of the research performed is outlined in Fig. 1.
Results and discussion
Influence of Ag-AQCs and Ag+ ions on the DNA–DOX interaction
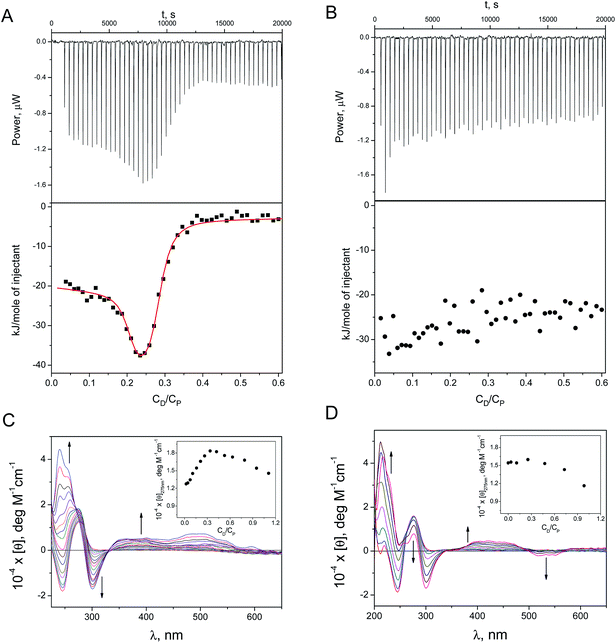
The study of ternary systems containing DOX, DNA and Ag-AQCs (or silver ions, used for comparison purposes) was conducted by isothermal titration calorimetry (ITC) and circular dichroism (CD) measurements. This physicochemical study will allow us to learn the effect caused by the cluster, once intercalated into the DNA, on different types of groove-binders. The study is composed of two types of experiments: (1) titration with DOX of a DNA sample previously incubated overnight with submicromolar amounts of Ag+ (CDNA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CAg+ = 200), and (2) identical conditions, replacing Ag+ by Ag-AQCs.
CAg+ = 200), and (2) identical conditions, replacing Ag+ by Ag-AQCs.
The ITC profile was generated by adding micromolar amounts of DOX (CD, molar concentration) to: (a) ct-DNA + Ag+ (Fig. 2A), and (b) ct-DNA + Ag-AQCs (Fig. 2B), (CP stands for the DNA concentration expressed as molar base pairs). Table 1 collects the thermodynamic binding constants, K1 and K2, obtained for DOX/(ct-DNA + Ag+) using a two-sites model; for comparison, the same data for the DOX/ct-DNA system is also included. The small diminution of the K1 and K2 values for DOX/(ct-DNA + Ag+) relative to DOX/ct-DNA indicates that the Ag+ ions exert only a small effect on the binding of DOX to ct-DNA. In contrast, a substantially different behavior was observed in the presence of Ag-AQCs (Fig. 2B); Ag-AQCs species are responsible for the observed full inhibition of the intercalation of DOX into ct-DNA, even for very small CDNA/CAg-AQCs ratios.
| K 1 (M−1) | K 2 (M−1) | K (M−1) | |
|---|---|---|---|
| a From ITC experiments; K1 and K2 for DOX–DNA taken from ref. 15. b From fluorescence experiments. | |||
| DOX/ctDNA | (2.3 ± 0.8) × 108 | (9.3 ± 0.7) × 105 | |
| DOX/(ctDNA + Ag+) | (4 ± 2) × 107 | (7 ± 1) × 105 | |
| DOX/(ctDNA + Ag-AQCs) | — | — | |
| (DOX + Ag-AQCs)/ctDNA | (1.0 ± 0.4) × 107 | (6 ± 1) × 104 | |
| DOX/Ag-AQC | (2.9 ± 0.1) × 108 | ||
Fig. 2 also shows the CD spectra recorded upon addition of micromolar amounts of DOX to (ct-DNA + Ag+) and to (ct-DNA + Ag-AQCs). The set of CD spectra and the molar ellipticity versus CD/CP plot recorded for DOX/(ct-DNA + Ag+) at λ = 275 nm (Fig. 2C, inset) and those for DOX/ct-DNA15 are very similar, meaning that the effect of the Ag+ ions on the CD spectra is trivial. In contrast, the pattern obtained with Ag-AQCs differs noticeably; disappearance of the positive bands at 240 and 500 nm, and the observed increase of the negative band at 540 nm both denote heavy structural distortion of the DNA. The molar ellipticity profile at λ = 275 nm (Fig. 2D, inset) reveals that, in the presence of Ag-AQC, the strong intercalated-groove complex is not formed for both DOX–DNA and DOX/(DNA + Ag+) systems, as stacking interactions are characterized by positive variation around 275 nm.15 However, the observed diminution of [θ] for CD/CP > 0.4 indicates that an external complex could be formed.
Likewise, formation of the non-fluorescent complex DOX/Ag-AQCs has been studied in the absence of DNA. Fig. 1SIA† shows the isotherm plot; fitting of the data pairs (see ESI†) yielded the value K = (2.9 ± 0.1) × 108 M−1. Regarding absorption measurements, the isosbestic points at 300 and 350 nm in the set of spectral curves of Fig. 1SIB† support interaction between DOX and Ag-AQCs; by contrast, lack of spectral changes in the titration of DOX with Ag+ reveals that no reaction is at work.
The ITC results have shown that the interaction with DNA of the species resulting from DOX + Ag-AQCs differs only slightly from the interaction of DOX with DNA (Fig. 2SI†); the constants obtained for DOX–DNA were only an order of magnitude greater than those obtained for (DOX + Ag-AQCs)/DNA (see Table 1).
In summary, so far we can conclude that Ag-AQCs intercalated into DNA are responsible for the observed non-intercalation of DOX into DNA and also that Ag-AQCs strongly interact with DOX in the absence of DNA. This study affords useful information to understand the mechanisms of inhibition of biological groove-binding agents, such as proteins, by Ag-AQCs.
Ag-AQCs affect the biological activity of DNA-binding proteins
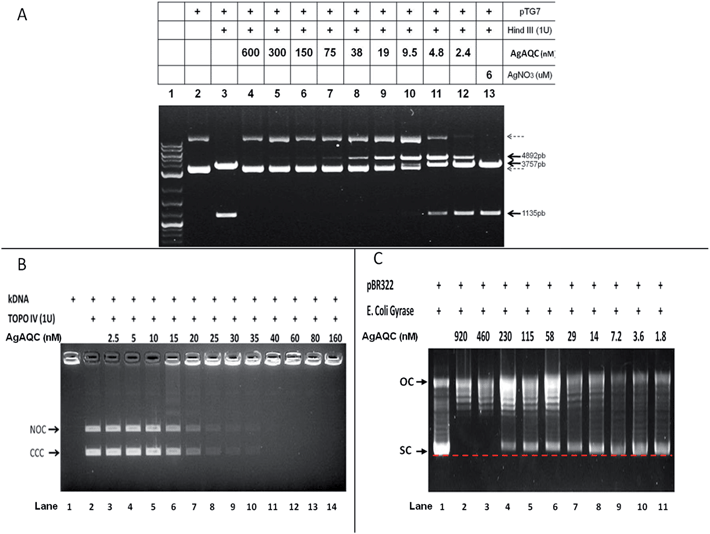
We also looked into the influence of Ag-AQCs on groove-binding agents such as DNA-binding proteins, for Ag-AQCs inhibit the binding of DOX to DNA. Initially, we focused on proteins that can bind to DNA in a dependent sequence mode, such as restriction enzymes; these enzymes recognize DNA with pronounced selectivity, as this is the basis to provide defense mechanism against invading foreign DNA. HindIII is a restriction enzyme that cuts pTG7 (4892 bp plasmid DNA), releasing two fragments of 3757 and 1135 bp (Fig. 3A, lane 3). Ag-AQCs at nanomolar levels inhibited HindIII restriction activity (Fig. 3A, lanes 4–12). On the other hand, Ag+ at 6 μM concentration, higher than any dose of Ag-AQCs used, produced no effect (Fig. 3A, lane 13). Thus, as occurs with DOX intercalation, the inhibitory action of the enzymatic cleavage is characteristic of Ag-AQCs, but not of Ag+ ions.DNA binding proteins can recognize DNA binding sites by two types of mechanisms: recognition of specific DNA sequence16 and recognition of DNA geometry such as type II topoisomerases.17 Bacteria require topoisomerases, that is, proteins that specifically alter the DNA topology to enable DNA replication. Therefore, we also studied the effects of Ag-AQCs on two members of bacterial type II topoisomerases, topoisomerase IV (Topo IV) and DNA gyrase.
The decatenation activity assay for Topo IV utilizes the kinetoplast DNA (kDNA), a large network of interlocked (catenated) circles, which cannot enter the agarose gel.18E. coli Topo IV decatenates the circles from the network (Fig. 3B, lane 1); the free circles are detected upon decatenation as discrete bands on the gel (Fig. 3B), nicked open-circular, and covalently closed-circular DNA (NOC and CCC, respectively). Ag-AQCs at nanomolar concentration levels inhibited the Topo IV activity (Fig. 3B, lanes 3–14) with an IC50 value of (roughly) 16 nM.
Topo IV, a type II enzyme with remarkable similarity in sequence to DNA gyrase, can relax but not introduce negative supercoils into DNA.19 The activity of E. coli gyrase can be assessed by a supercoiling based assay in which a relaxed plasmid DNA, pBR322, is treated with E. coli DNA gyrase. This enzyme converts the relaxed topoisomers to the SC form of the plasmid, which migrates faster on agarose gel. Also an upper band corresponding to the nicked OC DNA is visible, which was also present in the relaxed substrate, but co-migrates with some of the relaxed topoisomers (Fig. 3C, lane 1). Ag-AQCs at nanomolar levels inhibited the activity of E. coli gyrase (Fig. 3C, lanes 2–11). Notably, we found that SC DNA had less mobility in the presence of increasing Ag-AQCs concentrations (Fig. 3C, lanes 4–11, dotted red line) concurrent with the observation that the negative SC DNA may have slightly less mobility than normal, depending on the amount of intercalant bound to DNA.20 The different mobility of SC DNA clearly indicates that the Ag-AQCs inhibitory effect occurs by intercalation into DNA in a dose-dependent manner, the inhibition being greater the larger the number of Ag-AQCs intercalated into DNA.
Ag-AQCs bactericidal activity
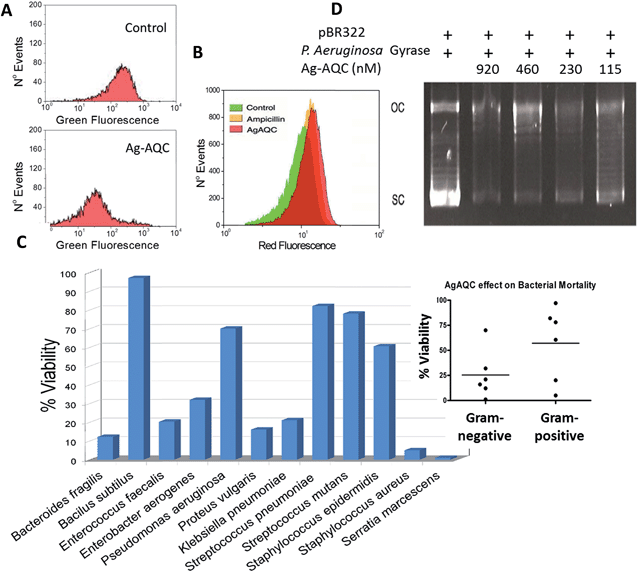
The topological consequences of intercalation are even more evident in bacteria, whose circular genomes exist in a tightly regulated topological state.21 Actually, DNA gyrase is affected in vitro by the presence of Ag-AQCs, therefore we explored whether it could also be affected in vivo. Fig. 4A shows that Ag-AQCs at low concentration display antimicrobial activity against E. coli. It has been reported that gyrase inhibitors induce oxidative damage cellular death in E. coli;22 hence, the effect of Ag-AQCs on the bacterial ROS levels was investigated. As shown in Fig. 4B, addition of Ag-AQCs to bacteria increases the superoxide levels over the controls, indicating that the bactericidal activity of Ag-AQCs could be partially mediated by oxidative damage.Ag-AQCs have a prominent inhibitory effect on bacterial DNA topoisomerases, that is, mediated by the distortion of the DNA topology. The DNA structure is evolutionarily well preserved, suggesting that any change affecting it could also affect multiple bacteria species. To probe this argument, we tested the activity of Ag-AQCs against randomly selected bacteria isolated from clinical samples representing various species. Interestingly, not all of the isolates were equally susceptible to the Ag-AQCs bactericidal action (Fig. 4C). After treatment with Ag-AQCs, five isolates showed strongest resistance to 50% viability. So far it is unknown whether this differential susceptibility is an outcome of the particular isolates or it is common to the species they belong to. However, our data point to bacterial species as the key factor in Ag-AQCs resistance. In fact, Gram-positive bacteria seem to be more resistant than Gram-negative (Fig. 4C, inset); actually, four out of five resistant strains were Gram-positive. The difference among Gram-positive and Gram-negative species is the presence of a thick peptidoglycan layer in the bacterial cell wall. The interaction of Ag-AQCs with DOX shown here (Fig. 1SI†) is an example of a more general interaction of Ag-AQCs with aromatic and cyclic compounds (unpublished results). Therefore, one can speculate that the increased resistance of Gram-positive bacteria could be explained by the action of the peptidoglycan of the wall cell that prevents Ag-AQCs from entering the bacteria. The only Gram-negative isolate displaying above 50% survival was Pseudomonas aeruginosa. This observation led us to consider whether P. aeruginosa gyrase could be less sensitive than E. coli gyrase to the action of Ag-AQCs, which could explain the increased resistance of P. aeruginosa. We assessed the activity of P. aeruginosa DNA gyrase in the presence of Ag-AQCs and found it as sensitive to the Ag-AQCs action as E. coli DNA gyrase (Fig. 4D and 3C, respectively). Therefore, we are inclined to believe that the resistance should be due to insufficient Ag-AQCs concentration inside the bacteria to kill them. Supporting this view, it has been described that most strains of P. aeruginosa produce diffusible cyclic pigments23 that may interact with Ag-AQCs, restraining its entry into the bacteria.
Human resistance to antibiotics is a worldwide problem.24 Use of antibiotics is the most important single event leading to resistance.25 Ag-AQCs is a new material whose beneficial and useful bactericidal action could provide an alternative to antibiotics that manifest resistance. Our preliminary results point in that direction and deserve further study. Finally, the interest of Ag-AQCs, which are able to interact with living organisms, should be highlighted, as this is an area that should be explored due to the potential consequences it may have, both beneficial and harmful. In a previous work11 we calculated theoretically that Ag3 intercalates, whereas Ag2 binds covalently to the DNA. However, so far obtaining sufficiently pure samples of Ag2 for biological assays has not been feasible. In view of the interesting biological activities showed by Ag3 it should be of interest to undertake a systematic study of the chemical and biological effects of silver clusters of different sizes. Such study will depend on future developments of synthetic strategies for the preparation of monodisperse cluster samples without strong binding ligands, which may affect their biological activities. This is currently a main challenge.
In summary, we have described for the first time that Ag-AQCs display bactericidal activity. These findings give way to new promising avenues to extend the understanding of the silver microbicidal properties, an important topic that has been discussed for centuries without having yet been resolved.
Experimental
Unless otherwise stated, all chemicals were obtained from Sigma-Aldrich. Stock solutions of DOX were prepared by dissolving weighed amounts at pH = 7.0 in a buffer containing 2.5 × 10−3 M sodium cacodylate (NaCaC), [(CH3)2AsO2Na]. ct-DNA was dissolved in water and sonicated employing a MSE-Sonyprep sonicator (20 cycles of 10 s each, with 20 s pause between cycles, 96 μm amplitude). The sonicator tip was introduced directly into the solution, which was kept in an ice bath to minimize thermal effects. Agarose gel electrophoresis tests showed that the polymer length was reduced to approximately 1000 base pair fragments. Aqueous solutions were prepared with doubly deionized water from a Puranity TU System with UV lamp and ultrafilter (VWR). Stock solutions were standardized spectrophotometrically using ε = 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 M−1 cm−1 at λ = 260 nm, ionic strength = 0.1 M (0.975 M NaCl + 0.025 M NaCaC) and pH = 7.0.
200 M−1 cm−1 at λ = 260 nm, ionic strength = 0.1 M (0.975 M NaCl + 0.025 M NaCaC) and pH = 7.0.
Ag3 clusters were obtained by an electrochemical procedure, as previously described.11 In a typical synthesis, a three-electrode chemical cell was used with two foil electrodes, Ag (working) and Pt (counter) electrodes, and a hydrogen reference electrode. 2 V constant voltage was applied for 1200 s in nitrogen deaerated MiliQ water at 25 °C. Prior to synthesis, the silver electrode was polished with sand paper (600 grid, Wolfcraft) followed by alumina (≈50 nm, Buehler), washed thoroughly with MiliQ water and sonicated. The platinum electrode was cleaned electrochemically by cyclic voltammetry in 1 M MeOH/1 M NaOH solution followed by cyclic voltammetry in 1 M H2SO4. After the synthesis, the remaining Ag+ ions were removed by addition of NaCl and subsequent precipitation and filtration. Purified samples (Ag-AQCs) were then concentrated at 35 °C using a rotary evaporator. Sample characterization was carried out by UV-Vis and fluorescence spectrometry, cyclic voltammetry, ESI-TOF mass spectrometry, X-ray absorption near edge structure (XANES) and atomic force microscopy, as previously described.11
Isothermal titration calorimetry
The ITC experiments were performed at 25 °C using a Nano ITC Instrument (TA, Waters LLC, New Castle, USA). To prevent formation of air bubbles, all of the solutions were degassed in a degassing station (TA, Waters LLC, New Castle, USA). DOX solutions were placed in a 50 μL syringe continuously stirred and 50 additions of 1 μL in 400 s intervals were injected into the sample cell containing the buffer or the ct-DNA solution previously incubated with Ag+ or Ag-AQCs. Control experiments were carried out to determine the contribution of the heat of dilution of both ct-DNA and DOX. The integration of the peaks, corrected by the dilution effect, yielded the binding isotherms (heat change versus CD/CP mole ratio). All of the data were analyzed using the NanoAnalyze software.Circular dichroism titration
The CD titrations were recorded on a MOS-450 Bio-Logic dichrograph (Claix, France) using 1.0 cm path-length cells. The titrations were carried out by injecting increasing DOX micro amounts into a known volume of the ct-DNA solution previously incubated with Ag+ or Ag-AQCs.Fluorescence titration
Fluorescence titrations were performed on a Shimadzu Corporation RF-5301PC spectrofluorometer (Duisburg, Germany) at λexc = 490 nm and λem = 555 nm by adding increasing amounts of Ag-AQC directly into the cell with the DOX solution.Topoisomerase IV decatenation assay
E. coli topoisomerase IV activity was assessed using a commercial kit (Inspiralis, UK). Briefly, 200 ng of kDNA were preincubated for 5 min at room temperature with Ag-AQCs at various concentrations in 40 mM HEPES–KOH (pH 7.6), 100 mM potassium glutamate, 10 mM magnesium acetate, 10 mM DTT, 1 mM ATP and 50 μg ml−1 albumin, in 30 μl total reaction volume. After that, 1U of Topo IV was added and incubation was continued for 30 min at 37 °C. The reaction was stopped by addition of 30 μl chloroform/iso-amyl alcohol and 6 μL of loading buffer 6×, vortexed and centrifuged briefly (5–10 seconds each) before being loaded on an agarose gel (1%: w/v) in TAE (40 mM Tris–acetate, 2 mM EDTA) buffer with ethidium bromide (0.5 μg ml−1). The IC50 for inhibition of decatenation was assessed using gel documentation software and statistical analysis of 3 independent experiments.Gyrase supercoiling assay
The activity of gyrase from E. coli and P. Aeruginosa were assessed using the gyrase supercoiling kit according to the manufacturer (Inspiralis, UK). Briefly, 0.5 μg of relaxed pBR322 DNA were preincubated for 30 min at room temperature with Ag-AQCs in 30 μl reaction under the following conditions: 35 mM Tris–HCl (pH 7.5), 24 mM KCl, 4 mM MgCl2, 2 mM DTT, 1.8 mM spermidine, 1 mM ATP, 6.5% (w/v) glycerol and 0.1 mg ml−1 BSA. After that, 1U of gyrase was added and incubation was continued for 30 min at 37 °C. The reaction was stopped by addition of 30 μl chloroform/iso-amyl alcohol and 6 μL of loading buffer 6× before being loaded on an agarose gel (1%: w/v) in TAE (40 mM Tris–acetate, 2 mM EDTA) buffer without ethidium bromide. At least three independent experiments were conducted for each gyrase.Assay of HindIII activity
pTG7, a 4892 base pairs plasmid, is cut by HindIII in two sites releasing a 1135 bp fragment. 300 ng of pTG7 was cut with 1 enzyme Hind III unit (New England Biolabs), 10 mM Tris–HCl, 50 mM NaCl, 10 mM MgCl2, 1 mM DTT at pH 7.9 in 25 μl final volume for 30 min at 37 °C. When required, Ag-AQCs were added at the beginning of the incubation at different concentrations. The reaction products were run in 1% agarose gel. At the end of the running the gel was stained in TAE buffer with 0.5 μg ml−1 ethidium bromide.Bactericidal assay
Briefly, E. coli (ATCC 25922) were picked from an agar plate and grown in Luria Bertani medium (LB) at 37 °C with 225 rpm constant agitation. Growth was monitored turbidimetrically and adjusted to achieve the standard McFarland 0.5 (OD 620 nm between 0.08 and 0.13), about 1 × 108 cfu ml−1. Bacteria (5 × 105 cfu ml−1) were incubated for 1/2 h at 37 °C in 60 μl final volume of PBS in the presence of different doses of Ag-AQCs, after which they were centrifuged and resuspended in LB medium and grown for 3 h. At the end of the incubation 50 μl of bacteria were mixed with 25 μl Syto 9 (Life Technologies) and green fluorescence was assessed by flow cytometry (Guava, EasyCyte, Merk-Millipore). Bacteria isolated from clinical samples were kindly provided by the Department of Microbiology, Faculty of Medicine, University of Santiago de Compostela, Spain. Briefly, bacteria were selected from an agar culture and grown in Mueller-Hinton broth (MHB) at 37 °C with 225 rpm constant shaking. Growth was monitored by turbidimetry and adjusted to the McFarland Standard no. 0.5 (OD 620 nm between 0.08 and 0.13), approximately 1 × 108 cfu ml−1. A bacterial inoculum of approximately 5 × 105 cfu ml−1 was incubated with different Ag-AQCs doses for 1 h at 37 °C in 60 μL final volume PBS. At the end of the incubation, bacteria were centrifuged and resuspended in MHB and grown for 20 h. Turbidity was observed by reading the absorbance at 600 nm on a plate reader. Additionally, we determined whether the treatment with the maximum concentration of Ag-AQCs could inhibit colony formation on agar plates.Detection of reactive oxygen species (ROS)
Dihydroethidium (DHE) (Life Technologies), by virtue of its ability to freely permeate cell membranes, is extensively used to monitor superoxide production. DHE essentially detects superoxide radicals. E. coli were grown in LB to a density equivalent to McFarland standard 0.5 (OD 620 nm between 0.08 and 0.13) and then diluted 10-fold to obtain an inoculum of approximately 1 × 107 cfu ml−1. Ag-AQCs, diluted in medium, were added and incubated for 3 h at 37 °C under 225 rpm constant stirring. To equalize the number of bacteria in all of the samples, at the end of the incubation the absorbance was read at 620 nm and adjusted to be the same in all samples before addition of DHE (5 μm/100 μl). After 20 min, samples were extensively washed with PBS and the fluorescence present in the bacteria was assessed by flow cytometry (Guava, EasyCyte, Merk-Millipore). A minimum of 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 events were captured on the region of interest.
000 events were captured on the region of interest.
Acknowledgements
This work was supported by Obra Social“la Caixa” (OSLC-2012-007), European Commission through FEDER program (0681 InveNNta 1 E); Ministerio de Ciencia e Innovación, Spain (MAT2010-20442, MAT2011-28673-C02-01); MINECO, Spain (MAT2012-36754-C02-01 and CTQ2014-58812-C2-2-R), Xunta de Galicia, Spain (GRC2013-044, FEDER Funds). C. P.-A. is grateful for the FPU grant from Ministry of Education, Culture and Sports, Madrid, Spain (FPU13/00180).Notes and references
- J. R. Morones, J. L. Elechiguerra, A. Camacho, K. Holt, J. B. Kouri, J. T. Ramirez and M. J. Yacaman, Nanotechnology, 2005, 16, 2346–2353 CrossRef CAS.
- M. I. Setyawati, X. Yuan, J. Xie and D. T. Leong, Biomaterials, 2014, 35, 6707–6715 CrossRef CAS.
- X. Yuan, M. I. Setyawati, D. T. Leong and J. Xie, Nano Res., 2014, 7, 301–307 CrossRef CAS.
- X. Yuan, M. I. Setyawati, A. S. Tan, C. N. Ong, D. T. Leong and J. Xie, NPG Asia Mater., 2013, 5, e39 CrossRef CAS.
- J. Y. Maillard and P. Hartemann, Crit. Rev. Microbiol., 2013, 39, 373–383 CrossRef CAS.
- D. Mandal, M. E. Bolander, D. Mukhopadhyay, G. Sarkar and P. Mukherjee, Appl. Microbiol. Biotechnol., 2006, 69, 485–492 CrossRef CAS.
- X. Jiang, T. Miclăuş, L. Wang, R. Foldbjerg, D. S. Sutherland, H. Autrup, C. Chen and C. Beer, Nanotoxicology, 2015, 9, 81–89 CrossRef.
- D. Nies, S. Silver and D. Nies, in Molecular Microbiology of Heavy Metals, Springer Berlin Heidelberg, 2007, vol. 6, pp. 117–142 Search PubMed.
- B. S. Gonzalez and M. A. Lopez-Quintela, in Functional Nanometer-Sized Clusters of Transition Metals: Synthesis, Properties and Applications, The Royal Society of Chemistry, 2014, pp. 25–50 Search PubMed.
- Y. A. Attia, D. Buceta, C. Blanco-Varela, M. B. Mohamed, G. Barone and M. A. López-Quintela, J. Am. Chem. Soc., 2014, 136, 1182–1185 CrossRef CAS.
- D. Buceta, N. Busto, G. Barone, J. M. Leal, F. Domínguez, L. J. Giovanetti, F. G. Requejo, B. García and M. A. López-Quintela, Angew. Chem., Int. Ed., 2015, 54, 7612–7616 CrossRef CAS.
- C. Carvalho, R. X. Santos, S. Cardoso, S. Correia, P. J. Oliveira, M. S. Santos and P. I. Moreira, Curr. Med. Chem., 2009, 16, 3267–3285 CrossRef CAS.
- G. Minotti, P. Menna, E. Salvatorelli, G. Cairo and L. Gianni, Pharmacol. Rev., 2004, 56, 185–229 CrossRef CAS.
- Y. Pommier, E. Leo, H. L. Zhang and C. Marchand, Chem. Biol., 2010, 17, 421–433 CrossRef CAS.
- C. Pérez-Arnaiz, N. Busto, J. M. Leal and B. García, J. Phys. Chem. B, 2014, 118, 1288–1295 CrossRef.
- D. Suman and G. S. Kumar, J. Mol. Struct., 2008, 872, 56–63 CrossRef.
- R. Rohs, S. M. West, A. Sosinsky, P. Liu, R. S. Mann and B. Honig, Nature, 2009, 461, 1248–1253 CrossRef CAS.
- A. K. McClendon, A. C. Gentry, J. S. Dickey, M. Brinch, S. Bendsen, A. H. Andersen and N. Osheroff, Biochemistry, 2008, 47, 13169–13178 CrossRef CAS.
- J. L. Nitiss, E. Soans, A. Rogojina, A. Seth and M. Mishina, Current Protocols in Pharmacology, 2012, unit 3.3 Search PubMed.
- A. D. Bates and A. Maxwell, DNA Topology, Oxford University Press, 2005 Search PubMed.
- Inspiralis, technical information, http://www.inspiralis.com/go/gyrase_supercoiling.php.
- F. Yang, S. S. Teves, C. J. Kemp and S. Henikoff, Biochim. Biophys. Acta, 2014, 1845, 84–89 CAS.
- D. J. Dwyer, M. A. Kohanski, B. Hayete and J. J. Collins, Mol. Syst. Biol., 2007, 3(91), 1–15 Search PubMed.
- D. Greenwood, M. Barer, R. Slack and W. Irving, Medical Microbiology, Churchill Livingstone, 18th edn, 2012 Search PubMed.
- WHO Antimicrobial resistance, Fact sheet No. 194, Updated April 2015, Centers for Disease Control and Prevention, Antibiotic resistance threats in the United States, 2013, http://www.cdc.gov/drugresistance/threat-report-2013/index.html.
Footnotes |
| † Electronic supplementary information (ESI) available: Fig. 1SI and 2SI and fitting procedure of eqn (1SI). See DOI: 10.1039/c5sc02022k |
| ‡ JN and CP-A equally contributed to this paper. |
| This journal is © The Royal Society of Chemistry 2015 |