Quantification of graphene and graphene oxide in complex organic matrices†
Kyle
Doudrick
*a,
Takayuki
Nosaka
b,
Pierre
Herckes
c and
Paul
Westerhoff
d
aDepartment of Civil and Environmental Engineering and Earth Sciences, University of Notre Dame, Notre Dame, IN 46556, USA. E-mail: kdoudric@nd.edu
bSchool for Engineering of Matter, Transport and Energy, Arizona State University, Tempe, AZ 85287-6106, USA
cDepartment of Chemistry and Biochemistry, Arizona State University, Tempe, AZ 85287-1604, USA
dSchool of Sustainable Engineering and the Built Environment, Arizona State University, Tempe, AZ 85287-5306, USA
First published on 3rd November 2014
Abstract
Interest is growing for graphene as a nanomaterial for electronic and composite applications. Increased production and use of graphene warrants development of strategies to detect and monitor its effect on human health and the environment. A quantification method using programmed thermal analysis (PTA) was developed for few-layer graphene (FLG) and graphene oxide (GO). FLG exhibited strong thermal stability, which allowed for easy detection in matrices consisting of thermally weaker background organic carbon. GO (50% oxygen content) exhibited a weaker thermal stability than FLG, making quantification more challenging in the presence of thermally similar background organic carbon. To resolve this, an in situ reduction method using a reducing agent (sodium borohydride) was developed to remove surface-bound oxygen from GO. This was used in combination with a digestate (SolvableTM) to create an optimized extraction method for recovering FLG and GO from complex organic matrices. FLG and GO will enter sewer systems due to their use by industry and in consumer products. We investigated the applicability of this method for quantifying FLG and GO in wastewater biomass because they are likely to accumulate in wastewater biosolids, as these are commonly the first exposure route for novel materials in the environment. Spiking 20 μg of FLG and GO into a 200 mg dried biomass L−1 wastewater solution resulted in recoveries of 52 ± 8% and 80 ± 6%, respectively. Results from this study can be applied to the development of extraction methods for graphene from similar complex organic matrices (e.g., lung tissue, in vitro/in vivo studies, algae, daphnia) to support a range of human and ecotoxicological studies.
Nano impactWith increased graphene use in consumer products there is a growing concern over its effect on human health and the environment. Quantification methods are needed to understand the risk associated with graphene. In this study, we describe a method for quantifying graphene in complex organic matrices. This method is useful for monitoring graphene in the environment and determining its impact on human health. Given graphene's likelihood to end up in wastewater treatment plants, we demonstrate the applicability of this method for wastewater biosolids. The results presented in this study will also be fundamental for the further development of methods for quantifying graphene in other complex matrices (e.g., sediment, tissue). |
Introduction
With the influx of graphene into the composite and electronic markets there is a growing concern about the risk of graphene to human health and the environment.1,2 Currently, the lack of established methods for quantifying graphene and the lack of reported methods for extracting graphene from complex matrices limits the ability to conduct appropriate human and eco-toxicity studies. The availability of quantification methods is important for developing reliable dose-response toxicity metrics and for monitoring workplace safety. In the environment, the same quantification methods are useful for determining exposure concentrations and assessing graphene fate and transport routes.Detection methods such as X-ray diffraction3 and Raman spectroscopy4 are useful for characterizing graphene, but they do not allow for appropriate quantitative analysis. Thermogravimetric analysis (TGA) and ultraviolet–visible (UV–vis) spectroscopy can also be used to detect graphene; they are quantitative methods, although limited in that respect. TGA is useful for determining the thermal stability of graphene and can also quantify purity (i.e., metal content),5 but it is limited to purer, dry samples rather than graphene in complex environmental or biological matrices. UV–vis has been used previously to characterize the dispersion state of graphene oxide (GO) in aqueous solutions,6 and it can be used as a means of quantifying GO in aqueous solutions, but only if the dispersion (i.e., aggregation) state stays constant. The UV–vis sensitivity becomes poor for graphene (stacked sheets in aqueous matrix) and GO in aqueous solution below approximately 1.5 mg L−1 and 75 μg L−1, respectively (Fig. S1† showing UV–vis spectrum for graphene and GO). In more complex matrices (e.g., surface water), quantifying graphene and GO will be more difficult due to different aggregation states and matrix interferences in the same wavelength range, which is especially true for GO (peaks between 220–250 nm; Fig. S1†). The lack of analytical methods for quantifying graphene in complex matrices signifies a need to develop robust analytical strategies that include both quantification and sample preparation.
We have previously developed a quantification method for carbon nanotubes (CNT),7 and have applied it to CNTs that were extracted from lung tissue with a high recovery.8 This quantification method, termed programmed thermal analysis (PTA), is an organic carbon/elemental carbon analysis that determines carbon mass and separates CNTs from other forms of carbon on the basis of the CNT's thermal stability. This separation is achieved using a time-dependent temperature ramp program; thermally weaker carbon compounds (e.g., tissue, bacteria) evolve early in the program while thermally stronger carbon compounds (e.g., CNT, graphene) evolve later. The ability to separate distinct forms of carbon is important for avoiding background interferences when quantifying carbonaceous nanomaterials in complex matrices containing organic carbon.
Before PTA can be used to quantify CNTs in complex organic matrices, CNTs must be extracted to separate them from excess carbonaceous material that could interfere with the analysis. With proper extraction methods in place, CNTs can be concentrated and then quantified using a number of methods (e.g., TGA-mass spectrometry,9 gel electrophoresis,10 infrared,11 radio-labeling,12 microwave,13 UV–vis,14 and inductively coupled plasma-mass spectrometry15). Given the physical and chemical similarities between graphene and CNTs, we hypothesize that the same approach can be used for extracting and quantifying graphene.
For PTA, oxygen functional groups on CNTs are problematic because they complicate separation of CNTs from organic carbon during analysis.7 Graphene is expected to be easily amenable to PTA because of its low oxygen content and consequently high thermal stability.16 Alternatively, GO tends to have a very high oxygen content, with a carbon to oxygen ratio (C![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) O) on the order of 1
O) on the order of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1; thus, its thermal behavior is similar to organic carbon. While the similar thermal behavior is not an issue for samples containing only GO (e.g., pure aqueous GO stock solutions), it interferes with analysis when quantifying GO in matrices containing organic carbon. GO can be transformed to “reduced graphene oxide” (RGO) using chemical reducing agents such as hydrazine17,18 or sodium borohydride.19–23 Removing oxygen makes graphene (oxide) more hydrophobic, which increases its tendency to aggregate and results in a more efficient separation and extraction. The key to any successful approach for environmental and biological samples will be doing this in situ (i.e., in a complex matrix) so that GO can easily be recovered.
1; thus, its thermal behavior is similar to organic carbon. While the similar thermal behavior is not an issue for samples containing only GO (e.g., pure aqueous GO stock solutions), it interferes with analysis when quantifying GO in matrices containing organic carbon. GO can be transformed to “reduced graphene oxide” (RGO) using chemical reducing agents such as hydrazine17,18 or sodium borohydride.19–23 Removing oxygen makes graphene (oxide) more hydrophobic, which increases its tendency to aggregate and results in a more efficient separation and extraction. The key to any successful approach for environmental and biological samples will be doing this in situ (i.e., in a complex matrix) so that GO can easily be recovered.
With the increase in graphene production and the advent of new graphene-containing products, graphene is likely to enter into wastewater treatment plants. Given graphene's similarity to CNTs, it will presumably end up in wastewater effluent or wastewater biosolids (treated sewage sludge containing living/dead microbes and inert solids).24 Of these exposure routes, biosolids seem to be the most appropriate end-point for graphene and GO.25–28
The aims of this study were to (1) develop a PTA quantification method for graphene and GO and (2) develop a method for recovering graphene and GO from complex organic matrices. We utilized few-layer graphene (10–20 nm thick) in place of single-layer graphene due to the problem obtaining an aqueous solution of single-layer graphene. Because of the difficulty extracting oxygenated carbonaceous nanomaterials (e.g., GO) from complex matrices, we applied an in situ reduction method to increase hydrophobicity and improve recovery. Given the likelihood of graphene to end up in wastewater biosolids, we demonstrated an extraction and quantification method for wastewater biosolids to assist with fate and transport studies. The results stemming from this research can be leveraged to develop extraction methods for graphene from other biological matrices (e.g., lung tissue, in vitro/in vivo studies, algae, daphnia).
Experimental methods
Materials
GO solution was used as received (TW Nano; manufacturer reported characteristics: 0.2 wt.%, >90% single layer, 0.5–20 μm in x–y, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.3 C
1.3 C![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) O ratio, >1200 m2 g−1). Graphene nanoplatelet powder was used as received (Angstron Materials, N006-P; manufacturer reported characteristics: >97% carbon, <1.5% oxygen, <1.5% ash, 10–20 nm thick, <14 μm in x–y direction, 21 m2 g−1). Graphene nanoplatelets, or few-layer graphene (FLG), are stacked graphene sheets and are used in place of graphene because pristine (i.e., no oxygen) single-layer graphene in aqueous solution is not achievable. GO and FLG consisted of flake like particles with dimensions similar to each other (Fig. S2a and c,† respectively). SEM images revealed the presence of rectangular plates, with small (x–y < 1 μm) and large (x–y ~ 5–10 μm) fractions for both GO and FLG. FLG was typically smaller than the maximum size listed by the manufacturer (average x–y from Fig. S2c† was approximately 4 × 2.5 μm).
O ratio, >1200 m2 g−1). Graphene nanoplatelet powder was used as received (Angstron Materials, N006-P; manufacturer reported characteristics: >97% carbon, <1.5% oxygen, <1.5% ash, 10–20 nm thick, <14 μm in x–y direction, 21 m2 g−1). Graphene nanoplatelets, or few-layer graphene (FLG), are stacked graphene sheets and are used in place of graphene because pristine (i.e., no oxygen) single-layer graphene in aqueous solution is not achievable. GO and FLG consisted of flake like particles with dimensions similar to each other (Fig. S2a and c,† respectively). SEM images revealed the presence of rectangular plates, with small (x–y < 1 μm) and large (x–y ~ 5–10 μm) fractions for both GO and FLG. FLG was typically smaller than the maximum size listed by the manufacturer (average x–y from Fig. S2c† was approximately 4 × 2.5 μm).
Sodium borohydride (99.99%, Sigma Aldrich, 480886), hydroiodic acid (57% in H2O, Sigma Aldrich, 210013), and ascorbic acid (reagent grade, Sigma Aldrich, A7506) were used as received. Solvable™ was obtained from Perkin Elmer. Solvable is a tissue solubilizer consisting of sodium hydroxide (≤2.5%), C10-16-alkyldimethyl, N-oxide (2–10%), and C11-15-secondary, ethoxylated alcohol (2.5–10%). Sodium hydroxide (97%, EMD SX0590), Tergitol 15-S-12™ (C12-14 secondary ethoxylated alcohol, CAS no. 84133-50-6, Dow Chemical Company), and N,N-dimethyldodecylamine, N-oxide (30% in H2O, Sigma Aldrich 40236) were obtained to examine the individual components of Solvable. Ultrapure water (18.2–18.3 MΩ cm) was used for all experiments.
Programmed thermal analysis
PTA was performed using an organic carbon/elemental carbon analyzer (Sunset Laboratory, Inc., Sunset, Oregon, USA). PTA was used to quantify graphene recovery, determine changes in graphene thermal stability after treatment, and quantify the biomass background carbon after treatment; PTA operation is described in detail elsewhere.7 Briefly, samples were heated using a graphene-specific temperature ramp program (Table S1†) in inert conditions (100% He) and then in oxidizing conditions (90% He/10% O2). The carbon that evolves during analysis is converted to methane and then detected using flame ionization detection (FID). This FID signal is calibrated with internal and external standards that are used to calculate the mass of carbon evolved. The graphene-specific program was designed to remove most of the background organic carbon during the initial inert phase and then transition into the oxidizing phase where the more stable background carbon is removed before evolution of graphene. PTA quantifies only the mass of carbon, so the oxygen mass is not considered for compounds like GO. The maximum temperature under inert conditions was set at 675 °C to avoid loss of oxygenated graphene. Samples were put onto a quartz-fiber filter (QFF; Pall Tissuquartz 2500 QAT-UP, 7204) designed for high temperatures (Fig. S3†) and then loaded into the PTA instrument for analysis.In situ reduction of graphene oxide
An in situ reduction method for GO was developed to overcome the difficulty in recovering hydrophilic carbonaceous nanomaterials from aqueous matrices. For reduction experiments, a specified amount (e.g., 0.4%, 2%) of NaBH4 was added to a mixture of GO solution and water or Solvable. Samples were then placed in a furnace at 60 °C for 2 h followed by centrifugal separation at 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 830 × g for 10 min and washed twice with water (additional washing causes poor pellet formation). Final pellets were collected and loaded onto a QFF for either Raman or PTA. Samples requiring a phase-separation were treated with NaBH4 for 36 h rather than 2 h.
830 × g for 10 min and washed twice with water (additional washing causes poor pellet formation). Final pellets were collected and loaded onto a QFF for either Raman or PTA. Samples requiring a phase-separation were treated with NaBH4 for 36 h rather than 2 h.
Extraction from biomass
Biomass was grown using a laboratory-scale sequencing batch reactor that was seeded using return activated sludge from a local (Mesa, AZ) full-scale wastewater treatment plant.27 1000 μg dry weight (~78 μL) of concentrated fresh biomass stock (12.8 g L−1) was added to 5 mL Solvable to obtain a biomass concentration of 200 mg L−1. GO or FLG (~20 μg) was then added. The ratio of carbon to biomass was ~0.02 μg C μg−1 dried biomass. Samples were placed in a furnace at 60 °C for 24 h to digest the biomass. After digesting, NaBH4 was added to begin the in situ reduction process. The treated samples were then centrifuged at 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 830 × g for 10 min. The pellet was twice washed with water followed by centrifuging each time. The final pellets were collected using a pipette and then loaded onto a QFF for PTA. Samples were prepared and analyzed in triplicate.
830 × g for 10 min. The pellet was twice washed with water followed by centrifuging each time. The final pellets were collected using a pipette and then loaded onto a QFF for PTA. Samples were prepared and analyzed in triplicate.
The method detection limit (MDL) for GO or FLG in 1000 μg dried biomass was calculated using a t-distribution with 99% confidence (one tail, seven replicates, 5 μg graphene).29 The 95% lower (LCL) and upper (UCL) confidence intervals were calculated as 0.64 × MDL and 2.20 × MDL, respectively.29
X-ray photoelectron spectroscopy
Surface elemental composition and chemical state were analyzed using X-ray photoelectron spectroscopy (XPS) performed on an ESCALAB 220i-XL (Vacuum Generators, U.S.) with a monochromatic Al Kα source at hν = 1486 eV, a base pressure of 7 × 10−10 mbar, and a spot analysis size of 500 μm. For GO and RGO solutions, powders were obtained by evaporating solutions in aluminum trays. The final dried product was crushed using an agate mortar and pestle. All samples were prepared for XPS by pressing the powder into a disk on clean indium foil. Peak fitting was performed manually using XPS peak analysis software (Casa XPS) on the basis of the theoretical atomic percentages calculated from the wide scan.Raman spectroscopy
Raman spectroscopy was used to determine the changes in the GO structure resulting from the in situ reduction. Raman was performed on a custom-built confocal instrument in 180° geometry. The sample was excited using a 532 nm laser with 100 mW maximum power, which was controlled using neutral density filters. The data were collected using an Acton 300i spectrograph and a back-thinned Princeton Instruments liquid nitrogen-cooled CCD detector with a spatial resolution <1 μm and spectral resolution of ~1 cm−1. Between 1300 and 1600 cm−1, there are two distinct peaks for graphene, called the D-band (1350 cm−1) and the G-band (1580 cm−1). The D-band is present because of defects or disorder (e.g., sp3 bonds) present within the graphene sample and increases in intensity with increasing disorder. The G-band is the graphitic band, and a higher, narrower peak indicates a more ordered graphene (i.e., sp2 bonds). The average ID/IG ratio was calculated from measurements taken at four different points for each sample.Scanning Electron Microscopy
Scanning Electron Microscopy (SEM) was completed using a Nova 200 FIB-SEM from FEI with a field-emission electron gun. SEM imaging was performed at 5 kV and 0.98 to 1.6 nA with dwell time between 0.3 and 3 μs.Ultraviolet–visible light spectroscopy
UV–vis absorption spectra of GO and FLG were investigated on a Hach DR5000. Serial dilutions were made from 2 g L−1 stock solutions and ultrapure water. All samples were scanned from 200 to 800 nm. For GO, no absorption occurred above 600 nm (brown color in solution) and it had two peaks at 238 and 300 nm. FLG (black color in solution) absorbed across all wavelengths with excellent calibration correlation (R2 > 0.99) and a broad peak at 227 nm.Results
Graphene detection
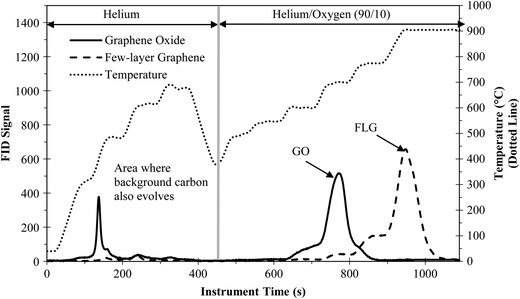
FLG and GO were quantified using PTA, which relies on separating carbon compounds on the basis of their thermal stability in inert (i.e., He) and combustion atmospheres (i.e., 90% He/10% O2). Weaker compounds and those with more oxygen will evolve during the inert phase and early in the oxidizing phase. Fig. 1 shows the PTA result for 20 μg of FLG and GO (run separately). Instrument detection was reliable with calibration data demonstrating a slope of 1.02 and an R2 of 1.00 for both FLG and GO (Fig. S4†). The majority of FLG evolved at high temperatures during the oxidizing phase, starting around 700 °C and peaking around 900 °C, with the strong thermal stability owing to the low defect density and low oxygen content. A small amount of FLG (~3%) evolved during the inert phase (i.e., where background organic carbon would evolve) and can be attributed to the oxygenated FLG. For GO, the high amount of oxygen resulted in a larger portion evolving during the inert phase (~20%), all of which would be lost in the background of a complex organic sample evolving at the same temperatures.7 Reducing or removing the oxygenated groups on graphene is key to improving the recovery of GO from complex organic matrices.Improving detection and extraction of FLG and GO through reduction
In order to improve GO detection and recovery, different reducing reagents (e.g., sodium borohydride (NaBH4), ascorbic acid, hydroiodic acid (HI)) were investigated to remove oxygen functionalities from GO. Ascorbic acid and HI were not ideal reagents, resulting in incomplete reduction of GO, the inability to fully aggregate GO, or GO adherence to the plastic vials (see ESI† for further discussion on failed reagents). NaBH4 emerged as the optimal reagent for GO reduction resulting in an increased thermal stability and hydrophobicity.XPS was used to investigate the C–C and C–O/C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bond content of GO. Fig. 2 shows the XPS analysis for GO in water and GO after treatment with NaBH4 in water (i.e., RGO). Two peaks were present, one at 284 eV, which is attributed to C–C/C
O bond content of GO. Fig. 2 shows the XPS analysis for GO in water and GO after treatment with NaBH4 in water (i.e., RGO). Two peaks were present, one at 284 eV, which is attributed to C–C/C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C, and the other at 286–290 eV, which coincides to a number of carbon and oxygen functionalities (mainly C–O and C
C, and the other at 286–290 eV, which coincides to a number of carbon and oxygen functionalities (mainly C–O and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O). The C–O/C–C ratio for GO was 1.1
O). The C–O/C–C ratio for GO was 1.1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, which agrees with the manufacturer's carbon to oxygen ratio of 1
1, which agrees with the manufacturer's carbon to oxygen ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. The C–O/C–C ratio for RGO was 5.6
1. The C–O/C–C ratio for RGO was 5.6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, an approximate 5-fold decrease in the number of C–O/C
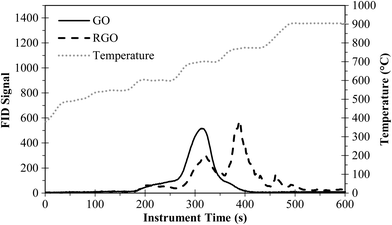
1, an approximate 5-fold decrease in the number of C–O/C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bonds. GO reduction also shifted the C–O peak to lower binding energies, indicating a change in the type of carbon–oxygen functionalities that remained on the GO. These results provide clear evidence that carbon–oxygen functionalities were removed by NaBH4 treatment. SEM images show that reduction of GO (Fig. S2a†) to RGO (Fig. S2b†) did not significantly alter the particle shape or x–y size (e.g., both large, 5–10 μm, and small (e.g., right side of Fig. S2b†), <1 μm, sheets were present), and stacked, plate-like structures were formed. Fig. 3 shows the PTA thermogram for RGO after treatment with 2% NaBH4 in water. Chemical reduction improved the thermal stability (i.e., peak shift to the right), providing additional evidence that oxygen functionalities were removed.
O bonds. GO reduction also shifted the C–O peak to lower binding energies, indicating a change in the type of carbon–oxygen functionalities that remained on the GO. These results provide clear evidence that carbon–oxygen functionalities were removed by NaBH4 treatment. SEM images show that reduction of GO (Fig. S2a†) to RGO (Fig. S2b†) did not significantly alter the particle shape or x–y size (e.g., both large, 5–10 μm, and small (e.g., right side of Fig. S2b†), <1 μm, sheets were present), and stacked, plate-like structures were formed. Fig. 3 shows the PTA thermogram for RGO after treatment with 2% NaBH4 in water. Chemical reduction improved the thermal stability (i.e., peak shift to the right), providing additional evidence that oxygen functionalities were removed.
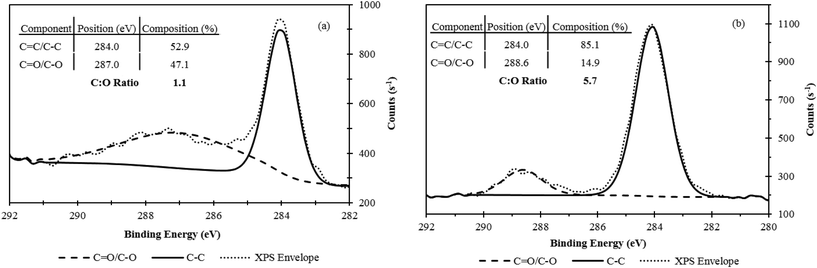 | ||
Fig. 2 XPS analysis of (a) GO and (b) RGO. The average position for C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C and C–C was 284.0 eV, and the average position for C C and C–C was 284.0 eV, and the average position for C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and C–O was 287.0 eV and 288.6 eV for GO and RGO, respectively. O and C–O was 287.0 eV and 288.6 eV for GO and RGO, respectively. | ||
Raman spectroscopy is used to determine the defect density of CNTs and graphene,4 defined as the ratio between the D-band (1350 cm−1) and G-band (1580 cm−1) (ID/IG). The defect density is an indication of the thermal stability7 and the degree of oxidation.30 We hypothesized NaBH4 reduction would decrease the defect density and result in an increase in the GO thermal stability due to a decrease in the number of oxygen functionalities. However, Raman results revealed that the ID/IG did not change significantly (>5%) after NaBH4 treatment. Although reduction of oxygen functionalities occurred (i.e., XPS and thermal stability results), the chemical reduction treatment did not heal defects. NaBH4 is known to reduce aldehydes and ketones into alcohols, and it is capable of reducing lactone and carbonyl groups to hydroxyl groups on functionalized CNTs.31 So, in the case of NaBH4 reduction of GO, presumably the GO functionalities are only being reduced as far as C–OH and C–H, and NaBH4 is not able to heal defects through C–C sp2 bond formation.
In water, NaBH4 enabled aggregation of GO, presumably a result of removing oxygenated functional groups, but separation via centrifugation was difficult (i.e., Fig. S5†). In a clean, aqueous matrix (i.e., only water and GO), filtration directly onto a quartz-fiber filter (QFF) is an option for separating the GO (e.g., 10–20 μm X–Y dimensions), but this is not an option for complex matrices because the filter will also collect interfering carbon compounds. For applications involving clean matrices free of carbon interferences, filtration may be an option; though retention using the QFFs, which are designed to function as air filters, may be poor for GO as observed for functionalized CNTs.7 Furthermore, if a different quantification method (e.g., electrophoresis, UV–vis) is used, the sample would need to be in a concentrated aqueous or powder form and not adhered to a filter.
In a Solvable matrix, which is the reagent used to solubilize organics (e.g., wastewater biomass (this paper), tissue8), GO aggregated and formed a very stable, compact pellet upon centrifugation. This is likely due to a combination of a high pH, double-layer compression from increased ionic strength, and the presence of two surfactants, which may cause a cloud-point like effect.32 The known individual components of Solvable were examined to determine the root of the effect. Both surfactants (10% concentration) alone and in combination caused aggregation while sodium hydroxide was not effective. Upon addition of NaBH4 to the surfactants, samples exhibited severe effervescence due to hydrogen generation, and GO was not easily recovered as it adhered to the vials, overflowed the vials along with the bubbles, or would not centrifuge into a pellet. However, adding sodium hydroxide to the two surfactants (individual or combined) curbed the effervescence. Therefore, the excellent performance of Solvable for extracting graphene can be attributed to a synergistic action of its components rather than a single species. Fig. 4 shows the percent recovery of RGO as a function of increasing NaBH4 concentration. Recovery with Solvable alone (i.e., no reducing agent) was 75 ± 0.5%. Adding low concentrations of NaBH4 (e.g., 0.04%) did not show improvement with an average recovery of 76 ± 3%. Increasing the NaBH4 concentration to 0.4% resulted in a slightly higher recovery (81 ± 3%), but recovery over 90% wasn't achieved until greater than 2% NaBH4 was used (95 ± 5%), with a maximum recovery of 97 ± 0.4% observed using 8% NaBH4. The improved physical recovery was attributed to a reduction in oxygen content, resulting in increased aggregation of the graphene particles. Removal of carbon–oxygen bonds shifts the hydrophilic nature of graphene to be more hydrophobic, resulting in improved aggregation during centrifugation. Reduction also decreases the amount graphene that would otherwise be lost in the organic carbon PTA background (i.e., during the inert phase as shown in Fig. 1).
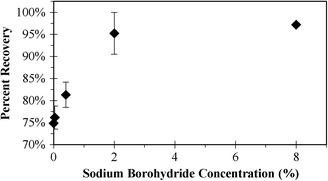 | ||
| Fig. 4 Percent recovery of RGO after GO reduction using various concentrations of NaBH4 (0, 0.04, 0.4, 2, 8%). Error bars indicate one standard deviation (each direction) for triplicate samples. | ||
When using PTA for quantifying graphene, the thermal stability (i.e., peak oxidizing temperature) is important for separating graphene from background organic carbon during analysis. Fig. 5 shows PTA mass loss curves under oxidizing conditions for GO using different extraction conditions. Surfactants have been previously shown to reduce the thermal stability of CNTs,7 and we observed the same effect for GO treated with Solvable, an alkali reagent containing surfactants. Solvable decreased the thermal stability of GO significantly, with an onset approximately 130 s earlier and 50 °C lower. Reduction of GO in water with NaBH4 improved the thermal stability (Fig. 3), so we hypothesized that this would improve the GO stability after Solvable treatment. Using low concentrations of NaBH4 (e.g., 0.04%) after the Solvable treatment only increased the thermal stability slightly (~20 s), but using a higher concentration of NaBH4 (>2%) returned the thermal stability close to the original (Fig. S6†). The improvement in the thermal stability may account for the improved recovery when using greater than 2% NaBH4 (i.e., Fig. 4). To achieve optimal extraction, a combination of Solvable and at least 2% NaBH4 is recommended.
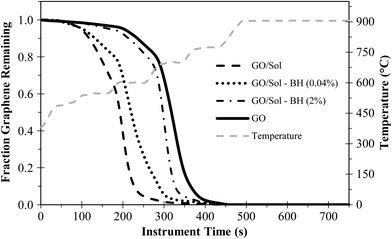 | ||
| Fig. 5 Mass loss curves for GO (~20 μg) under oxidizing PTA conditions using different extraction conditions. “Sol” is Solvable and “BH” is NaBH4. | ||
Recovery of GO and FLG from biomass
The ability to quantify GO or FLG in a complex organic matrix such as wastewater biosolids is important for assisting environmental studies. As such, we determined detection limits for GO and FLG in biomass as well as recovery of 20 μg of GO or FLG from 1 mg of clean, dried biomass (200 mg L−1). The MDL, LCL, and the UCL for GO in biomass were calculated to be 2.2, 1.4, and 4.9 μg, respectively. The MDL, LCL, and UCL for FLG in biomass were calculated to be 1.5, 0.93, and 3.2 μg, respectively. In comparison, the MDL for GO in a clean aqueous matrix (i.e., ultrapure water only) is 1.7 μg.Without Solvable treatment, FLG and GO detection in biosolids was not possible because the amount of biomass collected in the pellet overwhelmed PTA and resulted in indistinguishable peaks for graphene and biomass. Using the extraction method of Solvable and 2% NaBH4, GO/RGO and FLG (20 μg) recoveries from 1 mg dried biomass (0.02 μg graphene μg−1 dried biomass) were 80 ± 6% and 52 ± 8%, respectively. Although FLG is easier than GO/RGO to detect in a complex matrix using PTA because it is more thermally stable, physical separation from the biomass using centrifugation was less efficient, resulting in a lower recovery. We observed that FLG was very stable in Solvable (before and after biomass treatment), with little recovery occurring via centrifugation (<5%). Although FLG is already in a “reduced” form, adding NaBH4 helped to improve the FLG aggregation and extraction. We also examined nitric acid as a digesting agent in place of Solvable to determine if pH or surfactants were an issue. Like Solvable, FLG was more stable in nitric acid (pH < 0) than in ultrapure water (pH = 5.6), likely due to increased surface charge separation, but extraction was worse than with Solvable. This agrees with previous results showing Solvable to be optimal over common agents (e.g., nitric acid, hydrochloric acid, hydrofluoric acid, etc.) used for extracting CNTs from rat lung tissue.8
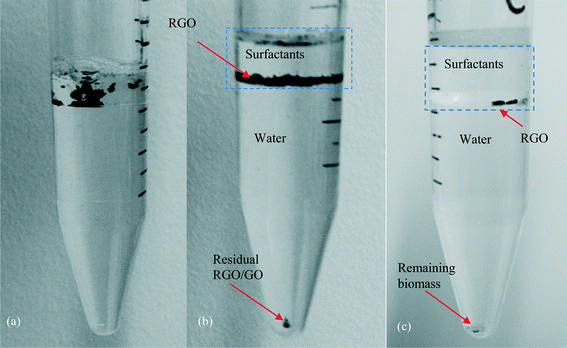
Solvable was efficient at dissolving the biomass, but a small amount of background carbon still remained and interfered with GO/RGO peaks (Fig. 6); no interference was observed for FLG. The interference for GO/RGO was consistent across triplicate samples with an average of 2.2 ± 1 μg. When the amount of biomass was increased to 5 mg (1 g biomass L−1), GO/RGO peaks were indistinguishable due to the false positive from interfering background carbon that remained after treatment. To improve the extraction for GO/RGO from high concentrations of biomass, we developed a phase-separation method by extending the heating time of the NaBH4 step to 36 h. This causes the water and surfactant phases of Solvable to separate (Fig. 7a), and after centrifugation, RGO remains mostly in the top surfactant phase (Fig. 7b). Similarly, when done in a wastewater matrix (i.e., 1 g biomass L−1), the undigested (interfering) biomass transfers into the water phase, and the RGO remains in the surfactant phase (Fig. 7c). This results in a physical separation of the RGO and the interfering background carbon, allowing for easy recovery of the RGO only. Note, control samples digested with Solvable for 36 h (i.e., no NaBH4) did not show any significant (<5%) additional removal of biomass interference. Therefore, using NaBH4 to separate the biomass and RGO into different phases is key for improving recovery in wastewater with a high biomass concentration. Using the phase-separation method, the recovery of RGO (20 μg) from 5 mg biomass was 110% ± 13%. Recovery greater than 100% is attributed to undigested biomass constituents interacting with RGO, causing the biomass to remain in the surfactant phase. This interaction is presumed to be adsorption of the biomass to RGO as no interfering background carbon from the biomass was observed in the surfactant phase for triplicate control samples that did not contain RGO. The phase-separation was not successful for FLG as the majority of the FLG transferred to the water phase along with the undigested biomass. The advantage of using the phase-separation method over the centrifugal separation method for GO is that larger amounts of biomass can be used while avoiding interferences from undigested biomass. However, in other instances, the centrifugal method is preferred because it is simpler, less time consuming, and useful for both graphene types. A detailed schematic of the two methods is shown in Fig. S7.†
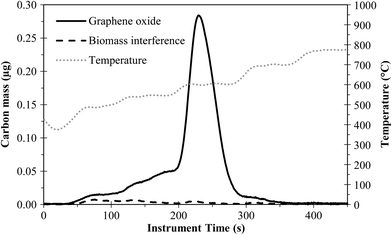 | ||
| Fig. 6 PTA thermogram showing biomass interference for GO in wastewater biosolids. Solvable and 2% NaBH4 treatment. | ||
Conclusion
We have successfully demonstrated an extraction and quantification method for graphene and GO using an in situ reduction method followed by detection with PTA. This method was demonstrated in biomass (200 mg L−1), resulting in recoveries for GO/RGO and FLG of 80 ± 9% and 52 ± 8%, respectively. A phase-separation method (similar to liquid–liquid extraction) was developed to improve the recovery of GO from more concentrated wastewater samples (e.g., 1 g biomass L−1). Although the phase-separation method is more complex than the centrifugal separation method, it is an intriguing technique that warrants further investigation for highly complex matrices (e.g., sediments). While FLG was easier to separate thermally using PTA, it was more difficult to physically recover using the extraction method. This was for a specific type of FLG, whereas other types from different manufacturers could behave differently. Further study is needed on FLG and single-layer graphene to determine if physical recovery differences exist between the varying types and if an additional processing step can improve the recovery.Reported recoveries are ideal as they were obtained using a lab-grown, clean biomass. When using biosolids obtained from a wastewater treatment plant, the recovery values are expected to increase due to presence of soot particulates, which behave thermally similar to graphene, thereby creating a false positive.7 Similarly, the presence of carbonaceous nanomaterials (e.g., CNTs, fullerenes) in environmental samples with graphene is possible,33 further complicating recovery when using PTA. While GO/RGO and FLG used herein can be separated and quantified using PTA (e.g., Fig. S7†), the presence of CNTs with a similar thermal stability as GO or FLG would be difficult to distinguish with PTA alone. Ideally, a graphene standard (similar to the NIST single-walled CNT standard reference material, SRM 2483) would be used to create a spike standard addition curve in order to quantify the amount of background soot (or CNTs) interfering with graphene. With the challenge of thermally similar carbonaceous materials present (e.g., soot) or predicted (e.g., CNT) in the environment, PTA and similar thermal methods alone are not currently suitable, and analytical advancements to these methods and more selective extraction methods are needed. However, PTA, in its early analytical development as described herein, is an excellent tool for monitoring the fate/transport and toxicity of graphene for model systems and organisms, respectively.
Acknowledgements
This research was partially funded by the Semiconductor Research Corporation (SRC, Task 425.040), National Science Foundation (CBET 1336542), US Environmental Protection Agency (RD83558001), and the NSF/ASEE Small Business Research Diversity Postdoctoral Fellowship program. Materials characterization was conducted through the Leroy Center for Solid-State Science. We would like to thank Dr. Yu Yang for providing biomass and the Dow Chemical Company for the Tergitol 15-S-12™.References
- X. Q. Guo and N. Mei, J. Food Drug Anal., 2014, 22, 105–115 CrossRef CAS PubMed.
- A. B. Seabra, A. J. Paula, R. de Lima, O. L. Alves and N. Duran, Chem. Res. Toxicol., 2014, 27, 159–168 CrossRef CAS PubMed.
- G. X. Wang, J. Yang, J. Park, X. L. Gou, B. Wang, H. Liu and J. Yao, J. Phys. Chem. C, 2008, 112, 8192–8195 CAS.
- M. S. Dresselhaus, A. Jorio and R. Saito, in Annual Review of Condensed Matter Physics, Annual Reviews, Palo Alto, 2010, vol. 1, pp. 89–108 Search PubMed.
- D. A. Dikin, S. Stankovich, E. J. Zimney, R. D. Piner, G. H. B. Dommett, G. Evmenenko, S. T. Nguyen and R. S. Ruoff, Nature, 2007, 448, 457–460 CrossRef CAS PubMed.
- D. Li, M. B. Muller, S. Gilje, R. B. Kaner and G. G. Wallace, Nat. Nanotechnol., 2008, 3, 101–105 CrossRef CAS PubMed.
- K. Doudrick, P. Herckes and P. Westerhoff, Environ. Sci. Technol., 2012, 46, 12246–12253 CrossRef CAS PubMed.
- K. Doudrick, N. Corson, G. Oberdörster, A. C. Eder, P. Herckes, R. U. Halden and P. Westerhoff, ACS Nano, 2013, 7, 8849–8856 CrossRef CAS PubMed.
- D. L. Plata, C. M. Reddy and P. M. Gschwend, Environ. Sci. Technol., 2012, 46, 12254–12261 CrossRef CAS PubMed.
- R. H. Wang, C. Mikoryak, E. Chen, S. Li, P. Pantano and R. K. Draper, Anal. Chem., 2009, 81, 2944–2952 CrossRef CAS PubMed.
- M. J. O'Connell, S. M. Bachilo, C. B. Huffman, V. C. Moore, M. S. Strano, E. H. Haroz, K. L. Rialon, P. J. Boul, W. H. Noon, C. Kittrell, J. P. Ma, R. H. Hauge, R. B. Weisman and R. E. Smalley, Science, 2002, 297, 593–596 CrossRef PubMed.
- E. J. Petersen, Q. G. Huang and W. J. Weber, Environ. Sci. Technol., 2008, 42, 3090–3095 CrossRef CAS.
- F. Irin, B. Shrestha, J. E. Canas, M. A. Saed and M. J. Green, Carbon, 2012, 50, 4441–4449 CrossRef CAS PubMed.
- S. Attal, R. Thiruvengadathan and O. Regev, Anal. Chem., 2006, 78, 8098–8104 CrossRef CAS PubMed.
- R. B. Reed, D. G. Goodwin, K. L. Marsh, S. S. Capracotta, C. P. Higgins, D. H. Fairbrother and J. F. Ranville, Environ. Sci.: Processes Impacts, 2013, 15, 204–213 CAS.
- J. F. Shen, Y. Z. Hu, M. Shi, X. Lu, C. Qin, C. Li and M. X. Ye, Chem. Mater., 2009, 21, 3514–3520 CrossRef CAS.
- S. Gilje, S. Han, M. Wang, K. L. Wang and R. B. Kaner, Nano Lett., 2007, 7, 3394–3398 CrossRef CAS PubMed.
- S. Dubin, S. Gilje, K. Wang, V. C. Tung, K. Cha, A. S. Hall, J. Farrar, R. Varshneya, Y. Yang and R. B. Kaner, ACS Nano, 2010, 4, 3845–3852 CrossRef CAS PubMed.
- D. C. Luo, G. X. Zhang, J. F. Liu and X. M. Sun, J. Phys. Chem. C, 2011, 115, 11327–11335 CAS.
- R. Muszynski, B. Seger and P. V. Kamat, J. Phys. Chem. C, 2008, 112, 5263–5266 CAS.
- Y. Si and E. T. Samulski, Nano Lett., 2008, 8, 1679–1682 CrossRef CAS PubMed.
- W. Gao, L. B. Alemany, L. J. Ci and P. M. Ajayan, Nat. Chem., 2009, 1, 403–408 CrossRef CAS PubMed.
- H. J. Shin, K. K. Kim, A. Benayad, S. M. Yoon, H. K. Park, I. S. Jung, M. H. Jin, H. K. Jeong, J. M. Kim, J. Y. Choi and Y. H. Lee, Adv. Funct. Mater., 2009, 19, 1987–1992 CrossRef CAS.
- B. Nowack, J. F. Ranville, S. Diamond, J. A. Gallego-Urrea, C. Metcalfe, J. Rose, N. Horne, A. A. Koelmans and S. J. Klaine, Environ. Toxicol. Chem., 2012, 31, 50–59 CrossRef CAS PubMed.
- P. K. Westerhoff, A. Kiser and K. Hristovski, Environ. Eng. Sci., 2013, 30, 109–117 CrossRef CAS.
- I. Chowdhury, M. C. Duch, N. D. Mansukhani, M. C. Hersam and D. Bouchard, Environ. Sci. Technol., 2013, 47, 6288–6296 CAS.
- Y. F. Wang, P. Westerhoff and K. D. Hristovski, J. Hazard. Mater., 2012, 201, 16–22 CrossRef PubMed.
- T. Y. Sun, F. Gottschalk, K. Hungerbuhler and B. Nowack, Environ. Pollut., 2014, 185, 69–76 CrossRef CAS PubMed.
- USEPA, in Part 136 Guidelines Establishing Test Procedures for the Analysis of Pollutants: Definition and Procedure for the Determination of the Method Detection Limit, USEPA, 2012.
- S. Osswald, M. Havel and Y. Gogotsi, J. Raman Spectrosc., 2007, 38, 728–736 CrossRef CAS.
- B. Scheibe, E. Borowiak-Palen and R. J. Kalenczuk, Mater. Charact., 2010, 61, 185–191 CrossRef CAS PubMed.
- J. F. Liu, J. B. Chao, R. Liu, Z. Q. Tan, Y. G. Yin, Y. Wu and G. B. Jiang, Anal. Chem., 2009, 81, 6496–6502 CrossRef CAS.
- F. Gottschalk, T. Y. Sun and B. Nowack, Environ. Pollut., 2013, 181, 287–300 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c4en00134f |
| This journal is © The Royal Society of Chemistry 2015 |