Automated system for extraction and instantaneous analysis of millimeter-sized samples†
Jie-Bi Hua,
Ssu-Ying Chena,
June-Tai Wubcd,
Yu-Chie Chen*ae and
Pawel L. Urban*ae
aDepartment of Applied Chemistry, National Chiao Tung University, Hsinchu, Taiwan. Fax: +886-3-5723764
bInstitute of Molecular Medicine, College of Medicine, National Taiwan University, Taipei, Taiwan
cDepartment of Medical Research, National Taiwan University Hospital, Taipei, Taiwan
dResearch Center for Developmental Biology and Regenerative Medicine, National Taiwan University, Taipei, Taiwan
eInstitute of Molecular Science, National Chiao Tung University, Hsinchu, Taiwan
First published on 5th February 2014
Abstract
Adequate sample treatment is critical when employing mass spectrometry (MS) in the analyses of complex biological matrices. Despite various improvements, it is generally difficult to automate the process of preparing solid biological samples for MS analysis. Here we demonstrate a facile approach for automation of the whole analysis (from an untreated sample to the final result). The proposed platform enables disruption and extraction of relatively small samples (individual fruit flies, fragments of tea leaves, powdered drug sampled with a cotton bud), and almost simultaneous analysis of the obtained extract within less than 10 min, and with very little intervention of the analyst. The operation is straightforward: once a sample is placed in the sample chamber, the analyst only needs to press a button on the touch screen of the user interface. The programmed open-source electronic device triggers addition of a small amount of solvent, and subsequent mechanical disruption/extraction of the specimen under controlled conditions (thermostated chamber). A small volume of the extract is directed to the ion source of the mass spectrometer incorporating a Venturi pump. During the operation of the instrument, fluorescence intensities (excitation wavelength windows: 320–380 and 460–500 nm) as well as MS extracted ion currents are recorded simultaneously. In the case of fruit fly samples, ∼70 signals were recorded in both modes while the analysis of green tea leaves yielded ∼30 signals. The resulting data reveal time-resolved extraction profiles characterizing every sample.
1. Introduction
In the beginning of the 21st century, automation of various daily life operations has gained popularity. In chemistry, automated systems are used to increase the efficacy, robustness, throughput, and reproducibility of routine analysis, but also to reduce the incidence of human errors.1 On the other hand, the new enabling analytical tools – including mass spectrometers – do require considerable involvement of the operators. It is anticipated that heavy automation and computerization of analytical chemistry may – one day – scrap the need for a large number of average-trained analysts, and leave the requirement for a few highly skilled engineers.Owing to its superior selectivity,2 sensitivity,3 and speed,4 mass spectrometry (MS) is regarded as one of the most comprehensive and universal chemical analytic tools available nowadays. In fact, its sensitivity – in some cases – approaches that of fluorescence detection.5 Moreover, unlike fluorescence detection, MS does not normally require labeling of the analyzed molecules, which greatly simplifies analytical workflows. On the other hand, sample preparation6,7 is considered to be a very important part of most protocols in analytical chemistry,8–10 including those employing MS as detection tool. It is usually required to develop and optimize appropriate sample treatment procedures for particular types of analyses. The success or failure of sample treatment can directly affect the subsequent analysis steps, and performance of the entire analytical routine. Various sample preparation techniques (e.g. solid-phase extraction,11 solid-phase microextraction,12 and automated 96-well extraction13) have been coupled with MS by using robotic handling systems. An interesting approach to sample preparation takes advantage of digital microfluidic devices.14 However, it is still challenging to automate preparation of solid samples for analysis by detection tools designed for liquid samples (e.g. electrospray ionization (ESI)-MS or similar approaches). For example, many biological species (e.g. insects, leaves) have cuticle layer, which makes it difficult to extract all embedded molecules. In most cases, the analysis of such samples requires mechanical processing. However, off-line sample treatment considerably lowers analytical throughput, and requires substantial effort. Furthermore, analyte losses during sample preparation and transfer pose a major concern when handling small (millimeter-sized) samples. Therefore, it is appealing to develop and implement new strategies for automated preparation of solid samples prior to the analysis by MS.
In this study, an automated microextraction system has been constructed and integrated with mass spectrometry as well as fluorescence detection (Fig. 1). Fluorescence and MS analysis of sample extracts was conducted in real time, immediately after the extraction took place. This dual detection scheme enables correlation of the fluorescence intensities of the obtained sample extracts with the intensities of mass spectral peaks. The proposed hyphenated system enables seamless chemical profiling of plant tissue samples such as tea leaves as well as small metazoans. Furthermore, by implementing a rapid and inexpensive sampling method (cotton swab sampling),15 one can use this strategy for analysis of small amounts of powders lifted from surfaces.
 | ||
| Fig. 1 Schematic diagram of the automated microextraction system integrated with a fluorescence microscope and a mass spectrometer. (For the wiring scheme, see Fig. S3;† for real image of the experimental setup, see Fig. S1 and S6;† for real image of the microcontroller device, see Fig. S4.†) | ||
2. Experimental section
2.1. Materials
Acetonitrile and water were purchased from Sigma-Aldrich (St Louis, MO, USA), and they were LC-MS grade. Green tea catechin mix standard was purchased from Cerilliant (Round Rock, TX, USA). The polyimide-coated fused silica capillary (inner diameter (ID) 150 μm, outer diameter (OD) 375 μm) was purchased from GL Science (Tokyo, Japan). The Tygon tubing with ID 0.25 and 0.38 mm (wall thickness, 0.91 mm) was purchased from IDEX Health & Science (Wertheim, Germany). Polyether ether ketone (PEEK) F-383X and Teflon FEP F-376X tubing sleeves (OD 1/32 inch) were purchased from IDEX Health & Science (Oak Harbor, WA, USA).2.2. Sample preparation device
The sample microextraction device (Fig. S1†) features a sample chamber made of 5 mm-thick acrylic, which is fixed to a high-density mini-breadboard (cat. no. MS12B/M; Thorlabs, Newton, NJ, USA). In order to ascertain constant conditions, the chamber was thermostated at ∼20 °C with two Peltier elements (13.5 V, 3 A; TEC1-12703). Thermostatting was enabled by the aluminum heat sinks and 12 V fans installed outside and inside the chamber to take away heat from the extraction chamber and distribute cold air inside of it, respectively. The vial cap (∅ = 9 mm, open top polypropylene, polytetrafluoroethene (PTFE)/silicone septum, cat. no. CHSC9-30; Thermo Fisher Scientific, Waltham, MA, USA) was installed near the ceiling of the acrylic box in such a way that the sample vial could readily be placed inside or taken out when required. Screw neck vials with integrated 200 μL volume glass inserts (Macherey-Nagel, Düren, Germany) were used.External to the box, a z-axis actuation system with a stepper motor was set up. This part had been taken out from a 24×-speed digital-video-disc (DVD) drive chassis, and fixed in vertical position on the high-density mini-breadboard using stainless steel metal posts. Laser optics present in the DVD holder was replaced with a custom-made Bakelite/steel holder of a 3 V direct current (DC) motor (cat. no. RF-3020; 38 mA; specified rotation speed ∼2 × 104 rpm). The rotor of the DC motor was fitted with a 3 cm piece of Tygon tubing (ID 0.38 mm) – tipped with a short section of PEEK tubing (∼5 mm) – acting as the spindle and used to crush solid samples during the extraction process. Due to the vertical movement enabled by the z-axis actuation system, the spindle could readily be inserted into the vial via a hole in the ceiling of the acrylic box. Two pieces of capillary tubing were also slid into the vial. They enabled delivery of extraction solvent (acetonitrile–water = 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30, v/v) and transfer of the extract to the mass spectrometer. A ∼2 cm section of PEEK tubing was used to deliver the extractant. It was connected to the peristaltic pump (Ismatec ISM936D, IPC Series, 8 channels; IDEX Health & Science, Oak Harbor, WA, USA) via Tygon tubing (ID 0.25 mm). Note well that when the microcontroller module (described below) primes the pump, the extraction solvent is delivered to the sample vial via this duct. In addition, a polyimide-coated fused silica capillary (30 cm) was slid through a Teflon fluorinated ethylene propylene (FEP) tubing sleeves, and fixed at the moving rod of a solenoid push-type linear actuator (RM-0730B; voltage: 24 V; current: 1 A; stroke: 10 mm; force equivalent: 80 g; SHRM, Shanghai Renmin Electrical Apparatus Works, Shanghai, China). Supplying electric potential to this electromagnet primes insertion of the capillary into the vial, so that its inlet section is dipped into the liquid extract present therein. This operation triggers the transfer of the extract from the extraction chamber to the mass spectrometer.
30, v/v) and transfer of the extract to the mass spectrometer. A ∼2 cm section of PEEK tubing was used to deliver the extractant. It was connected to the peristaltic pump (Ismatec ISM936D, IPC Series, 8 channels; IDEX Health & Science, Oak Harbor, WA, USA) via Tygon tubing (ID 0.25 mm). Note well that when the microcontroller module (described below) primes the pump, the extraction solvent is delivered to the sample vial via this duct. In addition, a polyimide-coated fused silica capillary (30 cm) was slid through a Teflon fluorinated ethylene propylene (FEP) tubing sleeves, and fixed at the moving rod of a solenoid push-type linear actuator (RM-0730B; voltage: 24 V; current: 1 A; stroke: 10 mm; force equivalent: 80 g; SHRM, Shanghai Renmin Electrical Apparatus Works, Shanghai, China). Supplying electric potential to this electromagnet primes insertion of the capillary into the vial, so that its inlet section is dipped into the liquid extract present therein. This operation triggers the transfer of the extract from the extraction chamber to the mass spectrometer.
The sampling capillary mentioned above (upstream section; length: 30 cm) was connected with another capillary (length: 25 cm) containing detection window for microscopy, and a 30 cm-long capillary (downstream section) coupled with the ion source of the mass spectrometer. Therefore, the total length of the sample line (ID 150 μm) was 85 cm. The ion source comprises a Venturi pumping spray16 assembled using a 1/8-inch stainless steel tee (Swagelok, Supelco; Solon, OH, USA). Application of the flow of nitrogen gas (∼2.1 × 105 Pa) coaxial to the fused silica capillary, evokes underpressure in the proximity of the sample capillary outlet (∼−1.4 × 104 Pa, calculated according to the Poiseuille equation based on the measured flow rate). This underpressure induces movement of the sample extract from the extraction chamber. The sample – aspirated by the Venturi pump16 – passes through the detection window (∼1 cm zone devoid of polyimide coating) positioned in the viewing area of the fluorescence microscope (Axio Imager M2; Zeiss, Göttingen, Germany). The flow rate of the sample extract was estimated to be ∼12 μL min−1, and the linear velocity was estimated to be ∼11 mm s−1.
The microscope is equipped with a motorized xyz-stage, motorized turret, and the AxioCam ICc 1 charge-coupled device (CCD) color camera (Zeiss); which enables efficient alignment of the observation window, and fast switching between various microscopy modes. The light source used for fluorescence measurements comprises a metal-halide lamp (X-Cite 120 Q; Lumen Dynamics, Ontario, Canada). An enhanced contrast objective (EC Plan-Neofluar 10× /0.3; Zeiss) was used. The imaging was controlled by AxioVision software (40 V 4.8.2.0, 2006–2010, Zeiss). It was conducted either in the bright field or fluorescence mode using the following excitation/emission filters: 320–380/435–485 nm (cat. no. 49000, Chroma, Bellows Falls, VT, USA); 460–500/510–560 nm (cat. no. 41001, Chroma, Bellows Falls, VT, USA). The exposure time was optimized separately for each mode (bright field: 5 ms; 320–380/435–485 nm: 150 ms; 460–500/510–560 nm: 200 ms). The duty cycle of imaging was 2.355 s per cycle (Fig. S2†).
2.3. Electronic microcontroller
The electronic system supporting the automated sample treatment prior to MS analysis is based on an open-source electronic platform – Arduino (Fig. S3 and S4†). All the main functions were programmed using a custom script (written in a C-derived language) loaded to an Arduino Mega 2560 microcontroller (Torino, Italy) printed circuit board (PCB), equipped with an ATmega chip (256 kB). The script was loaded via the universal serial bus interface from a Windows computer using the Arduino's integrated development environment (ver. 1.0.3). After uploading the script, the device is stand-alone, and can be controlled using a 2.4-inch thin film transistor touch screen (ITEAD Studio, Shenzhen, China) – connected to the Arduino Mega PCB and operated at 3.3 V level. Six output pins of the Arduino Mega PCB were connected to inputs of three relay PCBs (2 relays each, HL-52S V1.0). The relays control the motion of peristaltic pump (turn on/off, and change direction of flow – the function not used in this study), the spindle motor, triggering data acquisition by MS, and inserting capillary into the extraction vial (optional). Four pins were connected to an H-bridge stepper motor controller PCB (L9110S; Shenzhen LC Technology, Shenzhen, China). All the PCB modules were purchased on eBay.com. The connection with the Idex peristaltic pump was established through the analog interface (pins 1–5). The flow rate of the extraction solvent was set by applying a positive potential to the flow rate control pin (no. 5; analog control) of the 15-pin interface, which is typically 5 V. The communication with the mass spectrometer was achieved through its 37-pin auxiliary interface (pins 24, 25, and 28). When disconnected from the computer, the controller (Arduino Mega) was powered from a 9 V power supply (Kaming, Taipei, Taiwan). Another 3.3 V power supply (Kaming) was used to operate the stepper motor and the spindle motor. The same casing also houses a digital temperature controller (LW-801C; Shenzhen Lerway Technology, Shenzhen, China) connected to an external 12 V power supply (12.5 A, cat. no. NES-150-12; Mean Well, New Taipei City, Taiwan), Peltier elements, and the temperature sensor (NTC, 10K/3435).2.4. Mass spectrometry
The amaZon speed ion-trap mass spectrometer from Bruker Daltonics (Bremen, Germany) was used in this work. It enabled recording temporal traces in the alternating polarity mode allowing for registration of positive and negative ions in the same run. The voltage applied to the ion transfer capillary was 4500 or −4500 V in the negative- and positive-ion modes, respectively. The end-plate offset values were set to 500 or −500 V in the negative and positive-ion modes, respectively. The flow rate of dry gas was set to 5 L min−1, and the temperature of the heater was set to 250 °C. The mass range was set at m/z 100 to 1000. It is known that due to the space charge capacity of ion trap, when too many ions are handled at a time, the mass accuracy and resolution decrease. The use of the ion charge control (ICC) function mitigates this issue. In the experiments presented below, ICC was enabled to adjust and optimize the amount of ions injected to the mass analyzer in every MS duty cycle. The target amount of ions was set to 2.0 × 106, and the maximum accumulation time was set to 10 ms.2.5. Samples
Three kinds of samples (a metazoan, a plant tissue, and a drug powder) were chosen to evaluate the performance of the automated microextraction system. Canton-S fruit flies (Drosophila melanogaster; Bloomington Drosophila Stock Center at Indiana University, Bloomington, IN, USA) were cultured on a standard medium (water, agar, glucose, brown sugar, corn starch, yeast extract, p-hydroxy benzoic acid methyl ester, alcohol, propionic acid, 85% phosphoric acid) at room temperature. Carbon dioxide was used to anesthetize the flies before microextraction. After the insects stopped flying, they were transferred one-by-one to glass vials for weighing. Masses of empty vials were determined before the experiment, so that the net masses of the samples (individual fruit flies) could readily be computed. After weighing, the vials containing fruit flies were further transferred to the freezer (∼−20 °C), and stored there for at least ∼1 min.Green tea leaves (Camellia sinensis) from the Sun Moon Lake region were sampled randomly and cut into smaller pieces (size range, 1–3 mm) with scissors. The small pieces were then placed in screw-cap vials right before weighing. A mixture of chemical standards (∼10−5 M in acetonitrile–water = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20, v/v) was prepared to enable confirmation of the analyte identities by tandem MS.
20, v/v) was prepared to enable confirmation of the analyte identities by tandem MS.
Acetaminophen tablets were ground to powder, and transferred to glass vials (Kimble Chase, Rockwood, USA) for storage. Before the experiment, a small amount of powder (∼10 mg) was cast on a flat surface (Petri dish). Cotton swabs (Qian Qiao, Taoyuan, Taiwan) were used to sweep small amounts of this powder for qualitative analysis. The tip of the cotton swab (length, ∼6 mm) was then cut, and transferred to a screw neck vial with integrated 200 μL volume glass insert for microextraction.
2.6. Data treatment
Following the completion of analyses, fluorescence and mass spectrometry data were prepared for further treatment. We used DataAnalysis software (ver. 4.1, Bruker Daltonics) for the initial treatment of the collected mass spectra, plotting total ion currents (TICs) and extracted ion currents (EICs). We further exported these data to files formatted according to the American Standard Code for Information Interchange (ASCII). Every file contained two columns – one for time, and the other one for MS signal intensity. These ASCII files were imported to Excel (ver. 15, 2013; Microsoft, Redmond, WA, USA). In order to treat the micrographs, and to determine the levels of fluorescence at different excitation wavelengths, we used ImageJ software (ver. 1.47v, 2013; Wayne Rasband, National Institutes of Health, Bethesda, Maryland, MD, USA). We picked up fluorescence images corresponding to each excitation wavelength. In order to facilitate data processing we only measured mean brightness of a region (1388 × 100) corresponding to the center of the capillary duct, and then subtracted the mean brightness of a group of pixels (1388 × 100) outside the capillary duct (reference area) (cf. Fig. S5A;† 63 data points). In the case of the analysis of commercial tea leaves, average values were reported as the final result. Subsequent analysis (including correlations of sample mass, fluorescence, and MS peak intensities) was conducted using the OriginPro 8 software (ver. 8, 1991–2007; Northampton, MA, USA). After completing data treatment, all graphs were imported to CorelDraw X6 software (ver. 16, 2012; Corel Corporation, Mountain View, CA, USA), labeled, and prepared for the final display.3. Results and discussion
In order to facilitate chemical profiling of millimeter-sized samples, we have developed an automated microscale extraction device which enables unsupervised sample preparation, and on-line fluorimetric and mass spectrometric detection (see the ESI† sequence for reference). Coupling the automated microextraction system with fluorescence microscope (λex = 320–380 and 460–500 nm) enabled monitoring the extraction process and sample transfer from the extraction chamber to the mass spectrometer in real time (Fig. S6†). The extract was continuously aspirated from the sample chamber, and instantaneously transferred to the ion source of the ion-trap mass spectrometer due to the action of underpressure exerted by the Venturi pump. In order to characterize the hydrodynamic flow of the extract solution in the sampling capillary, Reynolds number (Re) – representing the ratio of inertial and viscous forces acting on fluid moving along channel17 – was estimated (see the ESI† for details). The low Re value (∼3) obtained for the sample transfer capillary points to the presence of laminar flow, and negligible influence of turbulence on the dispersion of the sample on its route from the sample chamber to the ion source. Although segmented flow sampling preserves the temporal characteristics better than continuous flow by reducing analyte spreading due to diffusion and advection,18–20 taking into account the complexity of the segmented flow setups, and the requirement for using immiscible carrier fluids (e.g. oil, fluorocarbon), we opted against flow segmentation in this study. This simplified operation of the system even though some influence of dispersion on the temporal resolution of extraction profiles cannot be avoided. To evaluate capabilities of the proposed automated system, we have tested it using three different kinds of samples: individual fruit flies, fragments of green tea leaves, and a powdered drug.3.1. Analysis of single fruit flies
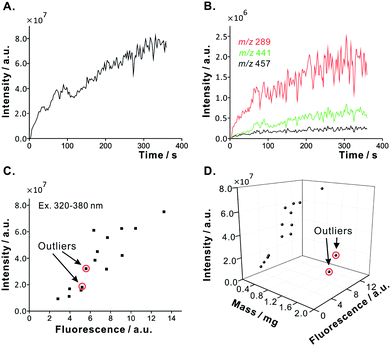
Fruit fly is one of the most widely studied and genetically characterized eukaryotic organisms;21 it is easy to rear, it has short life cycle, and simple anatomy.22 For these reasons, it has been chosen as a model sample to demonstrate the capabilities of the proposed automated analytical system. During the implemented routine, the ion-trap MS collects 6 min data sequences. In the experiment using fruit fly as a model sample, the data acquisition started 1 min before the motor spindle started to rotate. During this time, the target analytes, which were adsorbed on the surface of specimen, could diffuse to the extraction solvent. Switching the spindle motor on caused mechanical disruption of the sample, and liberation of biofluids. The analytes dissolved in the extraction solvent were transferred to the ion source. During the extraction process, ∼70 ion signals, corresponding to various metabolites, could be recorded in the positive (Fig. S7A†) and the negative (Fig. S7B†) ion modes (Table S1 and S2†). Relatively few peaks could be observed in the positive-ion mode; this may be due to the ion suppression caused by the presence of high-abundance ionic species (e.g. salts). We later focused on three selected mass spectral signals recorded in the negative-ion mode: adenosine monophosphate (AMP) (m/z 346), a putative gender marker (m/z 376), and a glycerophosphoinositol (m/z 860) (Fig. 2 and 3).The sample-related signals started to appear at ∼45 s after dipping the sampling capillary into the extraction chamber (Fig. 2A). As the extraction process went on, the spindle of the miniature motor exerted shear force on the biological samples, and caused their disintegration. The TIC, EICs, and the fluorescence signals consistently increased over time (Fig. 2). TIC and most EICs reached their plateaus after ∼260 s (black dashed line, Fig. 2B), while the fluorescence signal reached the plateau already at ∼220 s (black dashed line, Fig. 2C). Interestingly, there is no exact overlap of the profiles of some signals at different m/z values (Fig. S8†). This result suggests that the proposed system can be used to study temporal changes in the composition of extract during the extraction process. Further on, this indicates that various classes of metabolites are liberated at different times as different anatomical features of fruit fly sample are disrupted and extracted.
The flow velocity of the sample extract was 11 mm s−1 (based on an off-line measurement). The migration time of extract along the flow line (∼85 cm) was estimated to be ∼77 s. The time delay between the fluorescence detection window and the MS detector (distance, ∼42 cm) was estimated to be ∼40 s. Considering the migration time along the sampling capillary (∼77 s), it took at least ∼43 s to disintegrate and extract the fruit fly body.
Taking into account the delay time, one may regard the extraction process as complete after 180 s. During this time, the sampling capillary withdrew the extract, and the volume of the extract present in the sample vial decreased. Thus, the increase of the signal intensity in the course of real-time monitoring may be due to the progress of extraction but also due to the decreasing amount of solvent in the extraction chamber. However, according to the result shown in Fig. 2B, the intensities of the EICs at the m/z 346 and 376 increased, and the intensities of the EICs at the m/z 860 slightly decreased. The decrease of some signals may be due to the ion suppression related to the increasing concentration of suppressing species in the concentrated remainder of the extract.
It should be pointed out that many natural compounds, such as amino acids (e.g. tryptophan, phenylalanine, and tyrosine), peptides, and proteins, exhibit autofluorescence.22–25 Therefore, one could easily observe the increase of brightness of the fluorescence images recorded in the course of microextraction (Fig. 3A and B). It should also be noted that small particles originating from disrupted samples were occasionally aspirated by the sampling capillary (Fig. S9†). Even though this small debris did not seem to affect the MS signal, in some cases they caused the blockage of the sampling capillary. This issue may need to be addressed in future work.
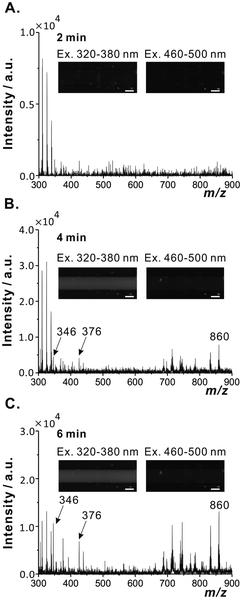 | ||
| Fig. 3 Mass spectra obtained in negative-ion mode and fluorescence images (λex = 320–380 and 460–500 nm) obtained during extraction of a male fruit fly specimen (mass: 0.85 mg); (A) 2 min; (B) 4 min; (C) 6 min. These data are the same as those presented in Fig. 2. Scale bars: 100 μm. | ||
3.2. Matching metabolic profiles with gender
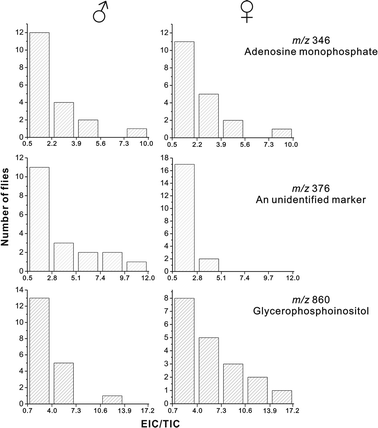
Further on, we attempted matching the metabolic profiles recorded during the time-resolved extraction experiments with gender. In fact, in various studies (e.g.26–28), the lipid composition of fruit fly was found to be different in male and female flies. In the current study, we could not find any qualitative differences in lipid composition between extracts obtained from male and female individuals. However, we observed the occurrence of a putative gender-related marker: it is represented by the peak at the m/z 376. This signal is more intense (relative to other signals) in the spectrum recorded for a male fly (Fig. 4B) than in the spectrum recorded for a female fly (Fig. 4D). Since the collected data points did not follow normal distribution, we chose a non-parametric statistical method – two-sample Kolmogorov–Smirnov test29 – to verify the null hypothesis that both populations are characterized with the same distribution. According to the result of this test, this null hypothesis had to be rejected in the case of m/z 376 (p = 0.0060), but it could not be rejected in the case of m/z 346 (p = 0.7415) and m/z 860 (p = 0.1161). Moreover, from the histograms, plotted separately for 19 flies male and 19 female flies, it is apparent that a large number of female fly samples produced spectra with a lower intensity of the signal at the m/z 376 (Fig. 5). On the other hand, the signal at the m/z 860 is higher in the spectra recorded for a large number of female flies as compared with male flies. The higher intensities of signals related to lipids in female fly samples may be related to a larger body size of female flies (0.92 ± 0.28 mg) as compared with male flies (0.78 ± 0.12 mg),30 a greater size of female reproductive organs, and the presence of unfertilized eggs in the abdomens of female flies.27,28 | ||
| Fig. 4 Temporal monitoring of the extraction of a male (mass: 0.85 mg) and a female (mass: 1.13 mg) fruit fly specimens (a comparison). Male fruit fly: (A) EICs for m/z 346, 376, and 860; (B) mass spectrum obtained in the negative-ion mode (end of extraction). The upper graphs ((A) and (B)) represent the same data as those displayed in Fig. 2B and 3C. Female fruit fly: (C) EICs for m/z 346, 376, and 860; (D) mass spectrum obtained in the negative-ion mode (end of extraction). | ||
Based on this proof-of-concept experiment, one may conclude that the automated microextraction system can enable analysis of microscale samples without laborious sample preparation. Although no absolute quantification was carried out, the automated analysis (incorporating microscale mechanical sample disruption and extraction followed by MS) revealed relative differences between various samples of fruit fly. This result suggests that the proposed method has some potential for practical application in biochemistry research.
3.3. Analysis of individual fragments of commercial tea leaves
Tea is one of the most widely consumed beverages in the world, and tea drinking is a remarkable part of cultural heritage in the Asian countries such as China, Japan, and Taiwan. Since the leaves of green tea are not subjected to exhaustive fermentation, chemical composition of green tea is very similar to that of fresh tea leaves.31 Due to the high price of some oriental teas, commercial products might occasionally be adulterated by mixing high-quality leaves with low-quality leaves. Therefore, it is appealing to develop analytical methods which would enable semi-quantitative profiling of individual tea leaves or their fragments. Along these lines, to further demonstrate capabilities of the proposed sample preparation system in the analysis of small solid samples, commercial tea leaves were cut into small pieces and analyzed separately.Based on the previous experiment, we were aware that the sample extract reaches the ion source region in 45 s after the inlet of the sampling capillary is dipped in the extract. Therefore, in the subsequent experiments, the sampling capillary was dipped into the sample vial ∼45 s before the ion-trap started collecting data. As expected, when the solvent was delivered to the sample vial, the intensity of TIC increased over time (Fig. 6A). The EICs – showing characteristic signals (e.g. catechin, m/z 289; epicatechin, m/z 289; epicatechin gallate, m/z 441; and epigallocatechin gallate, m/z 457), derived from green tea32–34 – could immediately be recorded (Fig. 6B). Fig. S10† shows the resulting mass spectrum, and Table S1† lists all the ions observed (∼30). The above assignment of MS signals was enabled by tandem MS analysis, and comparison with spectral signatures of standard compounds present in a standard mixture (cf. Fig. S11;† without microextraction system).
The small molecules trapped in thin pieces of tea leaves easily diffuse to the extraction solvent as soon as it is delivered into the sample vial (even without stirring) (Fig. 6A). This is contrary to the insect specimens, which have a hydrophobic cuticle layer preventing most metabolites from rapid liberation into the extraction solvent (Fig. 2A). This observation points to the advantage of the on-line monitoring of extraction by MS: on top of the information on sample composition, the results reveal “extractability” of the sample components (for example, intracellular metabolites) which may be trapped within the sample matrix.
In order to verify the anticipated correlation between sample mass and fluorescence, we further analyzed 15 pieces of tea leaves with different masses. Following the data treatment, it became apparent that both MS signal intensities (TIC) and fluorescence intensities of the sample extracts correlate with the sample mass (the Pearson correlation coefficient is ∼0.9903, including the outliers; Fig. 6C). Various compounds may contribute to the fluorescence intensity of tea extracts. The observed fluorescence is not derived from all the compounds detected by MS since not all the compounds detected by MS are fluorescent. On the other hand, not all the fluorescent compounds, present in tea extract, can easily be ionized by MS. However, we observed two outlier points (corresponding to relatively heavy samples, each ∼1.9 mg) which do not follow the correlation of fluorescence intensity with sample mass (Fig. 6D). The most likely explanation for the occurrence of such outlier points is the inherent chemical heterogeneity of tea leaves. Some fragments of tea leaves might have been overbaked, which could have led to degradation of fluorescent and ionogenic compounds. Alternatively, the tea leaf fragments corresponding to the outlier points in Fig. 6D might contain a significant amount of “dead” tissue, rich in lignin and cellulose, which does not contain large amounts of low-molecular-weight extractable metabolites.
3.4. Analysis of trace amounts of powdered drugs
To demonstrate another possible application of the proposed system, we implemented it in the analysis of powdered samples collected using cotton swabs. This enabled qualitative analysis of trace amounts of powder samples (acetaminophen, ∼0.7 mg) collected from a solid surface (Petri dish made of polystyrene) (Fig. 7A). In the positive-ion mode, we could observe the corresponding ions at the m/z 152 [M + H]+ and 174 [M + Na]+. Appearance of the acetaminophen signals can be clearly seen in the TIC and EIC traces (Fig. 7C and D). Thus, the cotton swab sampling method, used in conjunction with the automated microextraction system, can enable fast analysis of forensic samples. For example, trace amounts of powders found on crime scenes can be collected using cotton swabs, and the MS analysis can subsequently be conducted using the proposed approach without additional sample preparation. | ||
| Fig. 7 Demonstration of the cotton swab sampling method combined with the automated microextraction system (cf. Fig. 1) in the analysis of a powdered drug (acetaminophen). (A) Scheme of the sampling method. (B) Blank spectrum in the positive-ion mode (top); the spectrum of acetaminophen powder (∼0.7 mg) collected on cotton swab, and analyzed using the proposed system (bottom). Asterisk (*) indicates the signal of an unknown contaminant. (C) TIC. (D) EIC for m/z 152 [M + H]+. | ||
3.5. Final considerations
Since the times of Justus von Liebig, sample preparation for chemical analysis has undergone a remarkable transformation. Nowadays, many analyzed samples are small in size (millimeter-range dimensions and smaller). It is also desirable that analyses are conducted immediately after sampling and initial treatment (if any) in order to avoid major losses of analytes. In the 20th century, a number of instrumental techniques grounded their position in chemical laboratories. Although some analytical approaches enable analyses of unprocessed samples, in many cases, one cannot get away from sample preparation to obtain a satisfactory coverage of sample components. Automating sample preparation has been challenging, and hyphenating automated sample preparation with high-end detectors is still the dream of many analysts. The notion of open-source electronics provides ready solutions for the automation of daily life operations. In fact, there are dozens of open-source electronic circuits available on the market. These widely accessible electronic components can readily be implemented to augment protocols in chemical analysis. As shown above, these universal building blocks – along with facile programming tools – allow one construct simple but reliable automated systems for fast and effortless preparation of microscale samples.In the case of the proposed sample preparation/analysis system, on pressing one button on a touch screen interface, untreated microscale samples are processed without supervision, and the digital data deposited on the hard drive of a computer. This “single-button approach” decreases the time and effort requirements of multi-step analytical procedures involving sample treatment. The operation of the platform presented in this work does not require much training in chemistry, electronics, or programming. Therefore, the technical staff and the undergraduate students – who do not have much research experience – can readily embark on discovery-oriented projects (e.g. related to metabolomics) without being briefed on many technical details, and they can learn the intricacies of the analytical system step by step. In addition, such automated sample preparation tools can reduce the incidence of human errors, and they can give analysts more time to spend on other critical tasks such as data interpretation.
The proposed automated microextraction system greatly increases the throughput of the analyses conducted on microscale samples (e.g. single fruit fly, fragments of tea leaves, and trace amount of drugs). By recording temporal data (MS signal intensity or fluorescence vs. time), it provides information on the dynamics of the extraction process. However, to leverage the potential of this method in biochemistry, clinical analysis, and forensic science, it would be desirable to demonstrate its quantitative capabilities. This may be done, for example, by spiking the extraction solvent with isotopically labeled standards of the target analytes. It would also be interesting to couple the proposed automated system with a high-resolution mass spectrometer in order to facilitate identification of unknown analytes based on accurate mass matching.
Acknowledgements
This project was funded by the National Science Council of Taiwan, the National Chiao Tung University as well as the “Aiming for the Top University Program” of the National Chiao Tung University, and the Ministry of Education, Taiwan. We thank the fly core facility in the Medical College of the National Taiwan University for the provision of fruit fly stocks and food.Notes and references
- M. K. Bjørk, K. W. Simonsen, D. W. Andersen, P. W. Dalsgaard, S. R. Sigurðardóttir, K. Linnet and B. S. Rasmussen, Anal. Bioanal. Chem., 2013, 405, 2607 CrossRef PubMed.
- A. G. Marshall and C. L. Hendrickson, Annu. Rev. Anal. Chem., 2008, 1, 579 CrossRef CAS PubMed.
- M. Kandiah and P. L. Urban, Chem. Soc. Rev., 2013, 42, 5299 RSC.
- Y.-C. Chen and P. L. Urban, Trends Anal. Chem., 2013, 44, 106 CrossRef CAS PubMed.
- T. R. Northen, J.-C. Lee, L. Hoang, J. Raymond, D.-R. Hwang, S. M. Yannone, C.-H. Wong and G. Siuzdak, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 3678 CrossRef CAS PubMed.
- Comprehensive Sampling and Sample Preparation, ed. J. Pawliszyn, Academic Press, Oxford, 2012 Search PubMed.
- Handbook of Sample Preparation, ed. J. Pawliszyn and H. L. Lord, John Wiley & Sons, Hoboken, 2010 Search PubMed.
- K. Ridgway, S. P. D. Lalljie and R. M. Smith, J. Chromatogr. A., 2007, 1153, 36 CrossRef CAS PubMed.
- M. Trojanowicz, Mod. Chem. Appl., 2013, 1, 1000e113 Search PubMed.
- G. M. Whitesides, Nature, 2006, 442, 368 CrossRef CAS PubMed.
- M.-C. Hennion, J. Chromatogr. A., 1999, 856, 3 CrossRef CAS.
- H. Kataoka, Anal. Bioanal. Chem., 2002, 373, 31 CrossRef CAS PubMed.
- M. Jemal, Biomed. Chromatogr., 2000, 14, 422 CrossRef CAS.
- S. C. C. Shih, H. Yang, M. Jebrail, R. Fobel, N. McIntosh, O. Y. Al-Dirbashi, P. Chakraborty and A. R. Wheeler, Anal. Chem., 2012, 84, 3731 CrossRef CAS PubMed.
- I. A. Popov, H. Chen, O. N. Kharybin, E. N. Nikolaev and R. G. Cooks, Chem. Commun., 2005, 1953 CAS.
- V. G. Santos, T. Regiani, F. F. G. Dias, W. Romão, J. L. P. Jara, C. F. Klitzke, F. Coelho and M. N. Eberlin, Anal. Chem., 2011, 83, 1375 CrossRef CAS PubMed.
- T. M. Squires and S. R. Quake, Rev. Mod. Phys., 2005, 77, 977 CrossRef CAS.
- T. R. Slaney, J. Nie, N. D. Hershey, P. K. Thwar, J. Linderman, M. A. Burns and R. T. Kennedy, Anal. Chem., 2011, 83, 5207 CrossRef CAS PubMed.
- P.-H. Li, H. Ting, Y.-C. Chen and P. L. Urban, RSC Adv., 2012, 2, 12431 RSC.
- Z. Xiao, M. Niu and B. Zhang, J. Sep. Sci., 2012, 35, 1284 CrossRef CAS PubMed.
- E. C. Berglund, N. J. Kuklinski, E. Karagündüz, K. Ucar, J. Hanrieder and A. G. Ewing, Anal. Chem., 2013, 85, 2841 CrossRef CAS PubMed.
- Drosophila: Methods and Protocols, ed. C. Dahmann, Humana Press, Totowa, 2010 Search PubMed.
- J. M. Menter, Photochem. Photobiol. Sci., 2006, 5, 403 CAS.
- I. Georgakoudi, B. C. Jacobson, M. G. Müller, E. E. Sheets, K. Badizadegan, D. L. Carr-Locke, C. P. Crum, C. W. Boone, R. R. Dasari, J. V. Dam and M. S. Feld, Cancer Res., 2002, 62, 682 CAS.
- W. R. Zipfel, R. M. Williams, R. Christie, A. Y. Nikitin, B. T. Hyman and W. W. Webb, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 7075 CrossRef CAS PubMed.
- R. Shroff, L. Rulíšek, J. Doubský and A. Svatoš, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 10092 CrossRef PubMed.
- M. Parisi, R. Li and B. Oliver, BMC Res. Notes, 2011, 4, 198 CrossRef CAS PubMed.
- C. J. F. Scheitz, Y. Guo, A. M. Early, L. G. Harshman and A. G. Clark, PLoS One, 2013, 8, e72726 CAS.
- W. H. Press, S. A. Teukolsky, W. T. Vetterling and B. P. Flannery, Numerical Recipes in C: The Art of Scientific Computing, Cambridge University Press, Cambridge, 1992 Search PubMed.
- M. Carvalho, J. L. Sampaio, W. Palm, M. Brankatschk, S. Eaton and A. Shevchenko, Mol. Syst. Biol., 2012, 8, 600 CrossRef PubMed.
- D. Wang, J. Lu, A. Miao, Z. Xie and D. Yang, J. Food Compos. Anal., 2008, 21, 361 CrossRef CAS PubMed.
- D. D. Rio, A. J. Stewart, W. Mullen, J. Burns, M. E. J. Lean, F. Brighenti and A. Crozier, J. Agric. Food Chem., 2004, 52, 2807 CrossRef PubMed.
- B.-L. Lee and C.-N. Ong, J. Chromatogr. A, 2000, 881, 439 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: A video sequence showing the operation of the proposed system, explanatory notes, additional tables and figures. See DOI: 10.1039/c3ra48023b |
| This journal is © The Royal Society of Chemistry 2014 |