Analogues of the marine alkaloids oroidin, clathrodin, and hymenidin induce apoptosis in human HepG2 and THP-1 cancer cells†
Tihomir Tomašič a, Dominik Nabergoj ab, Sanja Vrbek ab, Nace Zidar a, Žiga Jakopin a, Aleš Žula a, Žiga Hodnik a, Marko Jukič a, Marko Anderluh a, Janez Ilaš a, Marija Sollner Dolenc a, Jean Peluso bc, Geneviève Ubeaud-Séquier bc, Christian D. Muller *bc, Lucija Peterlin Mašič *a and Danijel Kikelj a
aUniversity of Ljubljana, Faculty of Pharmacy, Aškerčeva 7, 1000 Ljubljana, Slovenia. E-mail: lucija.peterlin@ffa.uni-lj.si; Fax: +386-1-4258031; Tel: +386-1-4769635
bLaboratoire d'Innovation Thérapeutique, UMR 7200, Faculté de Pharmacie, Université de Strasbourg, 67401 Illkirch, France. E-mail: cdmuller@unistra.fr; Fax: +33-368854310; Tel: +33-688285839
cPlateforme eBioCyt, Faculté de Pharmacie & Féderation Translationnelle de Médecine, Université de Strasbourg, 67401 Illkirch, France
First published on 26th August 2014
Abstract
The marine alkaloids clathrodin, oroidin, and hymenidin, which were isolated from Agelas sponges, possess diverse biological activities. Herein, we describe the design of a library of their analogues and the evaluation of their apoptosis-inducing activities against the human hepatocellular carcinoma HepG2 and acute monocytic leukaemia THP-1 cell lines. The screening of the complete library of 96 compounds using the HepG2 cell line allowed us to determine key structural elements and physicochemical properties that are responsible for the apoptosis-inducing activity. The indole-based compounds 24c, 28c, 29c, and 34c were found to be the most potent inducers of apoptosis in HepG2 and THP-1 cell lines with EC50 values in the low micromolar range. Cell cycle analysis assays confirmed that compounds 24c, 28c, 29c, and 34c induce the apoptosis of THP-1 cells at 25 μM, which highlights these oroidin analogues as interesting candidates for further evaluation of their anticancer activity.
Introduction
Marine organisms, particularly sponges, have been recognised as a rich source of natural products that possess anticancer activity.1–3 Clathrodin, hymenidin, and oroidin (Fig. 1) are pyrrole-2-aminoimidazole4 alkaloids that were initially isolated from the sponges of the genus Agelas. Oroidin acts as a chemical defence agent against the predatory reef fish and can be considered a biogenetic precursor of a variety of secondary metabolites that exhibit great structural complexity and diversity and present a range of biological activities.3,5 The oroidin class of marine alkaloids has been reported to display modulatory activities on voltage-gated sodium6 and calcium7 channels, and oroidin and its analogues have also been extensively studied as inhibitors of bacterial biofilm formation.8 However, the apoptosis-inducing activity of the marine alkaloids clathrodin, hymenidin, and oroidin, and their synthetic analogues has not yet been evaluated.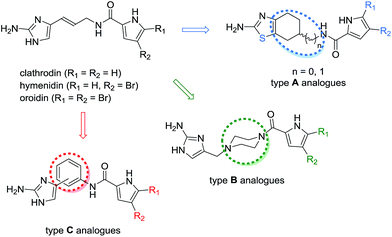 | ||
| Fig. 1 Structures of the pyrrole-2-aminoimidazole alkaloids clathrodin, hymenidin, and oroidin, and their structural modifications (type A–C analogues). | ||
The oroidin class of alkaloids is structurally relatively simple (Fig. 1) and possesses drug-like properties according to Lipinski's rule of five;9 thus, this class is well suited for use as a starting point for the design of novel analogues and mimetics that can be screened for their biological activities. Recently, inspired by the reported ability of clathrodin to block human voltage-gated sodium channels,6 we designed and synthesised a library of clathrodin and oroidin analogues and evaluated some of the resulting compounds to determine their modulatory activity on human voltage-gated sodium channels10,11 and their antibacterial activity.12
The clathrodin, hymenidin, and oroidin molecules possess a potentially unstable double bond in the linker between the 2-aminoimidazole and pyrrole moieties.13 Therefore, we designed and synthesised a library of their analogues by modifying the central part of the molecule (Fig. 1, type A to C analogues) to obtain more stable and conformationally restricted compounds as well as the western (Fig. 1, type A–C analogues) and eastern parts (type A–C analogues) of the molecule to enable structure–activity relationship studies. In the present work, we studied the apoptosis-inducing activity of clathrodin, oroidin, and hymenidin, and their type A–C analogues against HepG2 and THP-1 human cancer cell lines to evaluate their potential as anticancer agents against hepatocellular carcinoma and acute monocytic leukaemia.
Hepatocellular carcinoma is the most common type of liver cancer, and its high incidence has been attributed to persistent infection with hepatitis B or C virus, contact with hepatocarcinogens (e.g., aflatoxins), and cirrhosis. The development of drug resistance in hepatocellular carcinoma tumour cells after drug therapy indicates the important need for the discovery of novel anticancer agents for the successful treatment of liver cancer.14
Acute monocytic leukaemia, a type of acute myeloid leukaemia, is a hematopoietic cancer characterised by a disorder of hematopoietic progenitor cells, which lose their ability for normal differentiation and response to normal regulators of proliferation. Its incidence increases with age. Considering the aging population and the fact that acute myeloid leukaemia has the lowest survival rate of all leukaemias, new anticancer agents against acute myeloid leukaemia are urgently needed to fight this type of cancer in the future.15
Results and discussion
To evaluate the potential of the marine alkaloids clathrodin, oroidin, and hymenidin, and their type A–C analogues (a library of 96 compounds) to induce apoptosis in an in vitro human liver cancer model, we conducted a primary screening of all these compounds on the human hepatocellular carcinoma cell line HepG2 (ATCC® HB-8065™) using the annexin V/propidium iodide (PI) apoptosis assay with microcapillary flow cytometry (Guava EasyCyte™, Millipore/Merck, CA, USA) as the readout.16 Annexin V was used to assess the loss of membrane asymmetry, which is characterised by externalisation of phosphatidylserine, an early indicator of apoptosis.17 Propidium iodide, which is widely used as a DNA intercalating dye for the evaluation of cell viability or DNA content in cell cycle analyses, was used to determine the membrane integrity.18 Using this double-staining apoptosis assay, we were able to distinguish between living cells (annexin V-negative, PI-negative), early apoptotic cells (annexin V-positive, PI-negative) and late apoptotic/secondary necrotic cells (annexin V-positive, PI-positive).19 All the compounds were tested at 50 μM in four independent experiments (Tables S1–S4 in the ESI†). The HPLC purity of the tested compounds was above 95% monitored at 254 nm. Syntheses of the majority of compounds are reported elsewhere10–12,20,21 and their analytical data (1H- and 13C-NMR, mass spectra) can be found in the ESI.† Synthesis of the most active compounds 24c, 28c–30c and 34c is summarised in Scheme 1.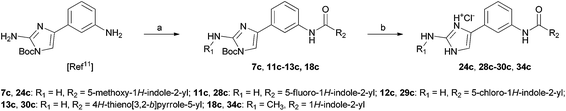 | ||
| Scheme 1 Synthesis of the most active compounds 24c, 28c–30c and 34c. Reagents and conditions: (a) corresponding carboxylic acid, TBTU, NMM, CH2Cl2, 35 °C, 24 h; (b) HCl(g), THF–EtOH, rt, 5 h. | ||
The marine alkaloids clathrodin, oroidin and hymenidin, which were synthesised in our laboratory,20 and their analogues 1–4 were found to possess only weak apoptosis-inducing activity in the HepG2 cell line with 25–38% apoptotic cells at 50 μM (Table S1†).
In the series of type A analogues (Fig. 1), conformational restriction was achieved by replacing the (E)-5-(3-aminoprop-1-enyl)-1H-imidazol-2-amine moiety by the 4,5,6,7-tetrahydrobenzo[d]thiazol-2-amine core in which the imidazole was isosterically replaced by the thiazole ring.10 However, type A analogues 1a–14a containing also various modifications in the central and eastern parts of the parent molecules (Table S2†) did not display improved apoptosis-inducing activity compared with oroidin (20–38% apoptotic HepG2 cells at 50 μM).
The type B analogues 1b–10b were obtained by modification of the central part through replacement of the 3-aminoprop-1-enyl linker between the 2-aminoimidazole and pyrrole moieties by the less flexible N-methylenepiperazine group (Fig. 1).21 Modification of the central part together with variations in the eastern and western parts of the parent molecules gave only weakly active compounds 1b–10b (Table S3,† 20–34% apoptotic HepG2 cells at 50 μM). In the type C analogues, the 1,3- or 1,4-disubstituted phenyl ring was incorporated in place of the prop-1-enyl linker to obtain a conformationally restricted central part of the molecule without changing the length of the molecule (Fig. 1).11 The analysis of the apoptosis-inducing activity of the type C analogues revealed a similar trend to those observed with the type A and B analogues. If the substituent in the eastern part of the molecule was a five-membered pyrrole (1c–3c, 14c, 19c, 20c, 31c, 37c, 38c, and 43c), pyrrolidine (4c, 21c, 40c, 41c, and 44c), furan (15c, 17c, and 32c), or imidazole (39c) ring, the compounds were only weakly active (Table S4, Fig. S1,† 13–44% apoptotic HepG2 cells at 50 μM). The only exceptions were the pyrrole-based compounds 35c and 36c, which contained a benzyl moiety on the imidazole ring nitrogen and induced apoptosis in 54% and 86% of HepG2 cells at a concentration of 50 μM (Table S4†), respectively. In contrast, a significant gain in the apoptosis-inducing activity was observed in the compounds containing an indole or substituted indole moiety in place of the pyrrole ring of the parent marine alkaloids.
The indole-based compounds 5c–12c, 16c, 18c, and 42c and 4H-thieno[3,2-b]pyrrole 13c containing a tert-butyloxycarbonyl (Boc) group on the imidazole N1 showed improved activity with 43–91% apoptotic HepG2 cells at 50 μM. A substitution at position 5 of the indole ring with a methoxy (7c), benzyloxy (8c), fluoro (11c), or chloro (12c) group resulted in improved activity compared to the non-substituted indoles 5c and 6c, whereas a hydroxy (9c) or trifluoromethyloxy (10c) substitution decreased the potency. A methyl substituent at the imidazole 2-amino group increased the percentage of apoptotic cells compared with those found with the indole-based compounds (5cvs.18c), whereas a reduction of the imidazole to obtain a 2-aminoimidazoline ring reduced the activity (18cvs.16c). Compound 42c with a 1,4-disubstituted phenyl ring in the central part and an indole moiety in the eastern part of the molecule was among the most active apoptosis-inducing compounds and was more active than its 1,3-phenylene counterpart 5c.
In general, the Boc-deprotected indole-based compounds 22c–29c, 33c, and 34c and the 4H-thieno[3,2-b]pyrrole-based compound 30c retained their apoptosis-inducing activity against the HepG2 cell line. Similar to their Boc-protected analogues, the compounds with the 6-fluoro- (28c), 6-chloroindole (29c), and 4H-thieno[3,2-b]pyrrole (30c) moieties were the most active with more than 90% apoptotic HepG2 cells at 50 μM.
We also evaluated the apoptosis-inducing activity of compounds 45c–65c (Table S4†), which were identified in the 3D similarity searching, based on the indole 22c, in the ZINC database of drug-like compounds.22 Interestingly, most of the indoles (45c, 53c, 55c, 57c, 62c, and 63c but not 58c and 64c) induced apoptosis in more than 50% of HepG2 cells at 50 μM, regardless of the ring type in the western part of the molecule, which indicates that the indole moiety is a crucial feature for significant apoptosis-inducing activity in type C oroidin analogues. Among compounds 45c–65c, only compound 52c showed improved apoptosis-inducing activity (92% apoptotic HepG2 cells at 50 μM) compared with the template compound 22c.
The molecular descriptor analysis of our library of oroidin analogues showed that most of the compounds possess drug-like properties according to Lipinski's rule of five9 (Fig. S2†). The presented charts show that most of the active compounds (>50% apoptotic HepG2 cells at 50 μM) are more lipophilic (log D values between 3 and 5) and have higher molecular weights (MW between 300 and 500) compared with their inactive counterparts (log D values between −1 and 5, MW between 200 and 500). In contrast, the number of hydrogen bond donors and acceptors is similarly distributed between the actives and inactives.
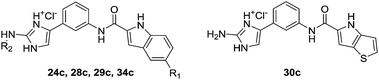
Of the 96 compounds screened, compounds 24c, 28c–30c, and 34c, which were among the most active type C oroidin analogues, were selected for further characterisation. First, the EC50 values for their apoptosis-inducing activity against the HepG2 cell line were determined using the annexin V/PI apoptosis assay (Table 1). 6-Fluoroindole 28c was found to be the most active compound with an EC50 value of 13 μM, followed by 6-chloroindole 29c (EC50 = 16 μM), 6-methoxyindole 24c (EC50 = 18 μM), indole 34c (EC50 = 20 μM), and 4H-thieno[3,2-b]pyrrole 30c (EC50 = 42 μM).
Considering the noteworthy HepG2 apoptosis-inducing activity of compounds 24c, 28c–30c, and 34c, these were further evaluated using the human monocytic leukaemia THP-1 cell line (ATCC® TIB-202™) (Table 1). The screening of these compounds at 50 μM against the THP-1 cell line showed that compounds 24c, 28c, 29c, and 34c induce apoptosis (87–97% apoptotic THP-1 cells), whereas compound 30c was found to be inactive (12% apoptotic THP-1 cells). The dose–response curves showed that compounds 24c, 28c, 29c, and 34c exhibited similar activities against the THP-1 cell line with EC50 values ranging from 20 μM to 24 μM (Table 1). Because activation of the apoptotic pathways is a key mechanism through which anticancer drugs kill tumour cells,23 it was important to confirm that compounds 24c, 28c, 29c, and 34cde facto exert their cytotoxic effect against the THP-1 cell line via the induction of apoptosis and not by necrosis. Hence, THP-1 cells were subjected to a cell cycle analysis after exposure to 24c, 28c, 29c, and 34c to determine the incidence of fragmented DNA (sub-G1 population) by PI staining of the nuclei.24 DMSO (0.25%), which was used as a negative control, did not affect the cell cycle in THP-1 cells (Fig. 2). The results of the cell cycle analysis after the incubation of THP-1 cells with compounds 24c, 28c, 29c, or 34c at 25 μM for 24 h and 48 h (Fig. 2 and 3 and Table S5†) show the presence of a sub-G1 cell population, which confirmed the presence of programed cell death, i.e. apoptosis. Compounds 28c and 29c showed similar potency: 41% and 40% of the cells were found in the sub-G1 peak after 48 h of treatment, respectively. Compounds 34c and 24c displayed more potent apoptosis-inducing activity with 49% and 60% of the cells in the sub-G1 population, although the EC50 values of all four compounds were very similar (20–24 μM).
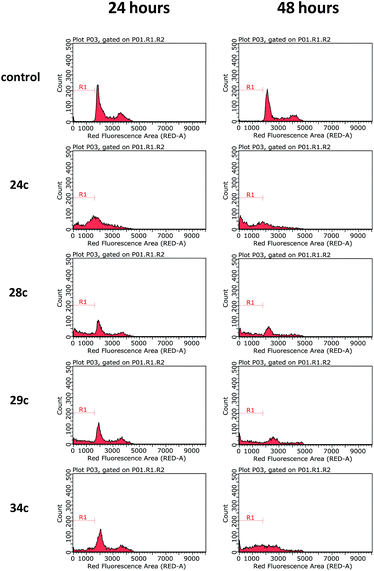 | ||
| Fig. 2 Cell cycle analysis of THP-1 cells after incubation with compounds 24c, 28c, 29c, and 34c at 25 μM for 24 h and 48 h. DMSO (0.25%) in culture medium was used as a negative control. Representative histograms of three independent experiments are displayed. | ||
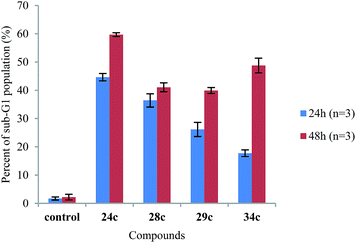 | ||
| Fig. 3 Comparison of the sub-G1 cell cycle population after the treatment of THP-1 cells with compounds 24c, 28c, 29c, and 34c at 25 μM for 24 h and 48 h (n = 3 independent experiments). | ||
Conclusions
In conclusion, the marine alkaloids clathrodin, oroidin, and hymenidin and a library of their synthetic analogues were tested to determine their apoptosis-inducing activities against the human hepatocellular carcinoma HepG2 cell line. Although the three tested marine alkaloids were found to be only weakly active, the library compounds, particularly their indole-based type C analogues, displayed promising activities. Compounds 24c, 28c–30c, and 34c, which were the most active compounds in the library, exhibited EC50 values between 13 μM and 42 μM against the HepG2 cell line and between 20 μM and 24 μM against the acute monocytic leukaemia THP-1 cell line. Through cell cycle analysis, it was confirmed that compounds 24c, 28c, 29c, and 34c induce apoptosis at 25 μM in THP-1 cells. These results render compounds 24c, 28c, 29c, and 34c interesting hits for further optimisation towards more potent oroidin, hymenidin and clathrodin analogues that induce apoptosis of human hepatic and blood cancer cells. Additional investigation of their mechanism of induction of apoptosis at the molecular level will be necessary to assess their potential for development towards new anticancer agents.Experimental section
Chemistry
Synthesis of compounds 24c, 28c–30c and 34c. A solution of Boc-protected starting compound (1 mmol) in a mixture of THF and EtOH = 1 : 2 (5 mL) was saturated with gaseous HCl and stirred at room temperature for 5 h. The solvent was removed under reduced pressure, and the solid was filtered off and washed with diethyl ether and dichloromethane, to afford the title compound.
2-Amino-4-(3-(5-methoxy-1H-indole-2-carboxamido)phenyl)-1H-imidazol-3-ium chloride (24c). Prepared from 7c according to General procedure A. Yield: 96%; off-white solid; mp 237–241 °C; IR (KBr) ν = 3301 (N–H), 3138 (C–H), 2955 (C–H), 2761 (C–H), 1673 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1653, 1625, 1585, 1541, 1452, 1418, 1336, 1281, 1238, 1208, 1177, 1153, 1132, 1116, 1022, 883, 839, 788, 755 cm−1. 1H NMR (DMSO-d6) δ 3.79 (s, 3H, OCH3), 6.89 (dd, 1H, 3J = 9.2 Hz, 4J = 2.4 Hz, Ar-H), 7.15 (d, 1H, 4J = 2.4 Hz, Ar-H), 7.33 (s, 1H, Ar-H), 7.37–7.49 (m, 6H, 4 × Ar-H, NH2), 7.69–7.72 (m, 1H, Ar-H), 8.08 (s, 1H, Ar-H), 10.42 (s, 1H, NH), 11.70 (s, 1H, NH), 12.16 (s, 1H, NH), 12.85 (s, 1H, NH); 13C NMR (DMSO-d6) δ 55.25 (OCH3), 102.02, 104.14, 109.43, 113.23, 115.19, 116.46, 119.74, 120.33, 126.39, 127.26, 128.12, 129.30, 131.54, 132.16, 139.45, 147.82, 153.84, 159.72; MS (ESI) m/z (%) = 348.2 ([M − Cl]+, 100). HRMS for C19H18N5O2: calculated, 348.1461; found, 348.1459. HPLC: Phenomenex Luna 5 μm C18 column (4.6 mm × 150 mm); mobile phase: 60–90% of MeOH in TFA (0.1%) in 20 min; flow rate: 1.0 mL min−1; injection volume: 10 μL; retention time: 3.029 min (98.2% at 254 nm, 98.7% at 280 nm).
O), 1653, 1625, 1585, 1541, 1452, 1418, 1336, 1281, 1238, 1208, 1177, 1153, 1132, 1116, 1022, 883, 839, 788, 755 cm−1. 1H NMR (DMSO-d6) δ 3.79 (s, 3H, OCH3), 6.89 (dd, 1H, 3J = 9.2 Hz, 4J = 2.4 Hz, Ar-H), 7.15 (d, 1H, 4J = 2.4 Hz, Ar-H), 7.33 (s, 1H, Ar-H), 7.37–7.49 (m, 6H, 4 × Ar-H, NH2), 7.69–7.72 (m, 1H, Ar-H), 8.08 (s, 1H, Ar-H), 10.42 (s, 1H, NH), 11.70 (s, 1H, NH), 12.16 (s, 1H, NH), 12.85 (s, 1H, NH); 13C NMR (DMSO-d6) δ 55.25 (OCH3), 102.02, 104.14, 109.43, 113.23, 115.19, 116.46, 119.74, 120.33, 126.39, 127.26, 128.12, 129.30, 131.54, 132.16, 139.45, 147.82, 153.84, 159.72; MS (ESI) m/z (%) = 348.2 ([M − Cl]+, 100). HRMS for C19H18N5O2: calculated, 348.1461; found, 348.1459. HPLC: Phenomenex Luna 5 μm C18 column (4.6 mm × 150 mm); mobile phase: 60–90% of MeOH in TFA (0.1%) in 20 min; flow rate: 1.0 mL min−1; injection volume: 10 μL; retention time: 3.029 min (98.2% at 254 nm, 98.7% at 280 nm). 2-Amino-4-(3-(5-fluoro-1H-indole-2-carboxamido)phenyl)-1H-imidazol-3-ium chloride (28c). Prepared from 11c according to General procedure A. Yield: 77%; off-white solid; mp 202–205 °C; IR (KBr) ν = 3443 (N–H), 3275 (N–H), 3145 (C–H), 2764 (C–H), 1662 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1628, 1607, 1544, 1486, 1449, 1411, 1327, 1287, 1244, 1231, 1204, 1145, 1103, 954, 840, 780, 752, 727 cm−1. 1H NMR (DMSO-d6) δ 7.11 (dt, 1H, 3J = 9.2 Hz, 4J = 2.0 Hz, Ar-H), 7.33 (s, 1H, Ar-H), 7.40–7.50 (m, 7H, 5 × Ar-H, NH2), 7.69–7.71 (m, 1H, Ar-H), 8.08 (t, 1H, 4J = 1.6 Hz, Ar-H), 10.49 (s, 1H, NH), 11.95 (s, 1H, NH), 12.14 (s, 1H, NH), 12.83 (s, 1H, NH); 13C NMR (DMSO-d6) δ 104.34 (d, 1C, 4JC–F = 5 Hz), 105.89 (d, 1C, 2JC–F = 23 Hz), 109.51, 112.65 (d, 1C, 2JC–F = 26 Hz), 113.63 (d, 1C, 3JC–F = 9 Hz), 116.52, 119.93, 120.39, 126.37, 127.00 (d, 1C, 3JC–F = 9 Hz), 128.17, 129.35, 132.96, 133.57, 139.28, 147.81, 157.20 (d, 1C, 1JC–F = 231 Hz), 159.47; 19F NMR (DMSO-d6) δ −123.59 (s, 1F); MS (ESI) m/z (%) = 336.1 ([M − Cl]+, 100). HRMS for C18H15N5OF: calculated, 336.1261; found, 336.1264. HPLC: Phenomenex Luna 5 μm C18 column (4.6 mm × 150 mm); mobile phase: 60–90% of MeOH in TFA (0.1%) in 20 min; flow rate: 1.0 mL min−1; injection volume: 10 μL; retention time: 3.585 min (99.4% at 254 nm, 99.1% at 280 nm).
O), 1628, 1607, 1544, 1486, 1449, 1411, 1327, 1287, 1244, 1231, 1204, 1145, 1103, 954, 840, 780, 752, 727 cm−1. 1H NMR (DMSO-d6) δ 7.11 (dt, 1H, 3J = 9.2 Hz, 4J = 2.0 Hz, Ar-H), 7.33 (s, 1H, Ar-H), 7.40–7.50 (m, 7H, 5 × Ar-H, NH2), 7.69–7.71 (m, 1H, Ar-H), 8.08 (t, 1H, 4J = 1.6 Hz, Ar-H), 10.49 (s, 1H, NH), 11.95 (s, 1H, NH), 12.14 (s, 1H, NH), 12.83 (s, 1H, NH); 13C NMR (DMSO-d6) δ 104.34 (d, 1C, 4JC–F = 5 Hz), 105.89 (d, 1C, 2JC–F = 23 Hz), 109.51, 112.65 (d, 1C, 2JC–F = 26 Hz), 113.63 (d, 1C, 3JC–F = 9 Hz), 116.52, 119.93, 120.39, 126.37, 127.00 (d, 1C, 3JC–F = 9 Hz), 128.17, 129.35, 132.96, 133.57, 139.28, 147.81, 157.20 (d, 1C, 1JC–F = 231 Hz), 159.47; 19F NMR (DMSO-d6) δ −123.59 (s, 1F); MS (ESI) m/z (%) = 336.1 ([M − Cl]+, 100). HRMS for C18H15N5OF: calculated, 336.1261; found, 336.1264. HPLC: Phenomenex Luna 5 μm C18 column (4.6 mm × 150 mm); mobile phase: 60–90% of MeOH in TFA (0.1%) in 20 min; flow rate: 1.0 mL min−1; injection volume: 10 μL; retention time: 3.585 min (99.4% at 254 nm, 99.1% at 280 nm). 2-Amino-4-(3-(5-chloro-1H-indole-2-carboxamido)phenyl)-1H-imidazol-3-ium chloride (29c). Prepared from 12c according to General procedure A. Yield: 71%; white solid; mp 201–204 °C; IR (KBr) ν = 3410 (N–H), 3260 (N–H), 3145 (C–H), 3032 (C–H), 2761 (C–H), 1693 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1667, 1610, 1542, 1485, 1442, 1412, 1326, 1301, 1275, 1245, 1224, 1190, 1124, 1056, 914, 854, 798, 782, 754, 725 cm−1. 1H NMR (DMSO-d6) δ 7.25 (dd, 1H, 3J = 8.8 Hz, 4J = 2.0 Hz, Ar-H), 7.33 (s, 1H, Ar-H), 7.40–7.51 (m, 6H, 4 × Ar-H, NH2), 7.69–7.71 (m, 1H, Ar-H), 7.79 (d, 1H, 4J = 2.0 Hz, Ar-H), 8.07 (s, 1H, Ar-H), 10.53 (s, 1H, NH), 12.05 (s, 1H, NH), 12.14 (s, 1H, NH), 12.84 (s, 1H, NH); 13C NMR (DMSO-d6) δ 103.87, 109.52, 114.01, 116.53, 119.98, 120.40, 120.82, 123.98, 124.42, 126.37, 127.98, 128.18, 129.36, 132.75, 135.21, 139.23, 147.80, 159.41; MS (ESI) m/z (%) = 352.1 ([M − Cl]+, 100). HRMS for C18H15N5OCl: calculated, 352.0965; found, 352.0959. HPLC: Phenomenex Luna 5 μm C18 column (4.6 mm × 150 mm); mobile phase: 60–90% of MeOH in TFA (0.1%) in 20 min; flow rate: 1.0 mL min−1; injection volume: 10 μL; retention time: 5.338 min (98.4% at 254 nm, 98.8% at 280 nm).
O), 1667, 1610, 1542, 1485, 1442, 1412, 1326, 1301, 1275, 1245, 1224, 1190, 1124, 1056, 914, 854, 798, 782, 754, 725 cm−1. 1H NMR (DMSO-d6) δ 7.25 (dd, 1H, 3J = 8.8 Hz, 4J = 2.0 Hz, Ar-H), 7.33 (s, 1H, Ar-H), 7.40–7.51 (m, 6H, 4 × Ar-H, NH2), 7.69–7.71 (m, 1H, Ar-H), 7.79 (d, 1H, 4J = 2.0 Hz, Ar-H), 8.07 (s, 1H, Ar-H), 10.53 (s, 1H, NH), 12.05 (s, 1H, NH), 12.14 (s, 1H, NH), 12.84 (s, 1H, NH); 13C NMR (DMSO-d6) δ 103.87, 109.52, 114.01, 116.53, 119.98, 120.40, 120.82, 123.98, 124.42, 126.37, 127.98, 128.18, 129.36, 132.75, 135.21, 139.23, 147.80, 159.41; MS (ESI) m/z (%) = 352.1 ([M − Cl]+, 100). HRMS for C18H15N5OCl: calculated, 352.0965; found, 352.0959. HPLC: Phenomenex Luna 5 μm C18 column (4.6 mm × 150 mm); mobile phase: 60–90% of MeOH in TFA (0.1%) in 20 min; flow rate: 1.0 mL min−1; injection volume: 10 μL; retention time: 5.338 min (98.4% at 254 nm, 98.8% at 280 nm). 4-(3-(4H-Thieno[3,2-b]pyrrole-5-carboxamido)phenyl)-2-amino-1H-imidazol-3-ium chloride (30c). Prepared from 13c according to General procedure A. Yield: 78%; off-white solid; mp 198–202 °C; IR (KBr) ν = 3241 (N–H), 3135 (C–H), 3047 (C–H), 2763 (C–H), 1677 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1625, 1541, 1488, 1460, 1385, 1348, 1308, 1231, 1191, 1115, 1084, 963, 877, 827, 748, 711 cm−1. 1H NMR (DMSO-d6) δ 7.03 (dd, 1H, 3J = 5.2 Hz, 4J = 0.8 Hz, Ar-H), 7.31 (s, 1H, Ar-H), 7.36–7.49 (m, 6H, 4 × Ar-H, NH2), 7.66–7.69 (m, 1H, Ar-H), 8.06 (t, 1H, 4J = 1.6 Hz, Ar-H), 10.24 (s, 1H, NH), 11.99 (s, 1H, NH), 12.14 (s, 1H, NH), 12.82 (s, 1H, NH); 13C NMR (DMSO-d6) δ 103.91, 109.43, 111.90, 116.27, 119.48, 120.15, 122.94, 126.45, 128.08, 128.28, 129.26, 130.49, 139.64, 141.32, 147.77, 159.41; MS (ESI) m/z (%) = 324.1 ([M − Cl]+, 100). HRMS for C16H14N5OS: calculated, 324.0919; found, 324.0911. HPLC: Phenomenex Luna 5 μm C18 column (4.6 mm × 150 mm); mobile phase: 60–90% of MeOH in TFA (0.1%) in 20 min; flow rate: 1.0 mL min−1; injection volume: 10 μL; retention time: 2.790 min (96.5% at 254 nm, 97.0% at 280 nm).
O), 1625, 1541, 1488, 1460, 1385, 1348, 1308, 1231, 1191, 1115, 1084, 963, 877, 827, 748, 711 cm−1. 1H NMR (DMSO-d6) δ 7.03 (dd, 1H, 3J = 5.2 Hz, 4J = 0.8 Hz, Ar-H), 7.31 (s, 1H, Ar-H), 7.36–7.49 (m, 6H, 4 × Ar-H, NH2), 7.66–7.69 (m, 1H, Ar-H), 8.06 (t, 1H, 4J = 1.6 Hz, Ar-H), 10.24 (s, 1H, NH), 11.99 (s, 1H, NH), 12.14 (s, 1H, NH), 12.82 (s, 1H, NH); 13C NMR (DMSO-d6) δ 103.91, 109.43, 111.90, 116.27, 119.48, 120.15, 122.94, 126.45, 128.08, 128.28, 129.26, 130.49, 139.64, 141.32, 147.77, 159.41; MS (ESI) m/z (%) = 324.1 ([M − Cl]+, 100). HRMS for C16H14N5OS: calculated, 324.0919; found, 324.0911. HPLC: Phenomenex Luna 5 μm C18 column (4.6 mm × 150 mm); mobile phase: 60–90% of MeOH in TFA (0.1%) in 20 min; flow rate: 1.0 mL min−1; injection volume: 10 μL; retention time: 2.790 min (96.5% at 254 nm, 97.0% at 280 nm). 4-(3-(1H-Indole-2-carboxamido)phenyl)-2-(methylamino)-1H-imidazol-3-ium chloride (34c). Prepared from 18c according to General procedure A. Yield: 95%; white solid; mp 190–194 °C; IR (KBr) ν = 3269, 3182, 3049, 2934, 2865, 2746, 1685, 1635, 1602, 1541, 1494, 1414, 1368, 1334, 1312, 1248, 1190, 1145, 1115, 1060, 1018, 989, 919, 971, 819, 796, 775, 750 cm−1. 1H NMR (DMSO-d6) δ 2.95 (d, 3H, 3J = 4.8 Hz, CH3), 7.09 (dt, 1H, 3J = 6.8 Hz, 4J = 1.2 Hz, indole-H), 7.24 (dt, 1H, 3J = 6.8 Hz, 4J = 1.2 Hz, indole-H), 7.43–7.50 (m, 5H, 5 × Ar-H), 7.68–7.72 (m, 2H, 2 × Ar-H), 7.84 (q, 1H, 3J = 4.8 Hz, NH), 8.11 (t, 1H, 4J = 2.0 Hz, Ar-H-2′), 10.44 (s, 1H, NH), 11.82 (s, 1H, NH), 12.40 (br s, 1H, NH), 12.62 (br s, 1H, NH); 13C NMR (DMSO-d6) δ 30.07 (CH3), 105.00, 110.20, 112.88, 117.28, 120.42, 120.67, 120.87, 122.25, 124.35, 127.43, 127.45, 128.69, 129.72, 131.80, 137.33, 139.87, 149.08, 160.25; MS (ESI) m/z (%) = 332 ([M − Cl]+, 100). HRMS for C19H18N5O: calculated, 332.1511; found, 332.1499. HPLC: Phenomenex Luna 5 μm C18 column (4.6 mm × 150 mm); mobile phase: 10–90% of MeOH in TFA (0.1%) in 20 min; flow rate 1.0 mL min−1; injection volume: 20 μL; retention time: 17.146 min (99.2% at 254 nm, 99.2% at 280 nm).
Acknowledgements
This work was supported by the Slovenian Research Agency (Grant no. P1-0208 and Grant no. Z1-5458), by the European Union FP7 Integrated Project MAREX: Exploring Marine Resources for Bioactive Compounds: From Discovery to Sustainable Production and Industrial Applications (Project no. FP7-KBBE-2009-3-245137) and by the COST CM1106 (Chemical Approaches to Targeting Drug Resistance in Cancer Stem Cells).Notes and references
- D. Sipkema, M. C. R. Franssen, R. Osinga, J. Tramper and R. H. Wijffels, Mar. Biotechnol., 2005, 7, 142 CrossRef CAS PubMed.
- R. Singh, M. Sharma, P. Joshi and D. S. Rawat, Anti-Cancer Agents Med. Chem., 2008, 8, 603 CrossRef CAS.
- A. Al-Mourabit, M. A. Zancanella, S. Tilvi and D. Romo, Nat. Prod. Rep., 2011, 28, 1229 RSC.
- A. Žula, D. Kikelj and J. Ilaš, Mini-Rev. Med. Chem., 2013, 13, 1921 CrossRef.
- B. Forte, B. Malgesini, C. Piutti, F. Quartieri, A. Scolaro and G. Papeo, Mar. Drugs, 2009, 7, 705 CrossRef CAS PubMed.
- A. L. Rentas, R. Rosa, A. D. Rodriguez and G. E. De Motta, Toxicon, 1995, 33, 491 CrossRef CAS.
- U. Bickmeyer, C. Drechsler, M. Kock and M. Assmann, Toxicon, 2004, 44, 45 CrossRef CAS PubMed.
- J. J. Richards, T. E. Ballard and C. Melander, Org. Biomol. Chem., 2008, 6, 1356 CAS.
- C. A. Lipinski, F. Lombardo, B. W. Dominy and P. J. Feeney, Adv. Drug Delivery Rev., 2001, 46, 3 CrossRef CAS.
- Ž. Hodnik, T. Tomašić, L. Peterlin Mašič, F. Chan, R. W. Kirby, D. J. Madge and D. Kikelj, Eur. J. Med. Chem., 2013, 70, 154 CrossRef PubMed.
- N. Zidar, Ž. Jakopin, D. J. Madge, F. Chan, J. Tytgat, S. Peigneur, M. Sollner Dolenc, T. Tomašić, J. Ilaš, L. Peterlin Mašič and D. Kikelj, Eur. J. Med. Chem., 2014, 74, 23 CrossRef CAS PubMed.
- N. Zidar, S. Montalvão, Ž. Hodnik, D. A. Nawrot, A. Žula, J. Ilaš, D. Kikelj, P. Tammela and L. Peterlin Mašič, Mar. Drugs, 2014, 12, 940 CrossRef CAS PubMed.
- T. Lindel, G. Breckle, M. Hochgurtel, C. Volk, A. Grube and M. Kock, Tetrahedron Lett., 2004, 45, 8149 CrossRef CAS PubMed.
- S. H. Henry, F. X. Bosch and J. C. Bowers, Adv. Exp. Med. Biol., 2002, 504, 229 CrossRef CAS.
- E. Estey and H. Dohner, Lancet, 2006, 368, 1894 CrossRef.
- M. van Engeland, L. J. Nieland, F. C. Ramaekers, B. Schutte and C. P. Reutelingsperger, Cytometry, 1998, 31, 1 CrossRef CAS.
- S. J. Martin, C. P. Reutelingsperger, A. J. McGahon, J. A. Rader, R. C. van Schie, D. M. LaFace and D. R. Green, J. Exp. Med., 1995, 182, 1545 CrossRef CAS.
- H. Lecoeur, Exp. Cell Res., 2002, 277, 1 CrossRef CAS PubMed.
- T. Vanden Berghe, S. Grootjans, V. Goossens, Y. Dondelinger, D. V. Krysko, N. Takahashi and P. Vandenabeele, Methods, 2013, 61, 117 CrossRef CAS PubMed.
- A. Žula, D. Kikelj and J. Ilaš, Tetrahedron Lett., 2014, 55, 3999 CrossRef PubMed.
- M. Jukič, R. Frlan, F. Chan, R. W. Kirby, D. J. Madge, M. Anderluh and D. Kikelj, Med. Chem. Res., 2014, Search PubMed accepted.
- T. Tomašić, B. Hartzoulakis, N. Zidar, F. Chan, R. W. Kirby, D. J. Madge, S. Peigneur, J. Tytgat and D. Kikelj, J. Chem. Inf. Model., 2013, 53, 3223 CrossRef PubMed.
- S. H. Kaufmann and W. C. Earnshaw, Exp. Cell Res., 2000, 256, 42 CrossRef CAS PubMed.
- C. Riccardi and I. Nicoletti, Nat. Protoc., 2006, 1, 1458 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental procedures, compound characterization data, 1H and 13C spectra of the most active compounds, analysis of molecular descriptors, biological activity data, and description of biological assays. See DOI: 10.1039/c4md00286e |
| This journal is © The Royal Society of Chemistry 2015 |