Open Access Article
Open Access ArticleDetecting intracellular translocation of native proteins quantitatively at the single cell level†
Zhenning
Cao
a,
Shuo
Geng
b,
Liwu
Li
b and
Chang
Lu
*ac
aSchool of Biomedical Engineering and Sciences, Virginia Tech, Blacksburg, VA 24061, USA. E-mail: changlu@vt.edu; Fax: +1 540 231 5022; Tel: +1 540 231 8681
bDepartment of Biological Sciences, Virginia Tech, Blacksburg, VA 24061, USA
cDepartment of Chemical Engineering, Virginia Tech, Blacksburg, VA 24061, USA
First published on 7th April 2014
Abstract
The intracellular localization and movement (i.e. translocation) of proteins are critically correlated with the functions and activation states of these proteins. Simple and accessible detection methods that can rapidly screen a large cell population with single cell resolution have been seriously lacking. In this report, we demonstrate a simple protocol for detecting translocation of native proteins using a common flow cytometer which detects fluorescence intensity without imaging. We sequentially conducted chemical release of cytosolic proteins and fluorescence immunostaining of a targeted protein. The detected fluorescence intensity of cells was shown to be quantitatively correlated to the cytosolic/nuclear localization of the protein. We used our approach to detect the translocation of native NF-κB (an important transcription factor) at its native expression level and examine the temporal dynamics in the process. The incorporation of fluorescence immunostaining makes our approach compatible with the analysis of cell samples from lab animals and patients. Our method will dramatically lower the technological hurdle for studying subcellular localization of proteins.
Introduction
Within eukaryotic cells, proteins efficiently and selectively transit between functionally distinct subcellular compartments including plasma membrane, cytosol, nucleus and other membrane-enclosed organelles. The subcellular localization of an intracellular protein or the change of it (i.e. intracellular translocation) is highly significant for several reasons. First, intracellular translocation can be a prerequisite for proteins to carry out their intended functions. For example, a transcription factor needs to move from the cytosol into the nucleus in order to regulate gene transcription and such events occur typically as a consequence of outside stimuli to the cell. Second, since translocation is often associated with modification and activation at the molecular level (e.g. phosphorylation and methylation), the subcellular localization of the protein molecule is often indicative of its state. In most cases, the proteins are only active at their intended subcellular location. Finally, subcellular mislocalization of proteins leads to diseases ranging from metabolic disorders to cancers.1,2 Mislocalization of Akt, NF-κB, FOXO, p27, and p53 have been well-documented as key features in a variety of cancers.2,3 Modulation of protein translocation is practiced as an important therapeutic approach for cancer treatment.1,2 The subcellular location of a target protein can also serve as a useful read-out for high-content screening of cancer drugs.2Conventionally intracellular translocations such as nucleocytoplasmic transport (i.e. the translocation between the nucleus and the cytosol4,5) have been evaluated using fluorescence microscopy or subcellular fractionation.6–13 However, there are important limitations with these approaches. Fluorescence microscopy (including total internal reflection fluorescence microscopy, or TIRFM13) typically analyzes a limited number of cells and does not provide information on the distribution of the cell population. Subcellular fractionation involves lysis and homogenization of cells and then separation of the materials from various subcellular compartments by centrifugation. The data obtained by subcellular fractionation reflect only the average properties of the cell population without revealing the heterogeneity that is often critically involved in cell signaling networks.14–19 For example, when cells show an all-or-none response to a particular stimulus (bistability), only a subset of cells respond to the signal.15,20–23 Thus high-throughput methods are desired to generate information on the translocation of a large number of cells with single cell resolution.
Laser scanning cytometry (LSC)24,25 and imaging flow cytometry26 which permit rapid acquisition of fluorescence images of solid phase or flowing cells, have been used to detect the subcellular localization of intracellular proteins. However, these instruments are very expensive and typically have only limited accessibility through large central facilities. More importantly, these imaging-based instruments require sufficient exposure time in order to generate enough spatial resolution for the analysis. This determines that their throughputs are no more than several hundred cells per second (compared to 104–105 cells per second by a conventional flow cytometer which detects only fluorescence intensity) and the complex algorithms used for image analysis often introduce errors and bias.27–29 In our previous work, we developed electroporative flow cytometry to examine protein translocations by adding electroporation-based protein release and flow cytometric screening.30,31 Unfortunately, electroporative flow cytometry requires special apparatus for combing electroporation and laser-induced fluorescence detection. More importantly, the approach requires proteins of interest tagged by fluorescent protein markers and does not allow examination of native proteins and primary cells isolated from animals and patients. We also explored using TIRF-based flow cytometry for probing protein translocation.32 However, its potential for high resolution recognition of nucleocytoplasmic translocation was limited.
In this work, we demonstrate a simple method that combines selective chemical release of cytosolic proteins and standard fluorescence immunostaining for detecting the translocation of native proteins at the native expression level with single cell resolution. We demonstrate the proof-of-principle by detecting nucleocytoplasmic transport of an important transcription factor NF-κB. NF-κB undergoes nucleocytoplasmic transport from the cytosol to the nucleus in order to regulate transcription and gene expression, upon extracellular stimuli (e.g. by TNFα).33,34 Briefly, we used saponin (i.e. a class of amphipathic glycosides) to selectively release cytosolic fraction of intracellular proteins by dissolving the cholesterol content and permeabilizing the plasma membrane.35 Such treatment was followed by fluorescence immunostaining of residual NF-κB. The cell population was then screened by a common flow cytometer for fluorescence intensity of each cell. We showed that the fluorescence intensity of a cell could be correlated to the subcellular localization of the protein. Taking advantage of common flow cytometry which is widely accessible, our approach detects the translocation of native proteins without imaging and with high throughput. Our approach is readily compatible with analysis of primary samples from animals and patients.
Results and discussion
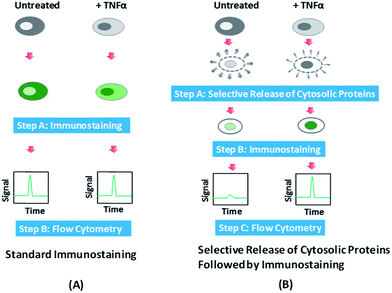
In Fig. 1, we outline the procedure and working principle of our protocol (i.e. selective release and immunostaining), in comparison to those of the standard immunostaining protocol, and how these procedures generate different results for single cell screening with standard flow cytometry.In our experiments, we applied TNFα to stimulate cells and produce NF-κB translocation from the cytosol to the nucleus. With standard fluorescence immunostaining, HeLa cells with NF-κB translocation (i.e. +TNFα) and those without NF-κB translocation (i.e. untreated) are crosslinked by paraformaldehyde and permeabilized by Triton X-100 before all intracellular NF-κB is fluorescently labeled by antibodies that specifically target NF-κB. The crosslinking by paraformaldehyde conserves all proteins in the cells and the permeabilization ensures the full access of the targeted protein (i.e. NF-κB in this study) by the antibodies for labeling. When these cells are subsequently screened by flow cytometry, which detects fluorescence from the entire cell, the translocation does not create difference in the fluorescence intensity detected because it only varies the subcellular localization of the protein, not the overall expression level (Fig. 1A). In comparison, in our method, we add a selective release step before immunostaining. During this step, saponin is used to treat the cells to dissolve cholesterol in the plasma membrane and make the membrane leaky.35–37 Saponin is known to primarily permeabilize the plasma membrane while keeping the cholesterol-poor internal membranes (e.g. the mitochondrial membrane and nuclear envelope) intact.35,38 Gentle treatment by saponin has also been shown to have minimal effects on cellular functions such as protein synthesis.39,40 Cytosolic proteins are preferentially released out of the cells and the nucleic proteins are largely unaffected in their amounts. Thus cells with NF-κB translocation (with NF-κB mostly in the nucleus) have far less decrease in the NF-κB amount due to the release, compared to the cells without NF-κB translocation (with NF-κB primarily in the cytosol). The selective release step is immediately followed by immunostaining that fixes the cells and labels all the remaining intracellular NF-κB. The flow cytometry results now are significantly different for cells with translocation and those without translocation, with the latter showing much smaller fluorescence intensity. Thus we are able to link the subcellular localization of the protein with the detected fluorescence intensity of a cell treated by our protocol.
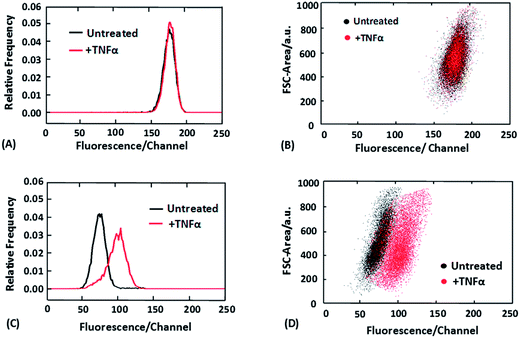
Fig. 2 shows the flow cytometry data obtained after standard immunostaining (Fig. 2A and B) and combined selective release and immunostaining (Fig. 2C and D).
Fig. 2A shows that as expected, with standard immunostaining the fluorescence intensity histogram generated by a cell population without NF-κB translocation overlaps with that generated by a cell population with the translocation (stimulated by TNFα for 30 min). In comparison, the two cell populations exhibit marked difference in the fluorescence intensity histogram when we had selective release of cytosolic proteins by saponin (0.05% saponin for 10 min) before the immunostaining (Fig. 2C). Furthermore, we also discovered that the two cell populations (processed with the selective release protocol) were even more distinct when two-dimensional dot plots were used to include information on the cell size (via detecting the forward scatter signal) (Fig. 2D). The two cell populations were entirely separated from each another in Fig. 2D. This improvement is attributed to the differentiation of large cells without translocation (NF-κB in the cytosol) and small cells with translocation (NF-κB in the nucleus). These two subpopulations may have similar fluorescence intensities after the selective release protocol but are very different in the cell size. The fluorescence images of cells after standard immunostaining and selective release/immunostaining were also collected (ESI Fig. S1†). The images confirm the proposed mechanism in Fig. 1. Our technique renders the fluorescence intensity different for cells with translocation and those without translocation, whereas standard immunostaining reveals the different localizations of the protein via imaging with the overall fluorescence intensity from whole cells being the same for the two types of cells.
We also optimized the selective release protocol in order to create the maximum differentiation of cells based on NF-κB subcellular localization. We varied the concentration and duration of saponin treatment for two cell populations (untreated cells and cells stimulated by 50 ng ml−1 TNFα for 30 min) and observed the difference in their fluorescence intensity histograms (ESI Fig. S2†). Fig. S2A† shows that with 10 min treatment time, the saponin concentration of 0.05% yielded the best separation between the two cell populations. The decreased differentiation at higher saponin concentrations (0.2–0.5%) presumably resulted from the release of both cytosolic and nucleic proteins. Fig. S2B† shows that with a fixed concentration of 0.05% for saponin, the optimal treatment time was in between 1 and 10 min. Exceedingly long treatment times (>10 min) also led to decreased separation, due to release of both cytosolic and nucleic proteins. Thus we determine that saponin treatment of 0.05% concentration and 1–10 min works the best as the selective release step for differentiation of cells with NF-κB in the nucleus and those with the same protein in the cytosol.
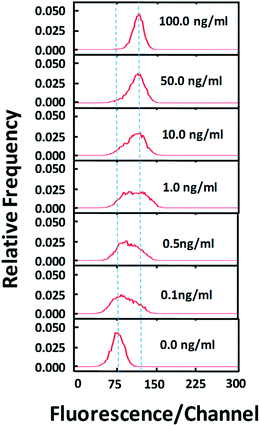
Using the optimized selective release protocol (in combination with immunostaining), we show that our method is effective for revealing various degrees of NF-κB translocation in the cell population. In Fig. 3, various concentrations of TNFα were used to stimulate the cell population for 30 min. With low concentrations (0.1–1.0 ng ml−1) of TNFα, the cell populations appear to have lower degree of NF-κB translocation overall and a subpopulation of cells have no translocation at all (based on the broad peak shape which is not Gaussian).
With high concentrations of TNFα (10–100 ng ml−1), the translocation occurs more completely for the cell population and the separation between the stimulated population and the control is increasingly complete.
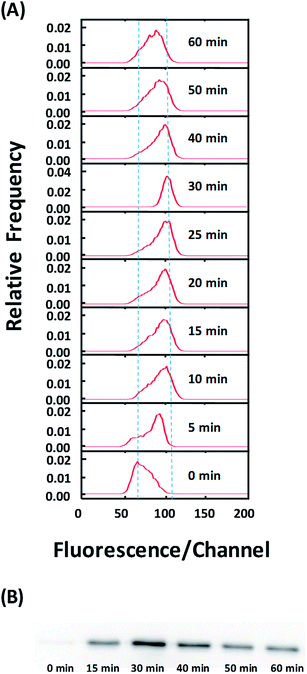
Finally, we used our approach to examine temporal dynamics in the cell population during NF-κB translocation. In Fig. 4A, the shift in the fluorescence intensity histogram suggests the movement of NF-κB from the cytosol to the nucleus after TNFα stimulation. The increase in the fluorescence intensity of the histogram indicates the increased occupation of the nucleic localization over time. The data show that the translocation occurs substantially within the first 5 min after simulation. There is increased translocation until 30 min after stimulation. Interestingly, translocation in the reverse direction (from the nucleus to the cytosol) occurs between 40–60 min. Such reverse translocation was previously reported in the literature41 and is due to re-inhibition of newly synthesized repressor IκB.8 The western blotting analysis (Fig. 4B) also corroborates these findings by our technique.
The resolution of the technology depends on several factors. First, the amount of the protein translocation between the nucleus and the cytosol affects the resolution. Based on the comparison of Fig. 4A and B, the translocation of ∼13% of the total NF-κB in the entire cell can be clearly resolved by the flow cytometry data. Second, the completeness and selectiveness of cytosolic release by saponin are critical for high resolution. As demonstrated in Fig. S2,† optimal treatment conditions need to be obtained for a particular cell/protein system in order to reach the best resolution. Third, the immunostaining after the selective release needs to be complete and yields strong fluorescence signal. This facilitates obtaining high quality flow cytometry data.
To conclude, by combining selective release of cytosolic proteins via chemical permeabilization with fluorescence immunostaining, we develop a protocol that links the fluorescence intensity of a single cell with the subcellular localization of a targeted protein. By screening the fluorescence emitted by single cells using common flow cytometry, we are able to detect the translocation quantitatively with single cell resolution. Because fluorescence immunostaining is ideally suited for studying cell samples from animals and patients, our approach provides a very simple route for examining protein translocation at the single cell level with direct biomedical relevance. We expect that this approach can be extended to a wide range of cell types and proteins.
Experimental section
Cell sample preparation
HeLa (CCL-2) cells were grown at 37 °C with 5% CO2 in Dulbecco's modified Eagle's medium (DMEM) (Mediatech, Herndon, VA) supplemented with 10% (v/v) fetal bovine serum (Sigma) and 1% penicillin (100 mg ml−1, Sigma). The cells were trypsinized and diluted at a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5–1
5–1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8 every 2 days to maintain the cells in the exponential growth phase. The harvested cells were centrifuged at 300g for 5 min and resuspended in DMEM culture medium at a final concentration of 1 × 106 cells per ml before experiments. To stimulate cells, cells (1 × 106 ml−1) were suspended in the culture medium with various concentrations of TNF-α (AbD Serotec, Raleigh, NC, USA) at 37 °C for various periods.
8 every 2 days to maintain the cells in the exponential growth phase. The harvested cells were centrifuged at 300g for 5 min and resuspended in DMEM culture medium at a final concentration of 1 × 106 cells per ml before experiments. To stimulate cells, cells (1 × 106 ml−1) were suspended in the culture medium with various concentrations of TNF-α (AbD Serotec, Raleigh, NC, USA) at 37 °C for various periods.
Standard fluorescence immunostaining
Fluorescence immunostaining was conducted following the literature with minor changes.42 The cell sample (∼1 × 106 cells) was fixed in 100 μl pre-warmed (at 37 °C) fixation buffer (4% paraformaldehyde in PBS buffer) for 10 min. Subsequently, the fixed cells were washed with a blocking buffer (1% BSA in PBS buffer) and permeabilized with 100 μl of a permeabilization buffer (0.2% Triton X-100 in PBS buffer). After 20 min incubation with the permeabilization buffer, the cells were centrifuged at 300g for 5 min to remove the permeabilization buffer and washed with the blocking buffer. The cells were then incubated with the blocking buffer containing a primary antibody [1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 dilution of NF-κB p65 (sc-8008, Santa Cruz Biotechnology, Dallas, Texas, USA)] for 1 h at room temperature. After incubation with the primary antibody, the cell sample was pelleted by centrifugation (300g, 5 min) and washed twice with the blocking buffer. Then, the cells were incubated (protected from light) in the blocking buffer containing fluorophore-conjugated secondary antibody [1
100 dilution of NF-κB p65 (sc-8008, Santa Cruz Biotechnology, Dallas, Texas, USA)] for 1 h at room temperature. After incubation with the primary antibody, the cell sample was pelleted by centrifugation (300g, 5 min) and washed twice with the blocking buffer. Then, the cells were incubated (protected from light) in the blocking buffer containing fluorophore-conjugated secondary antibody [1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 150 dilution of DyLight™ 488 Goat anti-mouse IgG1 antibody (409102, Biolegend, San Diego, CA)], which bound to the primary antibody, for 1 h at room temperature. The staining solution was then aspirated out and the labeled cells were washed twice with the blocking buffer to remove nonspecific binding. The cells were stored in PBS with 0.1% sodium azide at 4 °C, if not immediately analyzed by flow cytometry.
150 dilution of DyLight™ 488 Goat anti-mouse IgG1 antibody (409102, Biolegend, San Diego, CA)], which bound to the primary antibody, for 1 h at room temperature. The staining solution was then aspirated out and the labeled cells were washed twice with the blocking buffer to remove nonspecific binding. The cells were stored in PBS with 0.1% sodium azide at 4 °C, if not immediately analyzed by flow cytometry.
Selective release of cytosolic proteins followed by fluorescence immunostaining
The cell sample (∼1 × 106 cells) was incubated in 100 μl of the releasing buffer [0.05% (w/v) saponin (ID# 419-25A, Chem Service, West Chester, PA, USA) in DMEM] for 5 min. The processed cells were then pelleted by centrifugation at 300g for 5 min to remove excessive releasing buffer and then immediately fixed by the fixation buffer for 30 min. Subsequently, the fixed cells were permeabilized in 100 μl of the permeabilization buffer. After 5 min incubation in the permeabilization buffer, the cells were centrifuged at 300g for 5 min to remove excessive permeabilization buffer and washed once with the blocking buffer. The rest of the procedure involving labeling using primary and secondary antibodies was the same as that in “Standard fluorescence immunostaining”.Fluorescence microscopy
Immunostained cells were transferred to a 96 well plate and then centrifuged for 5 min at 300g to settle the cells to the bottom. Fluorescence images were taken by an inverted fluorescence microscope (IX-71, Olympus, Melville, NY) with a 20× dry objective (0.5 NA). The fluorescence excitation was provided by a 100 W mercury lamp. The excitation and emission were filtered by a fluorescence filter cube (exciter HQ480/40, emitter HQ535/50, and beam splitter Q505lp, Chroma Technology) for observing green fluorescence.Western blotting
Cellular protein samples were made using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Pierce Biotechnology, Rockford, IL, USA) following the manufacturer's recommendations. The total protein concentration for each sample was measured by Pierce BCA protein assay kit (Pierce Biotechnology, Rockford, IL, USA), and then equal amount of denatured proteins from each sample was separated by standard SDS-PAGE and analyzed by Western blotting. Briefly, samples were loaded into polyacrylamide gel for SDS-PAGE followed by transferring to a polyvinylidene fluoride (PVDF) membrane, which was blocked by 5% milk in TBST (Tris-Buffered Saline, 0.1% Tween-20) buffer. The membrane was stained with 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 diluted primary antibody (SC-8008, Santa Cruz Biotechnology) and secondary antibody [Rabbit anti-mouse horseradish peroxidase (HRP) (Pierce, Rockford, IL)] for 1 h each. Membranes were visualized by LAS-3000 luminescent image analyzer (Fujifilm, Hanover Park, IL, USA) after chemiluminescence treatment with Pierce ECL Western Blotting Substrate (Pierce Biotechnology, Rockford, IL, USA). The intensity of each band was quantified using ImageJ software.
1000 diluted primary antibody (SC-8008, Santa Cruz Biotechnology) and secondary antibody [Rabbit anti-mouse horseradish peroxidase (HRP) (Pierce, Rockford, IL)] for 1 h each. Membranes were visualized by LAS-3000 luminescent image analyzer (Fujifilm, Hanover Park, IL, USA) after chemiluminescence treatment with Pierce ECL Western Blotting Substrate (Pierce Biotechnology, Rockford, IL, USA). The intensity of each band was quantified using ImageJ software.
Flow cytometry analysis
Fluorescently stained cell samples were analyzed at medium flow rate by a FACS Canto II cytometer (BD, San Jose, CA, USA) equipped with 488 nm laser/filters for FITC and forward scatter (FSC) detection. For each histogram, 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000–20
000–20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 events were collected. The cytometer was routinely calibrated with Calibrite beads (BD). Stained cell samples may be stored for up to 24 h at 4 °C before analysis without a significant loss in fluorescence intensity. The data were processed by FlowJo and Origin 9.0.
000 events were collected. The cytometer was routinely calibrated with Calibrite beads (BD). Stained cell samples may be stored for up to 24 h at 4 °C before analysis without a significant loss in fluorescence intensity. The data were processed by FlowJo and Origin 9.0.
Notes and references
- J. R. Davis, M. Kakar and C. S. Lim, Pharm. Res., 2007, 24, 17–27 CrossRef CAS PubMed.
- T. R. Kau, J. C. Way and P. A. Silver, Nat. Rev. Cancer, 2004, 4, 106–117 CrossRef CAS PubMed.
- A. Bellacosa, C. Kumar, A. Di Cristofano and J. Testa, Adv. Cancer Res., 2005, 94, 29–86 CrossRef CAS.
- S. Nakielny and G. Dreyfuss, Cell, 1999, 99, 677–690 CrossRef CAS.
- E. A. Nigg, Nature, 1997, 386, 779–787 CrossRef CAS PubMed.
- H. Ma, T. M. Yankee, J. Hu, D. J. Asai, M. L. Harrison and R. L. Geahlen, J. Immunol., 2001, 166, 1507–1516 CrossRef CAS.
- H. Asanuma, T. Torigoe, K. Kamiguchi, Y. Hirohashi, T. Ohmura, K. Hirata, M. Sato and N. Sato, Cancer Res., 2005, 65, 11018–11025 CrossRef CAS PubMed.
- D. E. Nelson, A. E. C. Ihekwaba, M. Elliott, J. R. Johnson, C. A. Gibney, B. E. Foreman, G. Nelson, V. See, C. A. Horton, D. G. Spiller, S. W. Edwards, H. P. McDowell, J. F. Unitt, E. Sullivan, R. Grimley, N. Benson, D. Broomhead, D. B. Kell and M. R. H. White, Science, 2004, 306, 704–708 CrossRef CAS PubMed.
- F. Zhou, J. Hu, H. Ma, M. L. Harrison and R. L. Geahlen, Mol. Cell. Biol., 2006, 26, 3478–3491 CrossRef CAS PubMed.
- R. Cheong, C. J. Wang and A. Levchenko, Mol. Cell. Proteomics, 2009, 8, 433–442 CAS.
- S. Tay, J. J. Hughey, T. K. Lee, T. Lipniacki, S. R. Quake and M. W. Covert, Nature, 2010, 466, 267–271 CrossRef CAS PubMed.
- A. Hoffmann, A. Levchenko, M. L. Scott and D. Baltimore, Science, 2002, 298, 1241–1245 CrossRef CAS PubMed.
- X. Chen, W. Li, J. Ren, D. Huang, W. T. He, Y. Song, C. Yang, X. Zheng, P. Chen and J. Han, Cell Res., 2014, 24, 105–121 CrossRef CAS PubMed.
- J. E. Ferrell Jr, Curr. Opin. Biotechnol., 2002, 14, 140–148 CrossRef.
- J. E. Ferrell and E. M. Machleder, Science, 1998, 280, 895–898 CrossRef CAS.
- Y. Lu, J. J. Chen, L. Y. Mu, Q. Xue, Y. Wu, P. H. Wu, J. Li, A. O. Vortmeyer, K. Miller-Jensen, D. Wirtz and R. Fan, Anal. Chem., 2013, 85, 2548–2556 CrossRef CAS PubMed.
- Q. H. Shi, L. D. Qin, W. Wei, F. Geng, R. Fan, Y. S. Shin, D. L. Guo, L. Hood, P. S. Mischel and J. R. Heath, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 419–424 CrossRef CAS PubMed.
- K. J. Lee, S. M. Mwongela, S. Kottegoda, L. Borland, A. R. Nelson, C. E. Sims and N. L. Allbritton, Anal. Chem., 2008, 80, 1620–1627 CrossRef CAS PubMed.
- G. D. Meredith, C. E. Sims, J. S. Soughayer and N. L. Allbritton, Nat. Biotechnol., 2000, 18, 309–312 CrossRef CAS PubMed.
- S. Tay, J. J. Hughey, T. K. Lee, T. Lipniacki, S. R. Quake and M. W. Covert, Nature, 2010, 466, 267–271 CrossRef CAS PubMed.
- M. W. Covert, T. H. Leung, J. E. Gaston and D. Baltimore, Science, 2005, 309, 1854–1857 CrossRef CAS PubMed.
- T. K. Lee, E. M. Denny, J. C. Sanghvi, J. E. Gaston, N. D. Maynard, J. J. Hughey and M. W. Covert, Sci. Signaling, 2009, 2, ra65 Search PubMed.
- Y. Awwad, T. Geng, A. S. Baldwin and C. Lu, Anal. Chem., 2012, 84, 1224–1228 CrossRef CAS PubMed.
- M. M. Harnett, Nat. Rev. Immunol., 2007, 7, 897–904 CrossRef CAS PubMed.
- A. Deptala, E. Bedner, W. Gorczyca and Z. Darzynkiewicz, Cytometry, 1998, 33, 376–382 CrossRef CAS.
- P. V. Beum, M. A. Lindorfer, B. E. Hall, T. C. George, K. Frost, P. J. Morrissey and R. P. Taylor, J. Immunol. Methods, 2006, 317, 90–99 CrossRef CAS PubMed.
- D. A. Basiji, W. E. Ortyn, L. Liang, V. Venkatachalam and P. Morrissey, Clin. Lab. Med., 2007, 27, 653–670 CrossRef PubMed.
- J. Vigo, J. Salmon, S. Lahmy and P. Viallet, Anal. Cell. Pathol., 1991, 3, 145 CAS.
- U. G. Falkmer, Hum. Pathol., 1992, 23, 360–367 CrossRef CAS.
- J. Wang, N. Bao, L. L. Paris, H. Y. Wang, R. L. Geahlen and C. Lu, Anal. Chem., 2008, 80, 1087–1093 CrossRef CAS PubMed.
- J. Wang, B. Fei, Y. Zhan, R. L. Geahlen and C. Lu, Lab Chip, 2010, 10, 2911–2916 RSC.
- J. Wang, B. Fei, R. L. Geahlen and C. Lu, Lab Chip, 2010, 10, 2673–2679 RSC.
- A. Hoffmann and D. Baltimore, Immunol. Rev., 2006, 210, 171–186 CrossRef PubMed.
- S. Ghosh, M. J. May and E. B. Kopp, Sci. Signaling, 1998, 16, 225 CAS.
- M. Wassler, I. Jonasson, R. Persson and E. Fries, Biochem. J., 1987, 247, 407–415 CAS.
- M. C. Jamur and C. Oliver, in Immunocytochemical Methods and Protocols, Springer, 2010, pp. 63–66 Search PubMed.
- K. L. Goldenthal, K. Hedman, J. W. Chen, J. T. August and M. C. Willingham, J. Histochem. Cytochem., 1985, 33, 813–820 CrossRef CAS PubMed.
- B. S. Negrutskii and M. P. Deutscher, Proc. Natl. Acad. Sci. U. S. A., 1992, 89, 3601–3604 CrossRef CAS.
- B. S. Negrutskii, R. Stapulionis and M. P. Deutscher, Proc. Natl. Acad. Sci. U. S. A., 1994, 91, 964–968 CrossRef CAS.
- A. Hudder, L. Nathanson and M. P. Deutscher, Mol. Cell. Biol., 2003, 23, 9318–9326 CrossRef CAS.
- R. Cheong, A. Bergmann, S. L. Werner, J. Regal, A. Hoffmann and A. Levchenko, J. Biol. Chem., 2006, 281, 2945–2950 CrossRef CAS PubMed.
- H. M. Shapiro, Practical flow cytometry, Wiley-Liss, New York, 4th edn, 2003, pp. 293–345 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Supplementary Fig. S1 and S2. See DOI: 10.1039/c4sc00578c |
| This journal is © The Royal Society of Chemistry 2014 |