Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Simultaneous detection and quantification of three bacterial meningitis pathogens by SERS
Kirsten
Gracie
a,
Elon
Correa
b,
Samuel
Mabbott
a,
Jennifer A.
Dougan
a,
Duncan
Graham
a,
Royston
Goodacre
*b and
Karen
Faulds
*a
aCentre of Molecular Nanometrology, WestCHEM, Department of Pure and Applied Chemistry, University of Strathclyde, 295 Cathedral Street, Glasgow, G1 1XL, UK. E-mail: karen.faulds@strath.ac.uk
bSchool of Chemistry and Manchester Institute of Biotechnology, University of Manchester, 131 Princess Street, Manchester, M1 7ND, UK
First published on 20th December 2013
Abstract
Bacterial meningitis is well known for its rapid onset and high mortality rates, therefore rapid detection of bacteria found in cerebral spinal fluid (CSF) and subsequent effective treatment is crucial. A new quantitative assay for detection of three pathogens that result in bacterial meningitis using a combination of lambda exonuclease (λ-exonuclease) and surface enhanced Raman scattering (SERS) is reported. SERS challenges current fluorescent-based detection methods in terms of both sensitivity and more importantly the detection of multiple components in a mixture, which is becoming increasingly more desirable for clinical diagnostics. λ-Exonuclease is a processive enzyme that digests one strand of double stranded DNA bearing a terminal 5′-phosphate group. The new assay format involves the simultaneous hybridisation of two complementary DNA probes (one containing a SERS active dye) to a target sequence followed by λ-exonuclease digestion of double stranded DNA and SERS detection of the digestion product. Three meningitis pathogens were successfully quantified in a multiplexed test with calculated limits of detection in the pico-molar range, eliminating the need for time consuming culture based methods that are currently used for analysis. Quantification of each individual pathogen in a mixture using SERS is complex, however, this is the first report that this is possible using the unique spectral features of the SERS signals combined with partial least squares (PLS) regression. This is a powerful demonstration of the ability of this SERS assay to be used for analysis of clinically relevant targets with significant advantages over existing approaches and offers the opportunity for future deployment in healthcare applications.
Introduction
The detection of DNA sequences correlating to specific disease states has become vital in medical diagnostics. The majority of current methods of DNA detection involve the polymerase chain reaction (PCR) coupled with various fluorescence-based techniques.1,2 An alternative optical spectroscopy is surface enhanced Raman scattering (SERS) which is fast becoming a sensitive,3 highly molecularly specific technique with the ability to detect multiple analytes simultaneously due to the characteristic molecular fingerprint spectra obtained.4,5 As a result, numerous assays have been developed that involve the direct detection of DNA using SERS.6–11 The detection of many different disease states is possible but in order to demonstrate the potential of using SERS for analysis of disease, we opted for an infectious disease target since when the target is present, a positive result and identification is possible.Meningitis is an inflammation of the lining around the brain and spinal cord. Most cases of meningitis are caused by viruses such as herpes, mumps or measles.12–14 Unfortunately there are no effective therapies for most viruses that cause meningitis; therefore treatment is aimed at limiting the effects of the symptoms. Meningitis and septicaemia caused by bacteria are usually more serious compared to viral meningitis and require urgent medical attention with appropriate antibiotic therapy. Around 3400 people each year in the UK and Ireland are affected by bacterial meningitis, according to an investigation by the Meningitis Research Foundation.15,16 Three of the most common meningitis pathogens are Haemophilus influenzae, Streptococcus pneumoniae and Neisseria meningitidis. Most courses of treatment involve the administration of antibiotics, however this can obscure results from culture based methods, therefore making a positive diagnosis difficult. It is imperative that the correct pathogen is identified to allow for the most effective course of treatment. Results obtained from this new assay format are not affected by the presence of any antibiotics.
SERS is a promising technique for the detection of bacterial meningitis pathogens. SERS has previously been used to detect DNA due to the high discrimination achieved from the molecularly specific fingerprint spectra.17–19 Whilst SERS of individual DNA bases can be obtained, this does not allow identification of the order of bases and therefore is not sequence specific. As a result dye labels can be used to allow sequence specific detection of DNA using SERS.9 Using a label, such as a fluorescent dye attached to a specific sequence, the SERS signal of the label will be indicative of the presence of that specific sequence. SERS has been employed in various multiplex assays for the detection of several DNA sequences simultaneously. In 2002, Graham et al. were the first to use a SERS multiplex system for the detection of the cystic fibrosis trans-membrane conductance regulator (CFTCT) gene, which can be expressed in three forms.6 Further developments include an assay that reported the multiplex detection of DNA sequences coding for methicillin-resistant Staphylococcus aureus (MRSA).18 The assay was used for the detection of three genes associated with MRSA using three different fluorophores as the SERS labels. Vo-Dinh et al. developed a multiplex DNA detection assay using “molecular sentinels”, a structure that contains a looped sequence of DNA held together by two complementary sequences forming stems.20 “Molecular sentinels” have been used to achieve multiplexed detection of two genes that are biomarkers for breast cancer.
These examples highlight the use of SERS for clinically relevant targets however one of the main strengths of SERS is the ability to detect several analytes in a mixture and without separation steps. The simultaneous detection of five labelled oligonucleotides was reported by Faulds et al.4 Two different excitation wavelengths were used based on the responses the different dyes gave at different wavelengths. Five labelled oligonucleotides were mixed within a single sample and each of the dyes was successfully detected at either of the two wavelengths, without the need for any multivariate analysis. Following this success a 6-plex detection system was developed using one excitation wavelength and this time employed chemometrics,5 which is becoming increasingly popular for the analysis of Raman data.21,22 Each of the six dye labelled DNA sequences in the mixture were successfully identified using multivariate analysis. Zhong et al. then used the same 6-plex mixture and analysed the results using Bayesian methods, which gave a more accurate analysis of the various components within the mixture.23
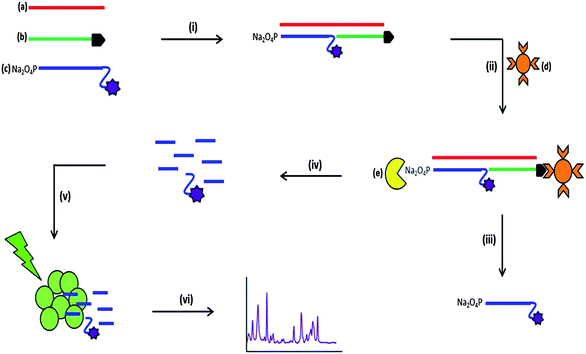
The assay format reported in this study involves the use of the enzyme λ-exonuclease (Fig. 1).57 It is a processive enzyme24–26 that digests one strand of 5′-phosphorylated double stranded DNA producing mononucleotides and leaving one DNA strand intact.27 The enzyme has a toroidal structure with three subunits. A central channel passes through the enzyme that decreases in size from 30 Å to 15 Å. Due to this feature, double stranded DNA can only enter the wider end of the channel and single stranded DNA is expelled from the narrow end.28 Only one of the three subunits is bound to the DNA substrate, the other two are present to ensure the enzyme moves in a processive manner along the full length of the DNA sequence and does not move backwards. When digestion occurs, a positively charged pocket within the subunit attracts the negatively charged phosphorylated DNA strand. The phosphodiester linkages are cleaved producing mononucleotides that leave through the narrow end of the central channel.29 λ-Exonuclease has previously been used in aptamer research30 and has been subject to many investigations into the kinetics and activity of the enzyme under different experimental conditions.31–34 The use of λ-exonuclease in this assay has the potential to exploit the signal amplification compared to target amplification methods such as PCR.35,36 The single strand of DNA that is left intact post-digestion is capable of undergoing successive digestion cycles, releasing the fluorescent dye into solution and amplifying the signal obtained.37
Here we report the combination of a new disease specific DNA-based SERS assay that demonstrates the simultaneous detection of three bacterial meningitis pathogens.38 Limits of detection that rival current fluorescent-based methods were obtained, however more importantly, by using chemometric analysis, each pathogen in the multiplex was successfully quantified.
Materials and methods
Colloid synthesis
Silver nanoparticles were synthesised using a modified Lee and Meisel method.39 Silver nitrate (90 mg) was dissolved in 500 mL distilled water. The solution was heated rapidly to boiling with continuous stirring. Once boiling, an aqueous solution of sodium citrate (1%, 10 mL) was added quickly. The heat was reduced and the solution was left to boil gently for 90 min with stirring. The colloid was then analysed by UV-vis spectroscopy and the λmax was 411 nm with the full width half-height (FWHH) measured to be 103 nm. The concentration of the colloid was calculated to be 0.3 nM.Oligonucleotides
The three target pathogen sequences were those used by Guiver et al. for their simultaneous detection using real-time PCR.38 Biotinylated DNA and unmodified target DNA sequences were purchased on a 0.01 μM scale with HPLC purification from Eurofins MWG (Ebersberg, Germany). 5′-Phosphate/dye modified DNA sequences were ordered on a 100 nM scale with HPLC purification from Integrated DNA Technologies (Leuven, Belgium) (Table 1).| Name | Sequence (5′–3′) | 5′ Modifications | 3′ Modifications |
|---|---|---|---|
| Streptococcus pneumoniae | TTCGAGTGTTGCTTATGGGCGCCA | — | — |
| Streptococcus pneumoniae “Capture Probe” | GCAACACTCGAA | — | Biotin |
| Streptococcus pneumoniae “Reporter Probe” | TGGCGCCCATAA | Phosphate | Spacer18-10A-TAMRA |
| Haemophilus influenzae | CCACGCTCATTCGTTTGATGAGTGGTG | — | — |
| Haemophilus influenzae “Capture Probe” | AACGAATGAGCGTGG | — | Biotin |
| Haemophilus influenzae “Reporter Probe” | CACCACTCATCA | Phosphate | Spacer18-10A-Cy3 |
| Neisseria meningitidis | ATGTGCAGCTGACACGTGGCAATG | — | — |
| Neisseria meningitidis “Capture Probe” | TCAGCTGCACAT | — | Biotin |
| Neisseria meningitidis “Reporter Probe” | CATTGCCACGTG | Phosphate | Spacer18-10A-FAM |
Buffer
Exonuclease buffer was supplied by New England Biolabs (Cambridge, UK) consisting of; 67 mM glycine–KOH, 2.5 mM MgCl2 and 50 μg mL−1 bovine serum albumin (BSA). The pH of the buffer was 9.4. Exonuclease buffer was supplied at 10× concentration and was diluted to 1× concentration when used in the assay.PCR
PCR was carried out using a Stratagene MX3005P fluorimeter and the commercially available Qiagen PCR reagents. Each reaction had a total volume of 25 μL; 2.5 μL of Qiagen reaction buffer (10×), 1 μL of MgCl2 solution (25 mM), 0.4 μL of deoxynucleoside triphosphates (10 mM), 1 μL of forward primer (10 μM), 1 μL of reverse primer (10 μM), 0.5 μL of Taq polymerase (5 U), 1 μL of template DNA (approx. 5 × 106 copies per μL) and made up to the total volume (25 μL) with DEPC (diethylpyrocarbonate) treated water. PCR was then carried out using the following thermal profile: 10 min at 95 °C, followed by 45 cycles of 95 °C for 30 s, 58 °C for 1 min then 72 °C for 1 min and a final extension for 1 min at 72 °C.Detection assay
A biotinylated modified sequence and a 5′-phosphate/fluorophore modified sequence underwent a sandwich hybridisation event with one complementary (target) sequence using a Minicycler PTC-150 system. An aliquot of each DNA sequence (10 μL, 1 μM) was added to a PCR tube containing phosphate buffered solution (70 μL, 0.3 M). The temperature was held at 90 °C for 10 min, and then was lowered to 10 °C for 10 min. For the no target control, the target DNA sequence was omitted and replaced with distilled water, and the non-complementary control used a sequence that was not complementary to either of the modified probes in place of the target sequence. Once the sequences were hybridised, 15 μL of streptavidin coated magnetic beads were added to the PCR tubes and left at room temperature for 30 min. Three washing steps were carried out using phosphate buffered solution (70 μL, 0.3 M). The beads were then resuspended in exonuclease buffer (30 μL, 1×) and lambda exonuclease was added (2 μL, 1 U) for digestion to occur for 90 min at 37 °C. SERS analysis was performed post-digestion.SERS analysis
For the limit of detection studies and multiplex assay experiments, SERS was carried out using an Avalon Instrument Ramanstation R3 (Belfast, UK), with an excitation wavelength of 532 nm from a diode laser. Disposable 1.5 mL PMMA semi-micro cuvettes were used. To the cuvette, 30 μL of λ-exonuclease digestion products was added to spermine hydrochloride (20 μL, 0.1 M), 250 μL distilled water and 150 μL silver citrate colloid. The sample was then mixed thoroughly and immediately analysed by SERS. Spectra were baseline corrected using a multipoint polynomial fit and level and zero mode in Grams software (AI 7.00). When carrying out limit of detection studies, 5 scans of 5 replicate samples at each concentration were analysed. The equation of the line obtained from the dilution studies was used to calculate the limits of detection. The limit of detection was calculated to be 3 times the standard deviation of the blank, divided by the gradient of the straight line. For the chemometric studies, SERS analysis was performed using an Avalon Plate Reader, with an excitation wavelength of 532 nm. A 96 well plate was placed onto a stage and the instrument's software was used to move the stage automatically so that a spectrum can be recorded from each well. The accumulation time was 0.01 s and each well was scanned 5 times. A single well can hold 300 μL. Each well contained 30 μL of dye-labelled oligonucleotide mixture (or post-assay mixture when testing the model), 20 μL spermine tetrahydrochloride, 100 μL distilled water and 150 μL silver citrate colloid. To train the PLS prediction model a suitable and reduced number of base line experiments (66 in total) were designed to be performed in the lab. The design of such experiments was based on the theory of Design of Experiments as previously described by Mabbott et al.40 Five scans of five replicates of each of the 66 different dye ratios were analysed and the spectra were averaged prior to model construction. Following this additional samples were generated to test the model and included: (i) additional dye-labelled oligonucleotide combinations and (ii) samples after having gone through the complete SERS assay; the latter were included as these are the closest to the clinical scenario.Chemometrics
Data were baseline corrected using the asymmetrical least squared smoothing method.41 Principal component analysis (PCA) was performed to assess the reproducibility of the data set using Matlab software version R2012a (The MathWorks, Natick, MA, USA). PCA reduces the dimensionality of the SERS data, making it easier to identify any variations in the spectra.42,43 PCA was carried out on four different data sets, three consisting of spectra obtained from single pathogen detection experiments and one data set obtained for the multiplex detection of the three pathogens. For quantification of the three pathogens in tertiary mixtures the supervised learning method of partial least squares (PLS) regression was employed. PLS is a multivariate calibration method that relates a set of independent variables X (Raman intensities) to a set of dependent variables Y (dye-labelled oligonucleotide concentrations). PLS projects the X and Y variables into sets of orthogonal latent variables, scores of X and scores of Y, so that the covariance between these two sets of latent variables is maximized.44 The purpose of PLS is to build a linear model Y = XB + E, where B is a matrix of regression coefficients and E represents the difference (error) between observed and predicted Y values.45 After calibration of the PLS models, validation data (new Raman spectra) not used in model building were used to challenge PLS. These included additional dye-labelled oligonucleotide combinations as well as samples that had gone through the whole SERS assay. If these predictions were correct then this would show that our approach could successfully and simultaneously quantify the three pathogens of bacterial meningitis. The PLS algorithm was run in Matlab version R2012a and the scripts are available from the authors on request.Results and discussion
The bacterial pathogens Neisseria meningitidis, Streptococcus pneumoniae and Haemophilus influenzae are the most common causes of acute bacterial meningitis, which is an extremely serious and in most cases fatal infection that affects the central nervous system.46,47 Traditional laboratory methods involve lengthy culture for the identification of bacterial meningitis pathogens, which can take up to 36 hours.48,49 It has also been reported that the administration of antibiotics prior to sample collection has a detrimental effect on the positive identification of the bacterial meningitis pathogens. Moreover, several studies have revealed that there is a marked discrepancy between the number of suspected cases of bacterial meningitis and confirmed cases of bacterial meningitis using culture methods.38,50,51 A solution to this problem was to use a non-culture method such as PCR, and this is now being used extensively and involves the amplification of pathogen DNA sequences and provides a universal detection technique for bacteria.52 However, this approach has been hindered due to the presence of residual bacterial DNA that affects any positive results for identification of the pathogens. The most effective course of patient treatment is determined by the results obtained from pathogen identification; therefore an ideal method of identification must be efficient in producing confirmative and more importantly reliable objective results. The novel detection assay described herein provides an ideal alternative to traditional laboratory methods, eliminating the use of DNA amplification and results obtained are not obscured by the administration of antibiotics prior to sample collection. Initial studies involved detecting one pathogen at a time using a SERS-based assay and determining the sensitivity of each pathogen target. Following these experiments, a triplex was designed to detect the three pathogens, N. meningitidis, S. pneumoniae and H. influenzae simultaneously, all of which are causes of bacterial meningitis. Chemometric analysis was then performed to quantify each pathogen in the multiplex after the detection assay was performed.Fig. 1 illustrates the multi-step process used for the detection of the bacterial meningitis pathogens. First, the synthetic pathogen sequence was hybridised to its two respective complementary modified synthetic DNA sequences. Streptavidin coated magnetic beads were then added, which retained only fully formed duplex DNA. With subsequent washing steps, any unhybridised or excess DNA was removed from the assay. λ-Exonuclease was then added and digestion of the 5′-phosphate modified probe that also possessed a fluorescent dye took place, which therefore liberated the dye from the duplex that was attached to streptavidin coated magnetic bead. The products of digestion were then added to a mixture of silver nanoparticles (capped with the citrate during reduction) and spermine hydrochloride; this was then followed by SERS analysis, which generated the characteristic SERS spectra of the fluorescent dye present on the “Reporter” probe. Citrate reduced silver nanoparticles have an overall negative charge on the metal surface, which repels the negatively charged phosphate backbone of DNA.53,54 The aggregating agent spermine hydrochloride is positively charged and is used to reduce the negative charge on the nanoparticle surface, allowing the DNA to come into close proximity with the metal surface.55 For SERS enhancement to be obtained the DNA must be either adsorbed or very close to the metal surface and using spermine hydrochloride facilitates the DNA to come in close proximity to the silver surface.56 As well as reducing the negative surface charge, spermine hydrochloride encourages the formation of nanoparticle clusters, which create “hotspots” such that when a laser is applied an enhancement in the unique vibrational spectrum is obtained.3 Along with the detection of single bacterial meningitis pathogens, two control experiments were undertaken each time the assay was performed, no target and non-complementary. The non-complementary control was different for each pathogen detection assay. When detecting S. pneumoniae pathogen, the synthetic sequence for N. meningitidis was present and treated as the non-complementary control. The H. influenzae sequence was the control when detecting N. meningitidis and for the detection of H. influenzae, the synthetic sequence coding for S. pneumoniae was the non-complementary control. No SERS peaks from the fluorescent dye labels should have been observed in either control as the synthetic pathogen target sequence is not present in the assay and therefore the dye labelled probe will be removed during the washing steps that were carried out prior to enzyme digestion.
Exact complement synthetic DNA sequences were used for the single pathogen detection experiments. Each synthetic pathogen target sequence had two complementary synthetic probe sequences, each complementary to half of the target sequence. One sequence was modified with a biotin group at the 3′ end of the strand and this was known as the “Capture” probe. The other sequence, referred to as the “Reporter” probe, was modified at the 5′ end with a phosphate group that is needed for enzyme digestion to occur; at the 3′ end there was a spacer-18, which is an 18-atom hexa-ethyleneglycol spacer that was used to stop digestion resulting in a sequence of 10 adenine bases and the fluorescent dye being left on the 3′ end that would then be free for SERS detection. The 10 adenine bases remained attached to the fluorescent dye post-digestion and previous studies have shown that this adenine “tail” enhances the SERS signal compared to free dye that does not have a DNA “tail”.11,57 For each of the three pathogens a different fluorescent dye was used to label each of the three target specific probes; when detecting S. pneumoniae the fluorescent dye TAMRA was present on the “Reporter” probe, for the detection of N. meningitidis FAM was the fluorescent dye monitored and finally for the detection of H. influenzae Cy3 was the chosen dye.
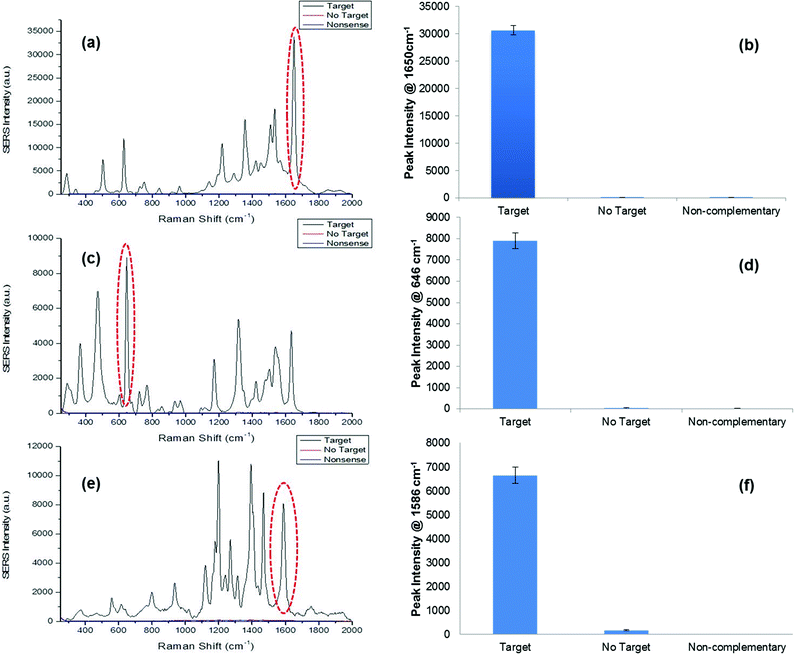
The synthetic DNA sequence representative of S. pneumoniae was successfully detected using the SERS assay. This was carried out by monitoring the characteristic TAMRA peaks in the SERS spectrum (Fig. 2a), as this was the dye modification on this “Reporter” probe. A prominent peak was observed at 1650 cm−1 that was present due to the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C bonds in the dye structure and the peaks observed around 1500–1560 cm−1 represented the aromatic ring vibrations.58 All peaks observed in the spectrum were attributed to the structure of TAMRA. As predicted the no target and non-complementary controls showed no SERS signal since the pathogen sequence had been omitted from the assay, therefore a fully formed duplex was not obtained, i.e. the “Reporter” probe was not present to facilitate SERS detection (Fig. 2b). N. meningitidis was detected successfully, using the respective synthetic DNA sequence, by monitoring the characteristic FAM peaks in the SERS spectrum as this was the dye present on the “Reporter” (Fig. 2c). The peak at 1625 cm−1 was a result of the C
C bonds in the dye structure and the peaks observed around 1500–1560 cm−1 represented the aromatic ring vibrations.58 All peaks observed in the spectrum were attributed to the structure of TAMRA. As predicted the no target and non-complementary controls showed no SERS signal since the pathogen sequence had been omitted from the assay, therefore a fully formed duplex was not obtained, i.e. the “Reporter” probe was not present to facilitate SERS detection (Fig. 2b). N. meningitidis was detected successfully, using the respective synthetic DNA sequence, by monitoring the characteristic FAM peaks in the SERS spectrum as this was the dye present on the “Reporter” (Fig. 2c). The peak at 1625 cm−1 was a result of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C vibrations, and similar to the SERS spectrum of TAMRA, peaks within the range 1400–1500 cm−1 represented CH2 and CH3 vibrations.58 Peaks between 1500 and 1600 cm−1 were from the aromatic rings within the structure of FAM. Again no target and non-complementary controls did not produce any SERS peaks as expected since the pathogen sequence was not present in the assay and further analysis of peak intensities at 648 cm−1 demonstrated that both controls showed no SERS intensity at this specific characteristic FAM peak (Fig. 2d). Finally, the synthetic DNA sequence of H. influenzae was also detected successfully using the novel SERS assay by monitoring the characteristic SERS peaks of the dye Cy3, which was chemically attached to this probe (Fig. 2e). Aromatic vibrations were observed between 1500 cm−1 and 1600 cm−1. The intense peak observed at 1200 cm−1 was due to the aliphatic chain in the dye structure.58 The non-complementary control gave no SERS intensity as expected, as the synthetic pathogen sequence was excluded from the assay. This was reaffirmed upon further analysis of the peak at 1586 cm−1. However, upon peak analysis for the no target control there was a small amount of SERS intensity observed. This can be attributed by the presence of a larger background in the SERS spectrum for this control compared to the non-complementary control and as it was the absolute intensity values used to calculate the peak heights, negligible SERS intensity could be observed (Fig. 2f). To summarise, when synthetic pathogen DNA was omitted or a non-complementary sequence was present in the assay, no SERS signal was obtained, therefore demonstrating the excellent robustness and strength of this assay for diagnostic purposes in clinical use.
C vibrations, and similar to the SERS spectrum of TAMRA, peaks within the range 1400–1500 cm−1 represented CH2 and CH3 vibrations.58 Peaks between 1500 and 1600 cm−1 were from the aromatic rings within the structure of FAM. Again no target and non-complementary controls did not produce any SERS peaks as expected since the pathogen sequence was not present in the assay and further analysis of peak intensities at 648 cm−1 demonstrated that both controls showed no SERS intensity at this specific characteristic FAM peak (Fig. 2d). Finally, the synthetic DNA sequence of H. influenzae was also detected successfully using the novel SERS assay by monitoring the characteristic SERS peaks of the dye Cy3, which was chemically attached to this probe (Fig. 2e). Aromatic vibrations were observed between 1500 cm−1 and 1600 cm−1. The intense peak observed at 1200 cm−1 was due to the aliphatic chain in the dye structure.58 The non-complementary control gave no SERS intensity as expected, as the synthetic pathogen sequence was excluded from the assay. This was reaffirmed upon further analysis of the peak at 1586 cm−1. However, upon peak analysis for the no target control there was a small amount of SERS intensity observed. This can be attributed by the presence of a larger background in the SERS spectrum for this control compared to the non-complementary control and as it was the absolute intensity values used to calculate the peak heights, negligible SERS intensity could be observed (Fig. 2f). To summarise, when synthetic pathogen DNA was omitted or a non-complementary sequence was present in the assay, no SERS signal was obtained, therefore demonstrating the excellent robustness and strength of this assay for diagnostic purposes in clinical use.
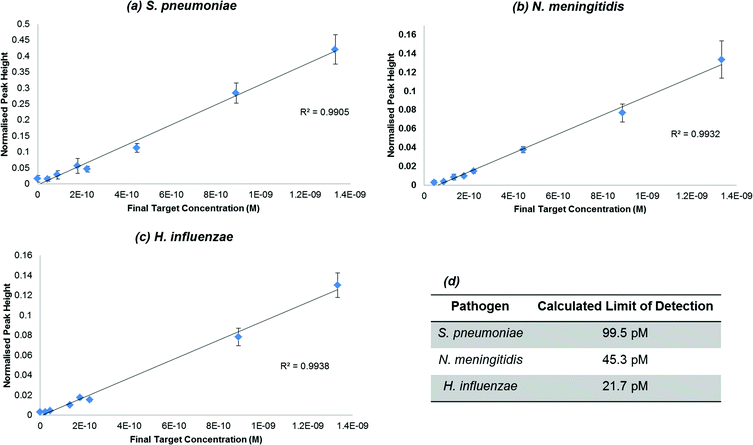
Once it was ascertained that a significant enhancement was observed in the presence of each of the chosen pathogens, a dilution series was carried out to determine the lowest concentration of synthetic pathogen DNA that could be detected using the assay. An initial concentration range of 6 × 10−8 M to 1 × 10−9 M of synthetic pathogen DNA was used throughout the SERS assay and for each study the limits of detection were determined using the equation of the line obtained from the dilution studies and calculating the limit of detection to be 3 times the standard deviation of the blank, divided by the gradient of the straight line (Fig. 3). The data were normalised using the intensity of the standard. This SERS assay has previously been used successfully for the detection of Chlamydia trachomatis with a reported detection limit of 77 pM.57 The calculated limits of detection for the synthetic DNA of each of the three pathogens, N. meningitidis, S. pneumoniae and H. influenzae, were 45.3 pM, 99.5 pM and 21.7 pM respectively. The detection limits obtained from this work are within the range of the limit obtained from previous work, which illustrates the robustness of this assay in that it can be applied to various classes of diseases and yield similar and successful results. Furthermore, with low detection limits in the pico-molar range it is a highly desirable detection method in clinical diagnostics where concentrations of disease DNA under analysis are very low and currently very difficult to measure using fluorescence. Indeed we have reported several times significantly lower detection limits of SERS compared to fluorescence of the same dye.3
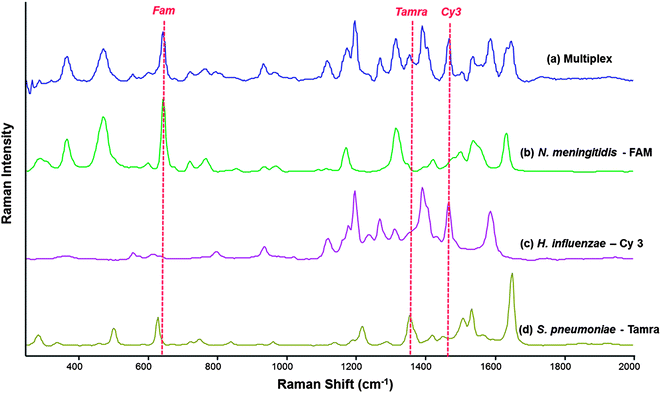
The SERS spectra of FAM and TAMRA are similar which is due to similarities in their chemical structures. By contrast, Cy3 has a markedly different SERS spectrum due to the difference in chemical structure compared to the other two dyes. FAM and TAMRA are xanthene based dyes compared to Cy3, which is an indole based cyanine. Despite the similarity in the SERS spectra of FAM and TAMRA, they can be used successfully in a multiplex system along with Cy3. Multiplex studies were carried out using both synthetic target pathogen DNA and PCR product, generated from the amplification of template sequences of each pathogen. Exact complement of synthetic DNA of each pathogen was used as initial proof-of-concept that the multiplexed SERS detection assay will work for these specific bacterial meningitis pathogens, this proved to be successful. Therefore, PCR product was then used to establish that the multiplexed assay could be used to analyse clinically relevant CSF samples. For this novel assay to be biologically relevant, it must perform well using PCR product of the pathogens. The assay was used in a multiplex scenario where PCR products of each pathogen were to be detected simultaneously. In the reaction mixture there were nine DNA sequences in total; three pathogen PCR product sequences, three complementary “Capture” probes and three complementary “Reporter” probes. Each step of the SERS detection assay was carried out in an identical fashion for single pathogen detection. As can be seen in Fig. 4, there is at least one identifiable peak of each dye associated with each of the three pathogens readily observed.
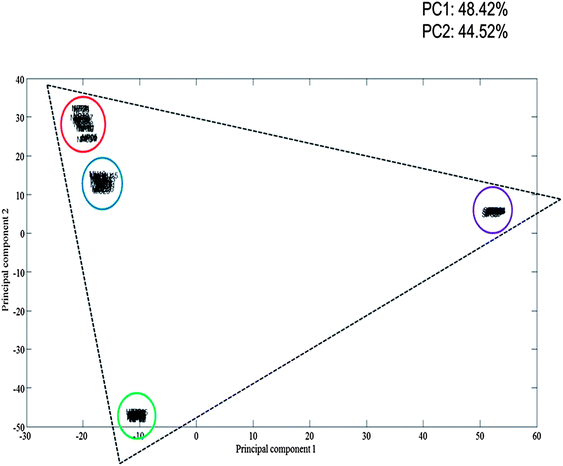
The multiplex spectra obtained are multivariate in nature, therefore it is difficult to analyse the spectra by eye alone. To overcome this, multivariate analysis in the form of principal component analysis (PCA) was performed on the multiplex SERS spectrum, as well as the three separate dye-labelled oligonucleotide SERS spectra, associated with the pathogens. PCA is an excellent approach to reduce the dimensionality of the SERS data.42 The resulting principal component (PC) scores plot (Fig. 5) clearly illustrates how strong the relationship was between the multiplex spectra and the pathogen spectra. As can be seen in Fig. 5, the dye-labelled oligonucleotide SERS spectra form the edges of a triangle. Five scans of the 5 replicate samples are shown in the PCA plot and each group is tightly clustered illustrating the excellent reproducibility of the SERS spectra. The SERS spectra from the multiplex sample are also clustered tightly together and are clearly separated and found towards the middle of all three dye-labelled oligonucleotide clusters. It is noteworthy that the multiplex cluster which contains a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixture of all three probes is positioned close to the N. meningitidis cluster, which suggests that it has stronger FAM signal compared to the other two dye-labelled oligonucleotides. This means that more powerful supervised learning method will be needed to effect accurate quantification of the three probes and hence the three bacteria in tertiary mixtures.
1 mixture of all three probes is positioned close to the N. meningitidis cluster, which suggests that it has stronger FAM signal compared to the other two dye-labelled oligonucleotides. This means that more powerful supervised learning method will be needed to effect accurate quantification of the three probes and hence the three bacteria in tertiary mixtures.
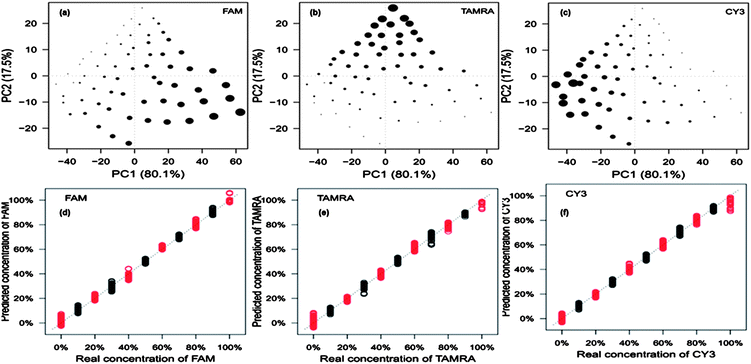
The crucial aspect of this work was that, based on the knowledge that all three pathogens are present in the multiplex, we wanted to determine the amount of each pathogen present within tertiary mixtures. This was performed by quantifying the three dye-labelled oligonucleotides present which will be expected to be proportional to the amount of target pathogen present in the assay. In order to quantify the dye-labelled oligonucleotides in the multiplex, fractional factorial design was used to determine the lowest number of experiments that test an equal combination of low, median and high levels for all 3 factors (dye-labelled oligonucleotides). This was calculated to be 66 different dye ratios. These were analysed by SERS. For ease of visualisation each set of replicates were averaged to provide 66 SERS spectra prior to PCA. This resulted in the generation of a single PCA scores plot that shows the quantitative relationship of each of the dye-labelled oligonucelotides within the 66 tertiary mixes. In Fig. 6a–c three PCA plots are produced which have identical PC scores locations. The size of the dot is proportional to the concentration of the dye-labelled oligonucleotide present in the mixture. The triangular shapes of the score plots are a very good indication that each dye-labelled oligonucleotide present can be quantified. Using these 66 data points, PLS regression models were constructed (Fig. 6d–f). The data points were spilt into two groups: one used to build the PLS model (black) and one used to test the model (red). The three PLS models all show an expected linear relationship (that is to say they fall on the expected y = x line), which further indicates excellent reproducibility and more importantly that the dyes can be quantified in a multiplex.
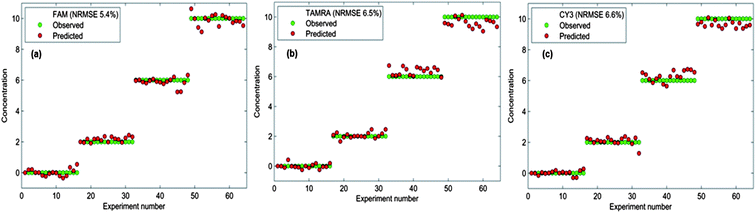
The model was originally built using only dye-labelled oligonucleotide mixtures; that is to say the detection assay was not performed in full. A second model was then generated that was scaled to quantify the dye-labelled oligonucleotides in the amounts that would be present in a sample post-assay. The re-scaled model was tested using “blind” samples, where the actual dye-labelled oligonucleotide concentrations were unknown to the analyst but not to the PLS model. The predicted results were compared to the actual values and a selection of these are shown in Table 2. Overall, the predicted results are in very good agreement with the actual concentrations of dyes added. A second batch of testing was performed where the samples used were all post-assay samples; this was the most important testing experiment as it mimicked a real clinical situation. Excitingly, very good agreement was also obtained for post SERS assay testing as can be seen in Table 3. The successful results obtained from the testing of the model shows that each of the three dye-labelled oligonucleotides can be quantified within the multiplex after the detection assay has been performed and regardless of whether the other two dye-labelled oligonucleotides are present. Error values were estimated for each dye-labelled oligonucleotide (Fig. 7) and the overlap between the actual and predicted values is excellent and indeed reflects a small error associated with the predictions obtained from the PLS model. This emphasises the accuracy and reliability of the chemometric model built to quantify the three target pathogens. Not only can the detection assay be used to detect three bacterial meningitis pathogens simultaneously, but each pathogen can be successfully quantified in the multiplex using chemometrics.
| Real dye % | Predicted dye % | |||||
|---|---|---|---|---|---|---|
| FAM | TAMRA | Cy3 | FAM | TAMRA | Cy3 | |
| 1 | 0 | 0 | 6.67 | 0 | 0 | 5.53 |
| 2 | 0 | 0 | 33.3 | 0 | 0 | 24.7 |
| 3 | 0 | 6.67 | 0 | 0 | 8.13 | 0 |
| 4 | 0 | 6.67 | 33.3 | 0 | 6.27 | 31.1 |
| 5 | 0 | 20 | 20 | 0 | 20 | 17.5 |
| 6 | 0 | 33.3 | 6.67 | 0 | 25.2 | 5.73 |
| 7 | 0 | 33.3 | 33. 3 | 0 | 32 | 30.2 |
| 8 | 6.67 | 0 | 0 | 5.33 | 0.2 | 0.07 |
| 9 | 6.67 | 0 | 33. 3 | 5.8 | 0 | 31.1 |
| 10 | 6.67 | 6.67 | 6.67 | 6.2 | 7.07 | 6.27 |
| 11 | 6.67 | 6.67 | 33.3 | 6.27 | 5.8 | 29.7 |
| 12 | 6.67 | 20 | 20 | 4.07 | 14.9 | 16.4 |
| 13 | 6.67 | 33. 3 | 0 | 6.27 | 25.4 | 0 |
| 14 | 6.67 | 33.3 | 20 | 4.93 | 28 | 18.8 |
| 15 | 6.67 | 33.3 | 33.3 | 6.13 | 29.9 | 26.7 |
| Real dye % | Predicted dye % | |||||
|---|---|---|---|---|---|---|
| FAM | TAMRA | Cy3 | FAM | TAMRA | Cy3 | |
| 1 | 6.67 | 6.67 | 6.67 | 7.67 | 7 | 6.2 |
| 2 | 6.67 | 6.67 | 0 | 6.4 | 6.87 | 0.07 |
| 3 | 6.67 | 0 | 6.67 | 5.87 | 0 | 5.93 |
| 4 | 0 | 6.67 | 6.67 | 0 | 7.27 | 6.4 |
| 5 | 6.67 | 0 | 0 | 6.47 | 0 | 0 |
| 6 | 0 | 6.67 | 0 | 0.8 | 7.53 | 2.07 |
| 7 | 0 | 0 | 6.67 | 3.27 | 0 | 8.07 |
| 8 | 0 | 0 | 0 | 0.07 | 0.07 | 0 |
Conclusions
A SERS assay has been successfully applied to the detection of three bacterial meningitis pathogens: Neisseria meningitidis, Streptococcus pneumoniae and Haemophilus influenzae. The assay detected all three pathogens simultaneously producing consistent and accurate results. More importantly, using chemometrics, each target pathogen could be quantified, post-assay, in the multiplex, which is extremely desirable in clinical diagnostics. This is the first time that a multiplex SERS assay has been carried out where each component (dye-labelled oligonucleotide) was successfully identified and quantified within the multiplex. This method of bacterial meningitis detection produces consistent results faster than conventional culture based methods. We believe this is a promising alternative to current PCR methods of detection and could be applied to the detection of a variety of bacterial, fungal and viral diseases.Acknowledgements
KF and KG wish to thank the RSC Analytical Trust Fund and EPSRC for funding through the award of DTG funding to KG. We wish to thank the EPSRC for funding; EP/F005407/1 for KF and JAD and P/K502303/1 for KF, DG, and SM. DG thanks the Royal Society for support from a Wolfson Research Merit award. EC is grateful to EU FP7 for the funding of COSMOS (grant agreement no. 312941), and RG is also indebted to BBSRC for financial funding.References
- M. J. Espy, J. R. Uhl, L. M. Sloan, S. P. Buckwalter, M. F. Jones, E. A. Vetter, J. D. C. Yao, N. L. Wengenack, J. E. Rosenblatt, F. R. Cockerill III and T. F. Smith, Clin. Microbiol. Rev., 2006, 19, 165–256 CrossRef CAS PubMed.
- R. T. Ranasinghe and T. Brown, Chem. Commun., 2005, 5487–5502 RSC.
- K. Faulds, R. P. Barbagallo, J. T. Keer, W. E. Smith and D. Graham, Analyst, 2004, 129, 567–568 RSC.
- K. Faulds, F. McKenzie, W. E. Smith and D. Graham, Angew. Chem., Int. Ed., 2007, 46, 1829–1831 CrossRef CAS PubMed.
- K. Faulds, R. Jarvis, W. E. Smith, D. Graham and R. Goodacre, Analyst, 2008, 133, 1505–1512 RSC.
- D. Graham, B. J. Mallinder, D. Whitcombe, N. D. Watson and W. E. Smith, Anal. Chem., 2002, 74, 1069–1074 CrossRef CAS.
- R. J. YunWei Charles Cao and C. A. Mirkin, Science, 2002, 297, 1536–1540 CrossRef PubMed.
- L. R. Allain and T. Vo-Dinh, Anal. Chim. Acta, 2002, 469, 149–154 CrossRef CAS.
- E. Papadopoulou and S. E. J. Bell, Angew. Chem., Int. Ed., 2011, 50, 9058–9061 CrossRef CAS PubMed.
- D. van Lierop, K. Faulds and D. Graham, Anal. Chem., 2011, 83, 5817–5821 CrossRef CAS PubMed.
- M. M. Harper, J. A. Dougan, N. C. Shand, D. Graham and K. Faulds, Analyst, 2012, 137, 2063–2068 RSC.
- L. Kupila, T. Vuorinen, R. Vainionpaa, V. Hukkanen, R. J. Marttila and P. Kotilainen, Neurology, 2006, 66, 75–80 CrossRef CAS PubMed.
- R. K. Gupta, J. Best and E. MacMahon, Br. Med. J., 2005, 330, 1132–1135 CrossRef PubMed.
- S. A. E. Logan and E. MacMahon, Br. Med. J., 2008, 336, 36–40 CrossRef PubMed.
- Meningitis Research Foundation, http://www.meningitis.org/, 2013.
- J. McVernon, C. L. Trotter, M. P. E. Slack and M. E. Ramsay, Br. Med. J., 2004, 329, 655–658 CrossRef PubMed.
- R. J. Stokes, A. Macaskill, P. J. Lundahl, W. E. Smith, K. Faulds and D. Graham, Small, 2007, 3, 1593–1601 CrossRef CAS PubMed.
- A. MacAskill, D. Crawford, D. Graham and K. Faulds, Anal. Chem., 2009, 81, 8134–8140 CrossRef CAS PubMed.
- K. K. Hering, R. Möller, W. Fritzsche and J. Popp, ChemPhysChem, 2008, 9, 867–872 CrossRef CAS PubMed.
- H. N. Wang and T. Vo-Dinh, Nanotechnology, 2009, 20, 065101 CrossRef PubMed.
- D. I. Ellis, D. P. Cowcher, L. Ashton, S. O'Hagan and R. Goodacre, Analyst, 2013, 138, 3871–3884 RSC.
- D. I. Ellis, V. L. Brewster, W. B. Dunn, J. W. Allwood, A. P. Golovanov and R. Goodacre, Chem. Soc. Rev., 2012, 41, 5706–5727 RSC.
- M. Zhong, M. Girolami, K. Faulds and D. Graham, J. Roy. Stat. Soc. C Appl. Stat., 2011, 60, 187–206 CrossRef.
- K. Subramanian, W. Rutvisuttinunt, W. Scott and R. S. Myers, Nucleic Acids Res., 2003, 31, 1585–1596 CrossRef CAS PubMed.
- K. R. Thomas and B. M. Olivera, J. Biol. Chem., 1978, 253, 424–429 CAS.
- N. G. Nossal and M. F. Singer, J. Biol. Chem., 1968, 243, 913–922 CAS.
- K. S. Sriprakash, N. Lundh, M. Mooonhuh and C. M. Radding, J. Biol. Chem., 1975, 250, 5438–5445 CAS.
- R. Kovall and B. W. Matthews, Science, 1997, 277, 1824–1827 CrossRef CAS.
- J. Zhang, K. A. McCabe and C. E. Bell, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 11872–11877 CrossRef CAS PubMed.
- M. Citartan, T.-H. Tang, S.-C. Tan and S. C. B. Gopinath, World J. Microbiol. Biotechnol., 2011, 27, 1167–1173 CrossRef CAS.
- G. Lee, J. Yoo, B. J. Leslie and T. Ha, Nat. Chem. Biol., 2011, 7, 367–374 CrossRef CAS PubMed.
- S. H. Kang, S. Lee and E. S. Yeung, Analyst, 2010, 135, 1759–1764 RSC.
- S. Lee, S. H. Kang and E. S. Yeung, Talanta, 2011, 85, 2135–2141 CrossRef CAS PubMed.
- R. S. Conroy, A. P. Koretsky and J. Moreland, Eur. Biophys. J. Biophys. Lett., 2010, 39, 337–343 CrossRef CAS PubMed.
- J. J. Li, Y. Chu, B. Yi-Hung Lee and X. S. Xie, Nucleic Acids Res., 2008, 36, e36 CrossRef PubMed.
- A. Barhoumi and N. J. Halas, J. Am. Chem. Soc., 2010, 132, 12792–12793 CrossRef CAS PubMed.
- H. Xu, L. Wang, H. Ye, L. Yu, X. Zhu, Z. Lin, G. Wu, X. Li, X. Liu and G. Chen, Chem. Commun., 2012, 48, 6390–6392 RSC.
- C. E. Corless, M. Guiver, R. Borrow, V. Edward-Jones, A. J. Fox and E. B. Kaczmarski, J. Clin. Microbiol., 2001, 39, 1553–1558 CrossRef CAS PubMed.
- P. C. Lee and D. Meisel, J. Phys. Chem., 1982, 86, 3391–3395 CrossRef CAS.
- S. Mabbott, E. Correa, D. P. Cowcher, J. W. Allwood and R. Goodacre, Anal. Chem., 2012, 85, 923–931 CrossRef PubMed.
- H. F. M. Boelens, P. H. C. Eilers and T. Hankemeier, Anal. Chem., 2005, 77, 7998–8007 CrossRef CAS PubMed.
- D.-H. Kim, R. M. Jarvis, Y. Xu, A. W. Oliver, J. W. Allwood, L. Hampson, I. N. Hampson and R. Goodacre, Analyst, 2010, 135, 1235–1244 RSC.
- S. Mabbott, A. Eckmann, C. Casiraghi and R. Goodacre, Analyst, 2013, 138, 118–122 RSC.
- H. Martens and T. Naes, Multivariate Calibration, Wiley, 1991 Search PubMed.
- V. Esposito Vinzi, W. W. Chin, J. Henseler and H. E. Wang, Handbook of Partial Least Squares, Springer, 2010 Search PubMed.
- P. B. McIntyre, K. L. O'Brien, B. Greenwood and D. van de Beek, Lancet, 2012, 380, 1703–1711 CrossRef.
- G. Tzanakaki, M. Tsopanomichalou, K. Kesanopoulos, R. Matzourani, M. Sioumala, A. Tabaki and J. Kremastinou, Clin. Microbiol. Infect., 2005, 11, 386–390 CrossRef CAS PubMed.
- X. Wang, M. J. Theodore, R. Mair, E. Trujillo-Lopez, M. du Plessis, N. Wolter, A. L. Baughman, C. Hatcher, J. Vuong, L. Lott, A. von Gottberg, C. Sacchi, J. M. McDonald, N. E. Messonnier and L. W. Mayer, J. Clin. Microbiol., 2012, 50, 702–708 CrossRef CAS PubMed.
- H. Sadighian and M. R. Pourmand, Iran. J. Public Health, 2009, 38, 60–68 CAS.
- P. Radstrom, A. Backman, N. Qian, P. Kragsbjerg and C. Pahlson, J. Clin. Microbiol., 1994, 32, 2738–2744 CAS.
- G. M. K. Abdeldaim, K. Stralin, J. Korsgaard, J. Blomberg, C. Welinder-Olsson and B. Herrmann, BMC Microbiol., 2010, 10 DOI:10.1186/1471-2180-10-310.
- R. Ghotaslou, S. Farajnia, F. Yeganeh, S. Abdoli-Oskouei, M. Ahangarzadeh Rezaee and M. Barzegar, Acta Med. Iran., 2012, 50, 192–196 Search PubMed.
- S. M. Heard, F. Grieser, C. G. Barraclough and J. V. Sanders, J. Colloid Interface Sci., 1983, 93, 545–555 CrossRef CAS.
- C. H. Munro, W. E. Smith, M. Garner, J. Clarkson and P. C. White, Langmuir, 1995, 11, 3712–3720 CrossRef CAS.
- D. Graham, W. E. Smith, A. M. T. Linacre, C. H. Munro, N. D. Watson and P. C. White, Anal. Chem., 1997, 69, 4703–4707 CrossRef CAS.
- K. Faulds, W. E. Smith and D. Graham, Anal. Chem., 2004, 76, 412–417 CrossRef CAS PubMed.
- J. A. Dougan, D. MacRae, D. Graham and K. Faulds, Chem. Commun., 2011, 47, 4649–4651 RSC.
- E. Smith and G. Dent, Modern Raman Spectroscopy: A Practical Approach, Wiley, 2005 Search PubMed.
| This journal is © The Royal Society of Chemistry 2014 |