Drug–tubulin interactions interrogated by transient absorption spectroscopy†
F. Boscáa,
G. Sastrea,
J. M. Andreub,
D. Jorneta,
R. Tormos*a and
M. A. Miranda*a
aDepartamento de Química/Instituto Universitario Mixto de Tecnología Química UPV-CSIC, Avenida de los Naranjos s/n, E-46022 Valencia, Spain. E-mail: rtormos@qim.upv.es; mmiranda@qim.upv.es
bCentro de Investigaciones Biológicas, CSIC, Ramiro de Maeztu 9, E-28040 Madrid, Spain
First published on 27th May 2015
Abstract
Colchicine (COL) is a bioactive molecule with antitumor properties. When COL binds to tubulin (TU), it inhibits microtubule assembly dynamics. We have investigated COL–TU interactions using laser flash photolysis (LFP) technique and performing fully flexible molecular dynamics simulations. Excitation of COL at 355 nm in aqueous medium did not lead to any transient absorption spectrum. By contrast, in the presence of TU a transient peaking at λmax ca. 420 nm was registered and assigned as triplet excited COL complexed with TU (3COL*@TU). In aerated medium, the lifetime was τ ca. 160 μs and the quantum yield was 0.138. Likewise, when the bicyclic COL analog MTC was submitted to LFP in the presence of TU, 3MTC@TU* was detected with a lifetime of ca. 62 μs and a quantum yield of 0.296, Aqueous solutions of MTC did not produce any signal in the microsecond timescale. The triplet energy of MTC was obtained by means of emission measurements and found to be ca. 200 kJ mol−1, a value that matches with that previously reported for COL (188 kJ mol−1). Molecular dynamic simulations, both with the ground and triplet excited state, reveal a strong interaction between COL and TU to give stabilized complexes with restricted mobility inside the protein binding site. These results demonstrate that LFP is a useful methodology to study the binding of COL derivatives to TU and open a new way to evaluate the interactions of non-fluorescent anticancer drugs with this protein.
Introduction
Tubulin (TU), the major component of microtubules, is a heterodimeric protein formed by α and β subunits that plays a crucial role in biological processes such as mitosis, intracellular transport or cell growth.1–4 This protein is the target for anticancer drugs, which inhibit or promote its assembly into microtubules.5–10Colchicine (COL, Chart 1) is a natural alkaloid obtained from meadow saffron (Colchicum autumnale).11 It is an important bioactive drug as well as a neurotoxin in animal models of Alzheimer's disease and epilepsy.12–14 Although COL has been shown to possess antitumor properties,15,16 its therapeutic use is limited by toxicity problems; this has led to the consideration of analogs.17–19 When COL binds to TU, it inhibits the assembly into microtubules and microtubule dynamics.20 The binding process is slow and strongly temperature-dependent,21 while dissociation is kinetically unfavorable due to its high activation energy.22 The binding site is mostly buried in the intermediate domain of the β subunit.6
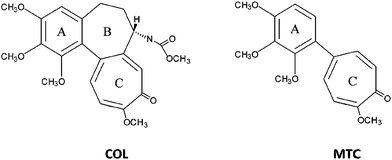 | ||
| Chart 1 Colchicine (COL) and 2-methoxy-5-(2′,3′,4′-trimethoxyphenyl)-2,4,6-cycloheptatrien-1-one (MTC) molecular structures. | ||
The structure of COL consists of three rings, the trimethoxybenzene ring (A), the methoxytropone (C) and the seven-membered ring with an acetamido substitution at C-7 (B).23,24 Interaction of COL with TU has been attributed to simultaneous binding of its A and C rings. A properly positioned oxygen atom in ring C is an essential feature for powerful inhibition of microtubule assembly.25–27 The middle connecting B ring, which interacts with the α subunit, is involved in the peculiar binding kinetics,23 but it is not an essential requirement for TU binding. Thus, the MTC analog (Chart 1) lacking the B ring binds rapidly and reversibly to the high affinity COL binding site of TU, thereby inhibiting microtubule assembly.28–30
The toxicity of COL has promoted computational studies in an attempt to find new TU binding agents with improved properties.6,31–33 Interestingly, using a combined computational and cytotoxicity study, the binding energies of several COL derivatives have been ranked according to systematic modifications of rings A–C and correlated with cytotoxicity.32
In general, protein binding parameters can be determined by a variety of techniques.34–41 Fluorescence has been found to be one of the most convenient methods, due to the dramatic enhancement of the emission quantum yield of COL upon TU binding.42 More recently, laser flash photolysis (LFP) has been introduced as a new tool to investigate drug–protein interactions and to determine binding parameters such as affinity constants or population of the binding sites.43,44 Interestingly, the absence of a triplet excited state detectable in LFP has revealed that COL does not bind to serum albumins.45 This technique is very sensitive and does not require separation of free and complexed drug. However, it has not yet been applied to TU binding studies.
With this background, the aim of the present work is to investigate COL–TU and MTC–TU interactions by means of a combined experimental and theoretical approach, using in parallel LFP and fully flexible molecular dynamics simulations. The obtained results confirm that the LFP methodology is indeed a powerful tool for interrogating the TU binding behavior of COL and its derivatives.
Experimental
General
COL was purchased from Sigma-Aldrich Chemical Company. MTC was a kind gift from T.J. Fitzgerald, Florida A&M University.46 TU was purified from bovine brain and stored as described.47 Before use, concentrated TU was diluted more than 50-fold into PG buffer to give the desired final concentration and employed within the next 4 h. PG buffer: 10 mM sodium phosphate and 0.1 mM GTP at pH 7.0.Absorption and emission spectra
Optical spectra in different media were measured on a Perkin-Elmer Lambda 35 UV/vis spectrophotometer. Phosphorescence measurements were recorded with a time-resolved spectrometer (TimeMaster fluorescence lifetime spectrometer TM-2/2003) from Photon Technology International. The spectra of the samples were recorded at 77 K in acetonitrile and in 1 mM PB aqueous solution using a single-cell Peltier cooler. The emission spectral band widths were set to 10 nm, and the excitations were performed at 350 nm. The phosphorescence emission spectra were recorded with a delay time of 0.5 ms and a total gate time of 10 ms.Laser flash photolysis experiments
A pulsed Nd:YAG laser was used for the excitation at 355 nm. The single pulses were ∼10 ns duration, and the energy was from 10 to 1 mJ per pulse. A pulsed xenon lamp was employed as detecting light source. The LFP apparatus consisted of the pulsed laser, the Xe lamp, a monochromator and a photomultiplier made up of a tube, housing and power supply. The output signal from the oscilloscope was transferred to a personal computer.The laser pulse was probed by a system containing a fiber that synchronizes the LFP with the digitizer operating in the pre-trigger mode. All transient spectra were recorded using 10 × 10 mm2 quartz cells with 4 mL capacity, and they were bubbled during 20 min with N2. All the experiments were carried out at room temperature. The solutions of COL and MTC were prepared at 5 × 10−5 M concentration.
| 3MTC* + β-Car → MTC + 3β-Car* | (1) |
Measurements were performed in deaerated acetonitrile solutions of MTC with and without the presence of β-Car (1 × 10−5 to 5 × 10−5 M). Then, the molar absorption coefficient (ε) of 3MTC* was calculated using eqn (2):
| k2/(k2 − k1) × ΔA (3β-Car* (520 nm)) × ε (3MTC* (400 nm)) = ΔA (3MTC* (400 nm)) × ε (3β-Car* (520 nm)) | (2) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 M−1 cm−1 (as described in toluene).49
000 M−1 cm−1 (as described in toluene).49
The intersystem crossing quantum yield (ΦISC) of MTC was obtained by the comparative method50 assuming that ε (3MTC*) is similar in all solvents. Hence, excitation of benzophenone (BP) and MTC was carried out separately using solutions with identical absorbance at the excitation wavelength (0.3 at 355 nm). Then eqn (3) was applied:
| ΦISC (MTC) = ΦISC (BP) × ΔA (3MTC* (400 nm)) × ε (3BP* (525 nm))/ΔA (3BP* (525 nm)) × ε (3MTC* (400 nm)) | (3) |
The 3MTC* quenching rate constants by oxygen and 3β-Car were determined using the Stern–Volmer eqn (4):
| 1/τ = 1/τ0 + k [quencher] | (4) |
Computational methods
With the aim of unveiling the dynamic behavior of the system COL–TU, molecular dynamics runs were performed using LAMMPS52 (Large-scale Atomic/Molecular Massively Parallel Simulator) software, which integrates Newton's equations of motion for all the atoms of the system, that interact via short- and long-range forces with an input consisting on the initial location of all the atoms. The TU geometry was taken from Ravelli et al.,6 where the TU structure is solved at 3.5 Å resolution in complex with COL. Starting from this initial geometry for the complex, 1000 water molecules were added and treated explicitly in the simulation using a known method,53 where the advantages of explicit versus continuum solvent models have been optimized with parallel techniques for computational efficiency. The molecular dynamics simulations have been performed within the NVT ensemble at 298 K and 1 atm for 1 ns with the explicit relaxation of all the atoms of the system using a timestep of 1 fs. The system was initially energy minimized at 10 K using the following algorithms: steepest-descent (50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 steps) and conjugated-gradients (100
000 steps) and conjugated-gradients (100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 steps). Then, three subsequent dynamic cycles of 10
000 steps). Then, three subsequent dynamic cycles of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 steps (at 98, 198, and 298 K) were performed for equilibration. Finally, 1 million steps of dynamics at 298 K allowed obtaining 1 ns of molecular dynamics behavior.
000 steps (at 98, 198, and 298 K) were performed for equilibration. Finally, 1 million steps of dynamics at 298 K allowed obtaining 1 ns of molecular dynamics behavior.
Periodic boundary conditions were implemented within a rectangular box of [230, 200, 120] Å in [x, y, z]. The force field employed was UFF,54 including an electrostatic part obtained from a MOPAC2012 (ref. 55 and 56) calculation using the minimized system as single point. For the short-range and long-range parts, a method with Lennard-Jones and Coulombics computed via damped shifted forces with a damping parameter 0.05 Å−1 and cutoffs of 10.0 Å (Lennard-Jones) and 12.0 Å (Coulombic) was employed.
Diffusion of the triplet excited state within TU has been considered by means of a two-steps strategy. Thus, the triplet geometry of COL and MTC has been calculated using first principles based on density functional theory (DFT). The geometries, as well as the corresponding charge distributions, have been compared with those of the ground states. Then, a reparameterization of UFF has been made so that the specific triplet geometries can be reproduced within this force field and the modified UFF has been employed to perform a new molecular dynamics run.
The first-principles geometry optimizations have also been performed using the PBE and ωB97X-D57 functionals, with TZVP58 basis set, obtaining very similar results. For calculation of excitation energies by means of DFT methods, the performance has been analyzed and rationalized according to five different categories of functional such as: LDA (local density approach), GGA (Generalized Gradient Approximation), meta-GGA (which includes not only the density and its first derivative in the exchange–correlation potential, as GGA, but also the second derivative); GH (global hybrids, which include a predetermined amount of exact exchange), and LCH (long-range-corrected hybrids, whose fraction of exchange depends on the interelectronic distance).59–61 The results of a GGA functional (PBE) and a LCH (ωB97X-D) have been compared giving similar results.
Results and discussion
Colchicine–tubulin interactions in the triplet excited state
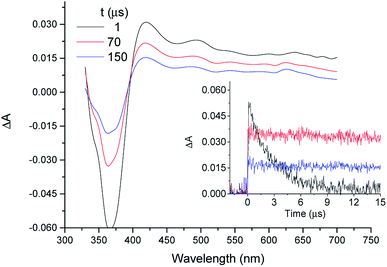
In organic solvents, LFP of COL gives rise to the triplet excited state (3COL*) as a well characterized transient with absorption maximum at λmax ca. 420 nm.45 In PG buffer, 355 nm laser excitation of 5 × 10−5 M COL, either under aerobic or anaerobic conditions, did not lead to any transient absorption spectrum, in agreement with previous results obtained in aqueous medium.45,62 As a control experiment, LFP of a TU solution gave no signal either. By contrast, when COL (5 × 10−5 M in GTP) was submitted to LFP in the presence of TU (0.5 and 1 × 10−5 M) a transient absorption spectrum peaking at λmax ca. 420 nm was indeed registered (see Fig. 1). This reveals generation of an intermediate detectable in the microsecond timescale. Its quenching by oxygen and its similarity to the transient absorption spectrum obtained for 3COL* in acetonitrile45 support the assignment as 3COL* complexed with TU (3COL*@TU). Moreover, in aerated medium, the determined lifetime (τ ca. 160 μs) was more than 500 times longer than that obtained for 3COL* in acetonitrile (τ = 0.29 μs), which agrees well with the markedly longer triplet lifetimes usually measured in the intraprotein microenvironment.43,44,63Comparison of the decay traces of 3COL* in acetonitrile and 3COL*@TU in aqueous solutions revealed that the initial intensity of the signal immediately after the laser pulse was higher in the organic solvent (Fig. 1, inset). A proper analysis of this observation should consider the following factors in aqueous medium: (a) the TU concentrations are five or ten times lower than those of COL, (b) the affinity constant between COL and TU is very high22 and (c) in the absence of TU, COL does not give rise to any observable signal in the microsecond timescale. Whereas (a and b) favor full complex formation, (c) implies that all the observed signal can be safely attributed to 3COL*@TU.
At this point, if it is assumed that the molar absorption coefficient of the complex is nearly the same as that reported for 3COL* in acetonitrile, the absorbance of the transient arising from the LFP of 5 × 10−5 COL should be five or ten times lower in the presence of 0.5 and 1 × 10−5 M TU than in acetonitrile. Since the real absorbance is close to four times higher than the expected value, the 3COL* quantum yield must be markedly higher inside the protein (0.138 vs. 0.037 in acetonitrile45). This enhancement suggests a restricted mobility in the protein microenvironment and consequently, limited degrees of freedom, leading to less efficient non-radiative decay pathways.
Interactions of MTC with tubulin
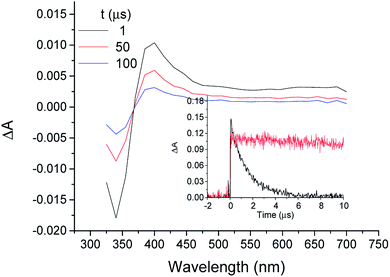
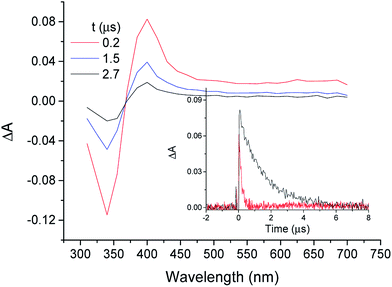
When MTC was submitted to LFP in the presence of TU, a transient absorption spectrum very similar to that of 3COL* with and a lifetime of ca. 62 μs was obtained (Fig. 2). In view of the parallel behavior of both substrates, it was inferred that the transient absorption species was the MTC triplet excited state (3MTC*). However, before tackling a detailed analysis of the interactions between MTC and TU, it was considered of interest to undertake a more detailed characterization of the transient. With this purpose, LFP experiments were performed with MTC (5 × 10−5 M) in acetonitrile, methanol, ethanol and H2O under aerated and deaerated conditions.As previously observed with COL, aqueous solutions of MTC did not produce any signal in the microsecond timescale, while a COL-like transient species with λmax ca. 400 nm was observed in the other solvents (Fig. 3). The initial absorption of this species decreased with the increasing protic character of the solvent. As expected for a triplet excited state, it was quenched by molecular oxygen in acetonitrile with rate constant of 3 × 109 M−1 s−1 (Fig. 3, inset).
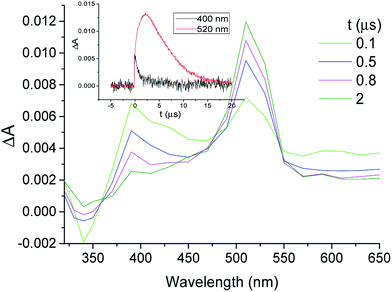
This assignment was confirmed by energy transfer to β-carotene (β-Car) in acetonitrile (eqn (1) in the Experimental section). Thus, the decay at λmax ca. 400 nm was concomitant with the growth of 3β-Car* at 520 nm (Fig. 4). This reaction was diffusion-controlled (kq ca. 2 × 1010 M−1 s−1), in agreement with the expectations based on the low 3β-Car* energy (79 kJ mol−1).48
The triplet molar absorption coefficient (ε) at 400 nm was found to be 31![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 M−1 cm−1 in acetonitrile, as indicated in the Experimental section (eqn (2)). When this parameter was used to determine the intersystem crossing quantum yield (ΦISC) of MTC (see eqn (3) in Experimental section), important solvent dependence was observed (Table 1). The hydrogen bond donating (HBD) ability of the solvents64 seems to be better correlated with the ΦISC changes than the polarity or the dielectric constant. Besides, the triplet lifetimes in all solvents were very similar (ca. 2 μs, Table 1).
200 M−1 cm−1 in acetonitrile, as indicated in the Experimental section (eqn (2)). When this parameter was used to determine the intersystem crossing quantum yield (ΦISC) of MTC (see eqn (3) in Experimental section), important solvent dependence was observed (Table 1). The hydrogen bond donating (HBD) ability of the solvents64 seems to be better correlated with the ΦISC changes than the polarity or the dielectric constant. Besides, the triplet lifetimes in all solvents were very similar (ca. 2 μs, Table 1).
| Solvent | Absorption (λmax, nm) | τT (μs) | ΦISC | Solvent dielectric constant | HBD ability |
|---|---|---|---|---|---|
| H2O | 343 | — | 0 | 78.3 | 1.19 |
| CH3OH | 341 | 2 | 0.020 | 32.6 | 0.98 |
| CH3CH2OH | 340 | 1.9 | 0.029 | 24.5 | 0.86 |
| CH3CN | 335 | 1.8 | 0.074 | 37.5 | 0.19 |
The triplet energy (ET) was obtained by means of emission measurements. Thus, the phosphorescence spectra of MTC in MeCN as well as in 1 mM PB aqueous medium at 77 K consisted of a structured band with two maxima at 615 and 670 nm. From these spectra, the ET was found to be ca. 200 kJ mol−1. This value matches with that obtained for COL in the same way and is close to that estimated for COL (188 kJ mol−1) in a previous study.65
After characterization of the transient, the interaction of MTC with TU was investigated. Assuming that the molar absorption coefficient for the 3MTC*@TU complex at 430 nm is similar to that determined for 3MTC* in acetonitrile, it is possible to estimate the percentage of MTC bound to TU. As in the case of COL, comparison of the maximum intensity of the decay signals in organic solution and in the presence of TU showed again that, at the employed concentrations, the value in acetonitrile was more than seven times higher than in the presence of the protein. Using the reported equilibrium constant (Ka ca. 5 × 105 M−1 at 25 °C)28,29 it was possible to establish that the degree of occupancy of the colchicine sites of TU by MTC at 25 °C is between 98 and 95%. Based on these results, and following the same reasoning as in the case of 3COL*@TU, the quantum yield of the 3MTC@TU* complex was found to be 0.296, ca. 4 times higher than that found for 3MTC* in acetonitrile (0.074).
Computational analysis
Molecular dynamics simulations at 1 atm and 298 K, allowed us to obtain the positions of all the atoms of the system during 1 ns. The possibility of extending the time window was considered, but this would only make sense to simulate the long trajectory from the bulk solution to the intraprotein microenvironment. This does not seem necessary when the ligand is initially placed in the known active site, as in our case. Once there, only minimal motion is anticipated, which requires a much shorter time window. As a matter of fact, extension of the simulation period up to 10 ns (see ESI†) did not lead to significant changes. Fig. 5 shows the positions of COL (red) as it moves around within TU, whose atoms are not displayed because the interest was focused on the relative diffusional paths. Thus, COL spends all the simulation time near the same area within a rectangular box of [Δx, Δy, Δz] = [3, 5, 4] Å; hence it is strongly adsorbed at its known binding site.6 A general view of the COL@TU complex, at the end of the simulation, is shown in Fig. 6.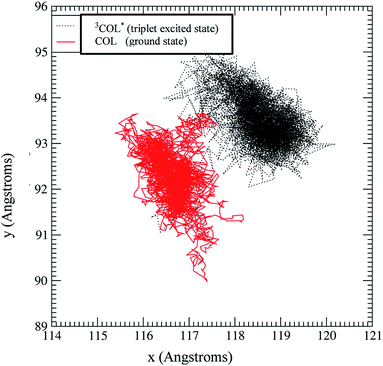 | ||
| Fig. 5 Trajectories across xy of the centre-of-mass coordinates of COL triplet excited state in TU obtained from the molecular dynamics run (500 ps) in which the force field has been adapted to reproduce the triplet excited state geometry as obtained from the DFT calculation (Fig. 8). In this graph, the corresponding trajectory (500 ps) of the COL ground state has been superimposed in order to show their similar diffusivity. | ||
The total energy of COL–TU and MTC–TU interactions (Fig. 7) can be split into its short-range (van der Waals) and long-range (Coulombic) contributions. These results explain the reduced mobility of COL (Fig. 5), and also its irreversible binding to TU, due to the strong adsorption energy (−330 kJ mol−1 on average).
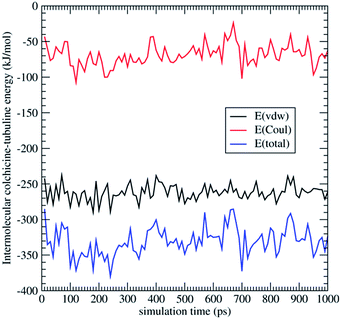 | ||
| Fig. 7 Intermolecular interaction energy and its components (short-range vdw and long-range Coulomb) corresponding to the molecular dynamics runs, COL@TU. | ||
Diffusion of the COL triplet excited state within TU is also shown in Fig. 5. This has been considered because lifetimes of this species inside TU are much longer than the 1 ns simulation time (in the order of 160 μs microseconds). Moreover, triplet excited states may show significant differences in geometry and/or polarity, which would exert a direct influence on the diffusivity inside TU.
While the previous molecular dynamics study based on the Universal force field (UFF) is capable to reproduce the geometries of the drugs in their ground state, this force field would be totally useless to predict and simulate excited state geometries. Hence, to study the diffusivity of excited triplet states a two-steps strategy have been employed. First, the triplet geometry of COL has been calculated using first principles based on density functional theory (DFT). This geometry, as well as the corresponding charge distribution, has been compared with those of their ground states to find the main differences that may result in specific triplet diffusivity. Secondly, a reparameterization of UFF has been made so that the specific triplet geometries can be reproduced within this force field. With the modified UFF, a molecular dynamics run has been performed in order to check whether there are significant differences between the COL@TU diffusivity in the ground and triplet states.
The first-principles geometry optimizations have also been performed using the PBE66,67 and ωB97X-D57 functionals, with TZVP58 basis set, obtaining very similar results. For calculation of excitation energies by means of DFT methods, the performance has been analyzed and rationalized according to five different categories of functional such as: LDA (local density approach), GGA (Generalized Gradient Approximation), meta-GGA (which includes not only the density and its first derivative in the exchange–correlation potential, as GGA, but also the second derivative); GH (global hybrids, which include a predetermined amount of exact exchange), and LCH (long-range-corrected hybrids, whose fraction of exchange depends on the interelectronic distance).59–61 The results of a GGA functional (PBE) and a LCH (ωB97X-D) have been compared giving similar results.
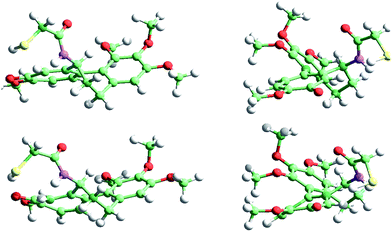
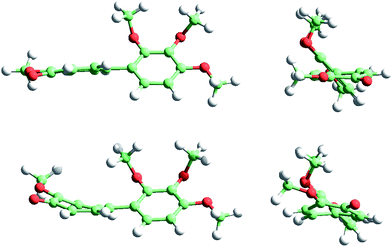
For the sake of brevity we only show the results obtained with ωB97X-D, which are more confident because of the improvements in the excitation energies when including exact exchange in the functional. In addition, ωB97X-D includes a dispersion term, which contributes to improve the optimized geometries of large molecules when there is a significant non-covalent short-range interaction. Fig. 8 shows the COL ground (top) and triplet (bottom) state geometries, whereas Fig. 9 shows the corresponding MTC geometries. In both cases the diffusional profiles are very similar and hence the small differences observed in the ground and triplet geometries are not expected to affect the diffusivity significantly. In other words, it can be expected that the mobility of COL and MTC within TU will not be strongly dependent on the electronic state of the molecule.
In order to check this hypothesis, a new molecular dynamics run of 500 ps was performed with COL@TU under the abovementioned conditions. With the COL triplet geometry, including the new values of the relevant distances, angles, etc., the UFF force field was modified and loaded as input of the LAMMPS simulation. Fig. 5 shows the new diffusivity of 3COL* superimposed to that of COL ground state. It can be seen that the COL-ground and triplet states spend most of their time in a very close location (ca. 2 Å), exhibiting roughly the same mobility. A comparable behavior of their corresponding states is expected for MTC, given their similar geometries (Fig. 9).
Conclusions
For the first time, laser flash photolysis has been applied as a new tool to study the interaction between tubulin and drugs such as COL or MTC. Laser excitation of COL in buffered aqueous solutions does not show formation of any short-lived species. However, in the presence of tubulin, COL gives rise to a triplet–triplet transient absorption spectrum very similar to that registered in organic solvents, revealing location of the drug in a hydrophobic region of the biomolecule. Complex formation is accompanied by a remarkable enhancement of the intersystem crossing quantum yields and triplet lifetimes compared to acetonitrile. Likewise, the triplet excited state of MTC has been characterized in several solvents; its quantum yield decreases in protic media (down to nearly zero in water), whereas its lifetime remains practically constant. Again, complexation of MTC with tubulin in aqueous solution leads to a triplet–triplet transient absorption spectrum similar to that obtained in organic solution. The lifetime of this species in the complex is much longer than that measured in organic solvents and the formation quantum yield is also markedly higher. Molecular dynamic simulations, both with the ground and triplet excited state, reveal a strong interaction between COL and tubulin to give stabilized complexes with reduced mobility inside the protein binding site. This effect justifies the enhanced quantum yields and lifetimes of the triplet excited states observed in the drug@protein complexes. These results demonstrate that laser flash photolysis is a useful methodology to study the binding of COL derivatives to tubulin and open a new way to evaluate the interactions of other drugs with this protein. The obtained results can be highly relevant to understand the primary event in the anticancer activity of drugs binding to the COL site of tubulin.Acknowledgements
Financial support from the Spanish Government (grants CTQ2010-19909; BFU2011-23416 and SEV 2012-0267), the Generalitat Valenciana (Prometeo II/2013/005) and Comunidad de Madrid (S2010/BMD-2353) is gratefully acknowledged. G.S. thanks ASIC-UPV for computing time.Notes and references
- T. Mitchison and M. Kirschner, Nature, 1984, 312, 237–242 CrossRef CAS PubMed.
- R. L. Margolis and L. Wilson, Cell, 1978, 13, 1–8 CrossRef CAS.
- A. Desai and T. J. Mitchison, Annu. Rev. Cell Dev. Biol., 1997, 13, 83–117 CrossRef CAS PubMed.
- J. Howard and A. A. Hyman, Nature, 2003, 422, 753–758 CrossRef CAS PubMed.
- M. A. Jordan and L. Wilson, Nat. Rev. Cancer, 2004, 4, 253–265 CrossRef CAS PubMed.
- R. B. Ravelli, B. Gigant, P. A. Curmi, I. Jourdain, S. Lachkar, A. Sobel and M. Knossow, Nature, 2004, 428, 198–202 CrossRef CAS PubMed.
- A. Cormier, M. Marchand, R. B. G. Ravelli, M. Knossow and B. Gigant, EMBO Rep., 2008, 9, 1101–1106 CrossRef CAS PubMed.
- A. E. Prota, K. Bargsten, J. F. Diaz, M. Marsh, C. Cuevas, M. Liniger, M. Neuhaus, J. M. Andreu, K. H. Altmann and M. O. Steinmetza, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 13817–13821 CrossRef CAS PubMed.
- A. E. Prota, K. Bargsten, D. Zurwerra, J. J. Field, J. F. Diaz, K. H. Altmann and M. O. Steinmetz, Science, 2013, 339, 587–590 CrossRef CAS PubMed.
- A. E. Prota, K. Bargsten, P. T. Northcote, M. Marsh, K. Altmann, J. H. Miller, J. F. Diaz and M. Steinmetz, Angew. Chem., Int. Ed., 2014, 53, 1621–1625 CrossRef CAS PubMed.
- F. Santavý, Collect. Czech. Chem. Commun., 1951, 16, 655–675 Search PubMed.
- A. Brossi, H. J. C. Yeh, M. Chrzanowska, J. Wolff, E. Hamel, C. M. Lin, F. Quin, M. Suffness and J. Silverton, Med. Res. Rev., 1988, 8, 77–94 CrossRef CAS PubMed.
- J. L. Weiner, A. V. Buhler, V. J. Whatley, R. A. Harris and T. V. Dunwiddie, J. Pharmacol. Exp. Ther., 1998, 284, 95–102 CAS.
- M. Imazio, R. Trinchero and Y. Adler, Future Cardiol., 2008, 4, 599–607 CrossRef CAS PubMed.
- R. de Vincenzo, G. Scambia, C. Ferlini, M. Distefano, P. Filippini, A. Riva, E. Bombardelli, D. Pocar, M. L. Gelmi, P. B. Panici and S. Mancuso, Anti-Cancer Drug Des., 1998, 13, 19–33 CAS.
- M. Fakih, A. Yagoda, T. Reploge, J. E. Lehr and K. J. Pienta, Prostate, 1995, 26, 310–315 CrossRef CAS PubMed.
- R. M. Lee and D. A. Gerwitz, Drug Dev. Res., 2008, 69, 352–358 CrossRef CAS PubMed.
- A. Abad, J. L. Lopez-Perez, E. del Olmo, L. F. Garcia-Fernandez, A. Francesch, C. Trigili, I. Barasoain, J. M. Andreu, J. F. Diaz and A. San Feliciano, J. Med. Chem., 2012, 55, 6724–6737 CrossRef CAS PubMed.
- R. Alvarez, P. Puebla, J. F. Diaz, A. C. Bento, R. Garcia-Navas, J. de la Iglesia-Vicente, F. Mollinedo, J. M. Andreu, M. Medarde and R. Pelaez, J. Med. Chem., 2013, 56, 2813–2827 CrossRef CAS PubMed.
- D. Panda, J. E. Daijo, M. A. Jordan and L. Wilson, Biochemistry, 1995, 34, 9921–9929 CrossRef CAS.
- A. Lambeir and Y. Engelborghs, J. Biol. Chem., 1981, 256, 3279–3282 CAS.
- J. F. Diaz and J. M. Andreu, J. Biol. Chem., 1991, 266, 2890–2896 Search PubMed.
- J. M. Andreu and J. M. Timasheff, Biochemistry, 1982, 21, 534–543 CrossRef CAS.
- L. Atwell, A. Brossi, M. A. Iorio, T. H. Williams, R. H. Sik and C. F. Chignell, J. Med. Chem., 1981, 24, 257–261 CrossRef.
- B. Pérez-Ramirez, M. J. Gorbunoff and S. N. Timasheff, Biochemistry, 1998, 37, 1646–1661 CrossRef PubMed.
- S. B. Hastie, R. C. Williams Jr, D. Puett and T. L. Macdonald, J. Biol. Chem., 1989, 264, 6682–6688 CAS.
- C. Dumortier, Q. Yan, S. Bane and Y. Engelborghs, Biochem. J., 1997, 327, 685–688 CAS.
- S. Bane, D. Puett, T. L. Macdonald and R. C. Williams Jr, J. Biol. Chem., 1984, 259, 7391–7398 CAS.
- J. M. Andreu, M. J. Gorbunoff, J. C. Lee and S. N. Timasheff, Biochemistry, 1984, 23, 1742–1752 CrossRef CAS.
- Y. Engelborghs and T. J. Fitzgerald, J. Biol. Chem., 1987, 262, 5204–5209 CAS.
- T. L. Nguyen, C. McGrath, A. R. Hermone, J. C. Burnett, D. W. Zaharevitz, B. W. Day, P. Wipf, E. Hamel and R. Gussio, J. Med. Chem., 2005, 48, 6107–6116 CrossRef CAS PubMed.
- J. T. Huzil, P. Winter, L. Johnson, A. L. Weis, T. Bakos, A. Banerjee, R. F. Luduena, S. Damaraju and J. A. Tuszynski, Chem. Biol. Drug Des., 2010, 75, 541–550 CAS.
- R. Cao, M. Liu, M. Yin, Q. Liu, Y. Wang and N. Huang, J. Chem. Inf. Model., 2012, 52, 2730–2740 CrossRef CAS PubMed.
- N. Laing, B. Dahlloef, B. Hartley-Asp, S. Ranganathan and K. D. Tew, Biochemistry, 1997, 36, 871–878 CrossRef CAS PubMed.
- K. K. Gireesh, A. Rashid, S. Chakraborti, D. Panda and T. Manna, Biochem. Pharmacol., 2012, 84, 633–645 CrossRef CAS PubMed.
- N. Gunasekera, G. Xiong, K. Musier-Forsyth and E. Arriaga, Anal. Biochem., 2004, 330, 1–9 CrossRef CAS PubMed.
- F. J. Medrano, J. M. Andreu, M. J. Gorbunoff and S. N. Timasheff, Biochemistry, 1991, 30, 3770–3777 CrossRef CAS.
- K. C. Morrison and P. J. Hergenrother, Anal. Biochem., 2012, 420, 26–32 CrossRef CAS PubMed.
- S. B. Hastie and R. P. Rava, J. Am. Chem. Soc., 1989, 111, 6993–7001 CrossRef CAS.
- B. Bhattacharyya, S. Kapoor and D. Panda, Methods Cell Biol., 2010, 95, 301–329 CAS.
- P. S. Sardar, S. S. Maity, L. Das and S. Ghosh, Biochemistry, 2007, 46, 14544–14556 CrossRef CAS PubMed.
- B. Bhattacharyya and J. Wolff, Proc. Natl. Acad. Sci. U. S. A., 1974, 71, 2627–2631 CrossRef CAS.
- V. Lhiaubet-Vallet, Z. Sarabia, F. Bosca and M. A. Miranda, J. Am. Chem. Soc., 2004, 126, 9538–9539 CrossRef CAS PubMed.
- I. Vaya, V. Lhiaubet-Vallet, M. C. Jimenez and M. A. Miranda, Chem. Soc. Rev., 2014, 43, 4102–4122 RSC.
- F. Bosca and R. Tormos, J. Phys. Chem. B, 2013, 117, 7528–7534 CrossRef CAS PubMed.
- T. J. Fitzgerald, Biochem. Pharmacol., 1976, 25, 1383–1387 CrossRef CAS.
- J. M. Andreu, Methods Mol. Med., 2007, 137, 17–28 CAS.
- S. L. Murov, I. Carmichael and G. L. Hug, Handbook of Photochemistry, Marcel Dekker, Inc., New York, 2nd edn, 1993 Search PubMed.
- J. N. Silva, F. Bosca, J. P. C. Tome, E. M. P. Silva, M. G. P. M. S. Neves, J. A. S. Cavaleiro, L. K. Patterson, P. Filipe, J.-C. Maziere, R. Santus and P. Morliere, J. Phys. Chem. B, 2009, 113, 16695–16704 CrossRef CAS PubMed.
- E. J. Land, Biochimie, 1980, 62, 207–221 CrossRef CAS.
- R. V. Bensasson and J. C. Gramain, J. Chem. Soc., Faraday Trans., 1980, 76, 1800–1810 CAS.
- S. J. Plimpton, J. Comp. Physiol., 1995, 117, 1–19 CAS.
- P. J. in't Veld, S. J. Plimpton and G. S. Grest, Comput. Phys. Commun., 2008, 179, 320–329 CrossRef PubMed.
- A. K. Rappe, C. J. Casewit, K. S. Colwell, W. A. Goddard III and W. M. Skiff, J. Am. Chem. Soc., 1992, 114, 10024–10035 CrossRef CAS.
- MOPAC2009, James J. P. Stewart, Stewart computational chemistry, version 13.207L; web: http://OpenMOPAC.net.
- J. D. Carvalho Maia, G. A. Urquiza Carvalho, C. Peixoto Mangueira, S. Ramos Santana, L. A. Formiga Cabral and G. B. Rocha, J. Chem. Theory Comput., 2012, 8, 3072–3081 CrossRef.
- J.-D. Chai and M. Head-Gordon, Phys. Chem. Chem. Phys., 2008, 10, 6615–6620 RSC.
- A. Schaefer, C. Huber and R. Ahlrichs, J. Chem. Phys., 1994, 100, 5829–5835 CrossRef CAS PubMed.
- D. Jacquemin, W. Wathelet, E. A. Perpete and C. Adamo, J. Chem. Theory Comput., 2009, 5, 2420–2435 CrossRef CAS.
- D. Jacquemin, E. A. Perpete, I. Ciofini and C. Adamo, J. Chem. Theory Comput., 2010, 6, 1532–1537 CrossRef CAS.
- M. J. G. Peach, P. Benfield, T. Helgaker and D. J. Tozer, J. Chem. Phys., 2008, 128, 044118 CrossRef PubMed.
- P. Bartovsky, R. Tormos and M. A. Miranda, Chem. Phys. Lett., 2009, 480, 305–308 CrossRef CAS PubMed.
- I. Vaya, C. J. Bueno, M. C. Jimenez and M. A. Miranda, Chem.–Eur. J., 2008, 14, 11284–11287 CrossRef CAS PubMed.
- Y. Marcus, Chem. Soc. Rev., 1993, 22, 409–416 RSC.
- A. L. P. Nery, F. H. Quina, P. F. Moreira Jr, C. E. R. Medeiros, W. J. Baader, K. Shimizu, L. H. Catalani and E. J. H. Bechara, Photochem. Photobiol., 2001, 73, 213–218 CrossRef CAS.
- J. P. Perdew, K. Burke and M. Ernzerhof, Phys. Rev. Lett., 1996, 77, 3865–3868 CrossRef CAS.
- J. P. Perdew, K. Burke and M. Ernzerhof, Phys. Rev. Lett., 1997, 78, 1396 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5ra05636e |
| This journal is © The Royal Society of Chemistry 2015 |