Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Molecular basis of Streptococcus mutans sortase A inhibition by the flavonoid natural product trans-chalcone†
Daynea J.
Wallock-Richards‡
a,
Jon
Marles-Wright‡
b,
David J.
Clarke
a,
Amarnath
Maitra
c,
Michael
Dodds
c,
Bryan
Hanley
d and
Dominic J.
Campopiano
*a
aEastChem School of Chemistry, The University of Edinburgh, David Brewster Road, Edinburgh, EH9 3FJ, UK. E-mail: Dominic.Campopiano@ed.ac.uk
bInstitute of Structural and Molecular Biology, The University of Edinburgh, Edinburgh, EH9 3BF, UK
cWm. Wrigley Jr. Company, 1132 W. Blackhawk Street, Chicago, IL 60642, USA
dThe Knowledge Transfer Network, The Roslin Institute, Easter Bush, Roslin, EH25 9RG, UK
First published on 18th May 2015
Abstract
Sortase A (SrtA) from Gram positive pathogens is an attractive target for inhibitors due to its role in the attachment of surface proteins to the cell wall. We found that the plant natural product trans-chalcone inhibits Streptococcus mutans SrtA in vitro and also inhibited S. mutans biofilm formation. Mass spectrometry revealed that the trans-chalcone forms a Michael addition adduct with the active site cysteine. The X-ray crystal structure of the SrtA H139A mutant provided new insights into substrate recognition by the sortase family. Our study suggests that chalcone flavonoids have potential as sortase-specific oral biofilm inhibitors.
Sortases are a family of membrane-associated transpeptidases which are highly conserved in Gram positive bacteria. They are a promising target for inhibitors which have the potential to be used as anti-infective therapies against pathogenic bacteria.1–3 Various sortase isoforms (SrtA, B, C) are responsible for covalently attaching surface proteins involved in adherence to host cells, iron acquisition, biofilm formation, invasion, signalling and pili formation on the bacterial cell wall.2,4,5 Sortase A (SaSrtA) was the first isoform of this family identified in Staphylococcus aureus by Schneewind and colleagues, who also elucidated the catalytic mechanism of the enzyme.6–10 The SrtA enzymes recognize a conserved five amino acid sequence (LPXTG) at the C-terminus of their protein substrates and cleave the amide bond between the threonine and glycine residues using a highly-conserved cysteine residue in the active site (Fig. S1, ESI†). The resulting thioacyl intermediate is then attacked by the nucleophilic pentaglycine side chain of lipid II, a cell wall precursor molecule, to form a C-terminally linked surface protein.1,2
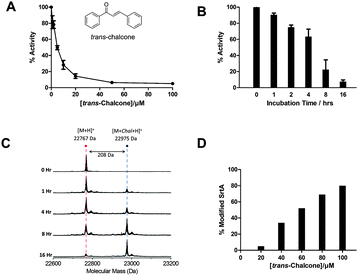
In addition to the conserved Cys residue in SrtA, two other active site residues have been identified as essential for optimal enzyme activity, a His side-chain which serves as a general base and an Arg side-chain which stabilises the oxoanion intermediate formed during the reaction.11–13 This catalytic triad (Cys–His–Arg) is highly conserved among the sortase A enzymes (Fig. S2, ESI†). Several classes of SrtA inhibitors have been identified and these include compounds from small-molecule synthetic libraries,9,11,14,15 rationally designed peptidomimetics14,16 and natural products.17–20 Flavonoids21 are polyphenolic plant natural products and two in particular, morin22 and curcumin,23,24 display good inhibition against Streptococcus mutans SrtA. However, the exact molecular basis of their activity has not been determined. Combining the knowledge gained from these previous inhibitor studies we hypothesized that trans-chalcone (Fig. 1A, inset), the precursor molecule of many flavonoids, would inhibit the S. mutans SrtA in vitro and possibly prevent bacterial biofilm growth.
The S. mutans SrtA enzyme was expressed in E. coli as an N-terminal 40 amino acid truncation to remove the predicted transmembrane domain (Fig. S3 and S4, ESI†) and the isolated SrtA had a mass of 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 767 Da ([M + H]+, Fig. S5, ESI†) which agrees well with the predicted value based on the sequence.25 The established FRET-based assay was used to measure the Km and Vmax values (90.4 ± 4.7 μM and 4.46 ± 0.11 × 10−4 μM s−1 respectively) for cleavage of the fluorogenic peptide substrate, dabcyl-QALPETGEE-edans (Fig. S6, ESI†).26 Using the published method,24 an IC50 value of 5 ± 0.6 μM was determined for trans-chalcone after incubating SrtA with varying concentrations of inhibitor overnight (Fig. 1A and Fig. S7, ESI†). The rate of inhibition was measured in a time dependent study by incubating 5 μM of the enzyme with 100 μM of the inhibitor for various times (1, 2, 4, 8 and 16 h, Fig. 1B). Analysis of the enzyme activity revealed that the inhibition of SrtA was slow, taking up to 16 h to reduce activity by 90%. Enzyme activity was not restored upon extensive dialysis of the enzyme (not shown). Taken together these observations suggested that trans-chalcone was a slow and tight binding inhibitor of SrtA.
767 Da ([M + H]+, Fig. S5, ESI†) which agrees well with the predicted value based on the sequence.25 The established FRET-based assay was used to measure the Km and Vmax values (90.4 ± 4.7 μM and 4.46 ± 0.11 × 10−4 μM s−1 respectively) for cleavage of the fluorogenic peptide substrate, dabcyl-QALPETGEE-edans (Fig. S6, ESI†).26 Using the published method,24 an IC50 value of 5 ± 0.6 μM was determined for trans-chalcone after incubating SrtA with varying concentrations of inhibitor overnight (Fig. 1A and Fig. S7, ESI†). The rate of inhibition was measured in a time dependent study by incubating 5 μM of the enzyme with 100 μM of the inhibitor for various times (1, 2, 4, 8 and 16 h, Fig. 1B). Analysis of the enzyme activity revealed that the inhibition of SrtA was slow, taking up to 16 h to reduce activity by 90%. Enzyme activity was not restored upon extensive dialysis of the enzyme (not shown). Taken together these observations suggested that trans-chalcone was a slow and tight binding inhibitor of SrtA.
This led us to postulate that a covalent adduct was formed between the inhibitor and the SrtA. To test this hypothesis, mass spectrometry was used to analyse both the concentration and time dependence of the reaction. From the mass spectrum we observed a peak of 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 767 Da for the mass of SrtA and an additional peak at 22
767 Da for the mass of SrtA and an additional peak at 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 975 Da (Δ208 Da) which corresponds to a single SrtA–chalcone adduct (Fig. 1C). The formation of the adduct was also dependent on the trans-chalcone concentration (Fig. 1D). Top-down fragmentation mass spectrometry analysis confirmed the site of modification on the enzyme to be Cys205, the only cysteine residue on SrtA (Fig. S8, ESI†). Incubation of the inhibited enzyme with 10 mM DTT overnight did not restore enzyme activity which suggested an irreversible modification of the enzyme by trans-chalcone (data not shown). Based on the mass increase and the site of modification of SrtA by trans-chalcone, the proposed mechanism for the reaction is via a Michael addition of trans-chalcone to the free thiol of Cys205 (Fig. S9, ESI†). This proposition was strengthened by analysing incubations of SrtA and dihydrochalcone (the saturated analogue of trans-chalcone) by enzyme assay and mass spectrometry. This chalcone neither inhibited the enzyme activity nor did we observe an additional SrtA peak of mass increase of +210 Da (Fig. S10, ESI†). Taken together these data highlight the requirement for the α,β-unsaturated system of trans-chalcone and support our proposed Michael addition mechanism rather than nucleophilic attack at the carbonyl carbon.
975 Da (Δ208 Da) which corresponds to a single SrtA–chalcone adduct (Fig. 1C). The formation of the adduct was also dependent on the trans-chalcone concentration (Fig. 1D). Top-down fragmentation mass spectrometry analysis confirmed the site of modification on the enzyme to be Cys205, the only cysteine residue on SrtA (Fig. S8, ESI†). Incubation of the inhibited enzyme with 10 mM DTT overnight did not restore enzyme activity which suggested an irreversible modification of the enzyme by trans-chalcone (data not shown). Based on the mass increase and the site of modification of SrtA by trans-chalcone, the proposed mechanism for the reaction is via a Michael addition of trans-chalcone to the free thiol of Cys205 (Fig. S9, ESI†). This proposition was strengthened by analysing incubations of SrtA and dihydrochalcone (the saturated analogue of trans-chalcone) by enzyme assay and mass spectrometry. This chalcone neither inhibited the enzyme activity nor did we observe an additional SrtA peak of mass increase of +210 Da (Fig. S10, ESI†). Taken together these data highlight the requirement for the α,β-unsaturated system of trans-chalcone and support our proposed Michael addition mechanism rather than nucleophilic attack at the carbonyl carbon.
Inhibition of SrtA should lead to reduced attachment of various cell surface proteins that effect biofilm formation so, with strong evidence that chalcone modifies SrtA in vitro, we tested the natural product using a S. mutans biofilm assay. The cells were assayed for growth on saliva-coated glass slides using the acta active adhesion (AAA) model by measuring quantitatively the biofilm biomass after 16–18 h of growth (Fig. S11, ESI†).27 The S. mutans cells were grown in different concentrations of trans-chalcone (0–500 μM) and the resulting biofilm biomass was measured. Morin, a known biofilm disruptor, was used as a positive control. We observed that biofilm formation was reduced in the presence of trans-chalcone in a concentration dependent manner up to 250 μM, with efficacy tailing off at higher concentrations (Fig. S11, ESI†). Since biofilm formation is a complex and dynamic process that involves changes in microbial metabolism and signaling, this encouraging result suggests that the exact mechanism of chalcone inhibition in vivo requires further in-depth study.22
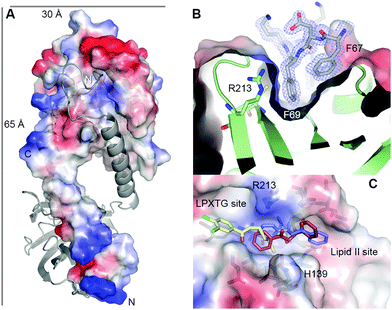
We attempted to gain structural insight into the SrtA:inhibitor interaction but we noticed that the wild-type SrtA enzyme was unstable during crystal trials due to auto-proteolysis (Fig. S12A, ESI†). To overcome this we produced and crystallised the more stable SrtA H139A mutant (Fig. S12B, ESI†) and, as expected, although this mutant was inactive in the in vitro fluorescent assay, we still observed the formation of the trans-chalcone adduct upon incubation of the SrtA H139A mutant enzyme with the inhibitor (Fig. S13, ESI†). This suggests that the mutant undergoes a similar Michael addition to the wild-type SrtA. Crystals of the SrtA H139A mutant defracted to 1.6 Å (Fig. S14, ESI†) and the crystal structure (PDB code: 4TQX) was determined by molecular replacement using the structure of the S. pyogenes SrtA as the starting model (PDB code: 3FN5).13 The electron density map at residues 49–53 at the N-terminus was ambiguous and suggests some degree of flexibility in this region; therefore these residues were omitted from the final model. The S. mutans SrtA has a canonical eight stranded β-barrel core (Fig. 2A, Fig. S15A, ESI†) which is highly conserved among the sortase superfamily.12
Our construct lacks the predicted N-terminal forty amino acids of the trans-membrane domain but the structure revealed a unique, extended N-terminal helix (residues 69–89, relative to the full-length enzyme) which is absent from all other sortase A structures present in the PDB (Fig. S15B, ESI†). This helix is extended away from the catalytic domain but connected to it by a short loop (residues 89–93). An N-terminal helix was observed in the crystal structure of S. aureus SrtB (PDB: 1NG5).28 However, in contrast to our structure, this helix is hinged at Asp41 in SrtB and places it in an equivalent position to the C-terminus of SrtA (Fig. S15A, ESI†). This is the first observation of the SrtA N-terminal domain and its structure suggests that it plays an important functional role in positioning the catalytic domain of sortases above the membrane where it can efficiently interact with it substrates.
The relative positions of the active site residues of SrtA (Cys205, Arg213) are highly conserved across the family, with no architectural compromises evident by introduction of the H139A mutation (Fig. S15C, ESI†). The only significant difference is observed between the cysteine residue of S. aureus SrtB (SaSrtB) and the other sortase enzymes. In SaSrtB, the cysteine residue points towards the histidine residue in the active site, whereas in the other sortases the cysteine residue points away from the His separated by a distance of ∼7 Å. In the crystal structure of the SrtA H139A mutant, Cys205, Ala139 and Arg213 are positioned on three neighboring β-strands, β-7, β-4 and β-8 respectively which form a tunnel-like hydrophobic pocket, similar to that described in other SrtA enzymes which are thought to adopt a conformation that is ideal for receiving the natural peptide substrate. Interestingly, when viewing the unit cell, the N-terminal α-helix (residues 69–89) from a symmetry-related molecule, makes extensive contacts with the active site cleft/binding groove of SrtA (Fig. 2A). These serendipitidous “non-physiological dimers” involving the active site offer a structural rationale as to why the wild type SrtA undergoes auto-proteolysis. Although it does not contain a canonical “sortase motif” it appears that the N-terminus of SrtA binds to the active site and this self-recognition leads to amide bond cleavage.
Although we prepared milligram quantities of chalcone-modified SrtA H139A it was difficult to handle at high concentrations and failed to crystallise. Recently, Schneewind and colleagues were able to identify a new class of mechanism-based inhibitors, aryl(β-amino)ethyl ketones (AAEK) from a screen of over 135![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 compounds which irreversibly modify the active site cysteine residue of SrtA and SrtB.15 Therefore, guided by the SaSrtA:LPETG peptide complex structure (PDB 1T2W),10 the B. anthracis SrtB-modified with aryl(β-amino)ethyl ketone (SrtB–AAEK, PDB: 2OQZ),15 and the positions of the bound Phe67 and Phe69 side-chains from the symmetry-related partner in our SrtA structure, we were able to construct models of the SrtA–chalcone bound adduct. We used the phenyl rings of both amino acids from the symmetry-related partner to place the two aromatic rings of the inhibitor covalently-bound to the thiol of Cys205. Interestingly, there is space to accommodate the adduct in the active site cleft in either the R- or the S-configuration (Fig. 2C). Of course there is free rotation about the C–S bond that could potentially swap the position of the rings. That said, by comparing the structure of the SrtB–AAEK adduct with our SrtA–chalcone adduct model, we note that the SrtA active site is open at both ends, in contrast to the SrtB-binding cleft which is blocked by Tyr235. These structural differences suggest a possible strategy for exploiting more structurally diverse chalcones drawn from the flavonoid family as isoform-specific sortase inhibitors.
000 compounds which irreversibly modify the active site cysteine residue of SrtA and SrtB.15 Therefore, guided by the SaSrtA:LPETG peptide complex structure (PDB 1T2W),10 the B. anthracis SrtB-modified with aryl(β-amino)ethyl ketone (SrtB–AAEK, PDB: 2OQZ),15 and the positions of the bound Phe67 and Phe69 side-chains from the symmetry-related partner in our SrtA structure, we were able to construct models of the SrtA–chalcone bound adduct. We used the phenyl rings of both amino acids from the symmetry-related partner to place the two aromatic rings of the inhibitor covalently-bound to the thiol of Cys205. Interestingly, there is space to accommodate the adduct in the active site cleft in either the R- or the S-configuration (Fig. 2C). Of course there is free rotation about the C–S bond that could potentially swap the position of the rings. That said, by comparing the structure of the SrtB–AAEK adduct with our SrtA–chalcone adduct model, we note that the SrtA active site is open at both ends, in contrast to the SrtB-binding cleft which is blocked by Tyr235. These structural differences suggest a possible strategy for exploiting more structurally diverse chalcones drawn from the flavonoid family as isoform-specific sortase inhibitors.
In summary, our study provides strong evidence that trans-chalcone inhibits S. mutans SrtA by covalently modifying its target and also reveals novel aspects of the structure and mechanism of the sortase family. Biofilm inhibition data also lends support for further efforts into potential uses of natural products to control the growth of oral flora.
This work was supported by Wm. Wrigley Jr. Company (DJW-R) and the University of Edinburgh (DJW-R, JM-W, D. J. Clarke).
Notes and references
- A. W. Maresso and O. Schneewind, Pharmacol. Rev., 2008, 60, 128–141 CrossRef CAS PubMed.
- L. A. Marraffini, A. C. Dedent and O. Schneewind, Microbiol. Mol. Biol. Rev., 2006, 70, 192–221 CrossRef CAS PubMed.
- O. Schneewind and D. M. Missiakas, Philos. Trans. R. Soc., B, 2012, 367, 1123–1139 CrossRef CAS PubMed.
- K. W. Clancy, J. A. Melvin and D. G. McCafferty, Biopolymers, 2010, 94, 385–396 CrossRef CAS PubMed.
- T. Spirig, E. M. Weiner and R. T. Clubb, Mol. Microbiol., 2011, 82, 1044–1059 CrossRef CAS PubMed.
- O. Schneewind, A. Fowler and K. F. Faull, Science, 1995, 268, 103–106 CAS.
- H. Ton-That, G. Liu, S. K. Mazmanian, K. F. Faull and O. Schneewind, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 12424–12429 CrossRef CAS.
- H. Ton-That, S. K. Mazmanian, L. Alksne and O. Schneewind, J. Biol. Chem., 2002, 277, 7447–7452 CrossRef CAS PubMed.
- H. Ton-That and O. Schneewind, J. Biol. Chem., 1999, 274, 24316–24320 CrossRef CAS PubMed.
- Y. Zong, T. W. Bice, H. Ton-That, O. Schneewind and S. V. Narayana, J. Biol. Chem., 2004, 279, 31383–31389 CrossRef CAS PubMed.
- B. C. Chenna, B. A. Shinkre, J. R. King, A. L. Lucius, S. V. Narayana and S. E. Velu, Bioorg. Med. Chem. Lett., 2008, 18, 380–385 CrossRef CAS PubMed.
- U. Ilangovan, H. Ton-That, J. Iwahara, O. Schneewind and R. T. Clubb, Proc. Natl. Acad. Sci. U. S. A., 2001, 98, 6056–6061 CrossRef CAS PubMed.
- P. R. Race, M. L. Bentley, J. A. Melvin, A. Crow, R. K. Hughes, W. D. Smith, R. B. Sessions, M. A. Kehoe, D. G. McCafferty and M. J. Banfield, J. Biol. Chem., 2009, 284, 6924–6933 CrossRef CAS PubMed.
- B. A. Frankel, M. Bentley, R. G. Kruger and D. G. McCafferty, J. Am. Chem. Soc., 2004, 126, 3404–3405 CrossRef CAS PubMed.
- A. W. Maresso, R. Wu, J. W. Kern, R. Zhang, D. Janik, D. M. Missiakas, M. E. Duban, A. Joachimiak and O. Schneewind, J. Biol. Chem., 2007, 282, 23129–23139 CrossRef CAS PubMed.
- C. J. Scott, A. McDowell, S. L. Martin, J. F. Lynas, K. Vandenbroeck and B. Walker, Biochem. J., 2002, 366, 953–958 CAS.
- S. S. Kang, J. G. Kim, T. H. Lee and K. B. Oh, Biol. Pharm. Bull., 2006, 29, 1751–1755 CAS.
- S. H. Kim, D. S. Shin, M. N. Oh, S. C. Chung, J. S. Lee, I. M. Chang and K. B. Oh, Biosci., Biotechnol., Biochem., 2003, 67, 2477–2479 CrossRef CAS PubMed.
- I. Oh, W. Y. Yang, S. C. Chung, T. Y. Kim, K. B. Oh and J. Shin, Arch. Pharmacal Res., 2011, 34, 217–222 CrossRef CAS PubMed.
- B. S. Park, J. G. Kim, M. R. Kim, S. E. Lee, G. R. Takeoka, K. B. Oh and J. H. Kim, J. Agric. Food Chem., 2005, 53, 9005–9009 CrossRef CAS PubMed.
- N. C. Veitch and R. J. Grayer, Nat. Prod. Rep., 2011, 28, 1626–1695 RSC.
- J. A. Lemos, R. G. Quivey, H. Koo and J. Abranches, Microbiology, 2013, 159, 436–445 CrossRef CAS PubMed.
- P. Hu, P. Huang and M. W. Chen, Arch. Oral Biol., 2013, 58, 1343–1348 CrossRef CAS PubMed.
- P. Hu, P. Huang and W. M. Chen, Appl. Biochem. Biotechnol., 2013, 171, 396–402 CrossRef CAS PubMed.
- T. Igarashi, E. Asaga and N. Goto, Oral Microbiol. Immunol., 2003, 18, 266–269 CrossRef CAS.
- H. Ton-That, S. K. Mazmanian, K. F. Faull and O. Schneewind, J. Biol. Chem., 2000, 275, 9876–9881 CrossRef CAS PubMed.
- R. A. Exterkate, W. Crielaard and J. M. Ten Cate, Caries Res., 2010, 44, 372–379 CrossRef CAS PubMed.
- R. G. Zhang, R. Y. Wu, G. Joachimiak, S. K. Mazmanian, D. M. Missiakas, P. Gornicki, O. Schneewind and A. Joachimiak, Structure, 2004, 12, 1147–1156 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Includes kinetics data, mass spectrometry analysis, crystallographic data and biofilm analysis. See DOI: 10.1039/c5cc01816a |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2015 |