Curcumin, a promising anti-cancer therapeutic: a review of its chemical properties, bioactivity and approaches to cancer cell delivery
Melessa Salem
a,
Sohrab Rohani
b and
Elizabeth R. Gillies
*bc
aBiomedical Engineering Graduate Program, The University of Western Ontario, 1151 Richmond St., London, Canada N6A 5B9
bDepartment of Chemical and Biochemical Engineering, The University of Western Ontario, 1151 Richmond St., London, Canada N6A 5B9. E-mail: egillie@uwo.ca
cDepartment of Chemistry, The University of Western Ontario, 1151 Richmond St., London, Canada N6A 5B7
First published on 4th February 2014
Abstract
The development of new anti-cancer treatments with greater efficacy and fewer side effects remains a significant challenge of modern scientific and medical research. Curcumin, a natural polyphenol found in the dietary spice turmeric, has been demonstrated to inhibit cancer cell survival and proliferation, and to induce apoptosis without promoting the development of side effects. However, due to its sparing solubility and low bioavailability, curcumin has not yet been clinically used to treat cancer. This review describes the main physicochemical properties of curcumin, including its chemical structure, stability, and degradation products as a function of pH and temperature. It also describes the proposed mechanisms by which curcumin exhibits anti-cancer activity. Finally, we review the various approaches that have been studied to enhance the solubility and bioavailability of curcumin, including the preparation of co-crystals, and the development of delivery systems based on liposomes, micelles, exosomes, nanoparticles and dendrimers.
Introduction
For the past few decades there has been rapid growth in the interest of using natural products for therapeutic applications. Although this idea seems recent, our ancestors frequently used natural compounds to combat illness. The development of synthetic pharmaceuticals revolutionized modern medicine over the last century, but the undesirable properties and side effects of these drugs has inspired a search for natural approaches to disease prevention and treatment with the hope that naturally occurring compounds may be better tolerated than their synthetic counterparts. Of particular interest is curcumin, the principal active ingredient in the traditional dietary spice turmeric, responsible for its colour, taste, and most of its chemical and biological properties. Curcumin is derived from Curcuma longa, a plant of the ginger family. The other two curcuminoids present in turmeric in lower concentrations are demethoxycurcumin, and bis-demethoxycurcumin.While curcumin gained immense attention as a medicinal drug in modern medical applications only a few decades ago, it has been used for hundreds of years in some areas of the world. In Asia, specifically India and China, turmeric has been used as a drug for more than two thousand years.1 Ayurveda, an ancient medicinal system practiced in India, incorporated the use of natural herbs to treat various illnesses. Amongst the commonly used herbs, turmeric was used most abundantly due to the medical effects of curcumin. In the traditional Ayurvedic approach, turmeric was crushed into a paste for the treatment of eye infections, burns, bug bites, and any skin related diseases. Furthermore, new mothers in India are given a drink containing turmeric paste, honey, ginger, and milk to drink daily following child birth.1 Turmeric is also used in different forms to cure cough and respiratory complications, along with dental diseases, flatulence, and indigestion. Curcumin was also used as a medicine in ancient Polynesian culture. It has been noted in historical documents that the Polynesian people carried turmeric with them during long voyages to Hawaii.2 Today, Hawaiians utilize curcumin for various medicinal purposes. It is known to them as Olena.
Curcumin was not introduced to Western cultures until the 14th century when European explorers were introduced to Asian countries. In particular, Marco Polo was introduced to curcumin during his visit to the Asian continent in 1280 AD.2,3 It was noted in his writings that the native people of India and China used a vegetable that contained all the properties of saffron, but this spice was not quite saffron. In addition, the Portuguese sailor, Vasco de Gama, sailed to India approximately 500 years ago specifically in search of spices for their medicinal use.4 It was not until his return to Europe that curcumin was truly introduced into Western cultures. Despite curcumin's extensive history, scientists were not able to isolate the curcumin molecule until the 1800s. In 1870, scientists obtained the crystalline form of curcumin, and elucidated its overall structure in 1910.3 Its chemical structure is responsible for its unique physicochemical and biological properties.
Physicochemical properties
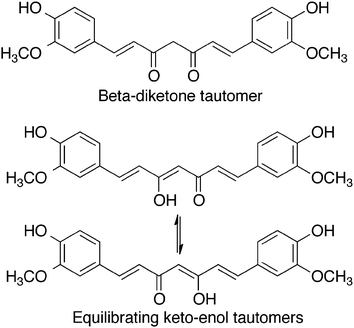
Curcumin, 1,7-bis(4-hydroxy-3-methoxyphenyl)-1,6-heptadiene-3,5-dione, is a hydrophobic polyphenol. It is a golden-yellow solid, with a molecular weight of 368 g mol−1 and a melting point of 183 °C. It is often used as a dye owing to its vibrant colour. As shown in Fig. 1, curcumin can exist in different tautomeric forms. The aromatic rings are functionalized with methoxy and hydroxy groups in an ortho position with respect to one another. The aromatic rings are connected to one another via a seven-carbon spacer that contains two α, β-unsaturated carbonyl groups. As a result of this structure, a beta-diketone and equilibrating keto-enol tautomeric forms of curcumin are possible.5Effect of pH
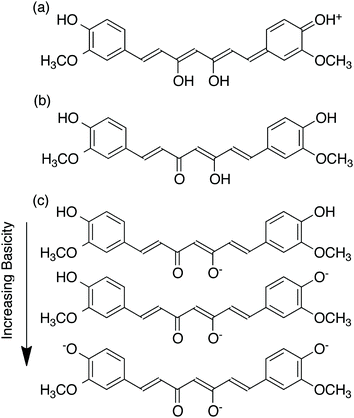
Among the factors that affect the properties of curcumin is pH. Although curcumin has been shown by X-ray diffraction analysis to exist in the keto-enol form in the solid state,6 there has been controversy concerning whether curcumin exists in its beta-diketo or keto-enol form at neutral and slightly acidic pH.5 For example, Jovanovic et al. have proposed that the beta-diketone form of curcumin predominates in mildly acidic aqueous solution and in cell membranes.7 They have also suggested that the central methylene group of the diketone form can act as a potent hydrogen atom donor in radical reactions, potentially mediating its biological activity. However, Payton et al. have recently performed detailed NMR studies supporting that curcumin exists as keto-enol tautomers in a range of hydrophobic and hydrophilic solvents, over a pH range from 3–9.5 These results are in agreement with density functional theory calculations by Shen and Li,8 which correlated the calculated absorption wavelength and oscillator strength of curcumin with the experimental values, supporting the predominance of the keto-enol tautomers. The keto-enol form was calculated to be more stable than the diketone form by 7.75 kcal mol−1. This was attributed to the planar structure of the enol form, allowing for resonance stabilization to occur, while that of the ketone form was twisted.As shown in Fig. 2, at pH less than 1, a protonated form of curcumin is observed and the molecule can adopt a fully conjugated protonated form.9 Fig. 2 also illustrates the proposed deprotonated forms of curcumin that can occur as the pH is increased.9 As a consequence of deprotonation, curcumin exhibits increased aqueous solubility in alkaline solutions while it is practically insoluble in water at acidic and neutral pH. However, curcumin has been demonstrated to undergo degradation under alkaline conditions, as demonstrated by Tonnesen et al. and Wang et al.9,10
Effect of temperature
Solid curcumin is generally stored at −20 °C, though there is no evidence that curcumin is not stable at somewhat higher temperatures. Wang et al. studied the stability of curcumin at body temperature.10 In this study, curcumin was incubated under various physiological conditions at 37 °C to measure its stability. Curcumin was incubated in 0.1 M phosphate buffer solution, culture medium, culture medium with 10% serum, and human blood. Samples were analyzed by high performance liquid chromatography (HPLC) after one, four, and eight hours of incubation. It was noted that the rate of curcumin degradation was dependent on the conditions. When no serum was present, curcumin underwent degradation more rapidly than in the medium with serum or in human blood. This difference was accounted for by the stabilization of curcumin by the serum and it was suggested that care must be taken in the manipulation of curcumin under serum-free conditions, particularly in the interpretation of the results of biological studies, where the presence of degradation products such as vanillin should be taken into account.In another study, Gopinath et al. investigated curcumin incorporated collagen films (CICFs) for dermal wound healing.11 CICFs were prepared by dissolving a weighed amount of curcumin in ethanol, and mixing this solution with 50 mL of collagen/acetic acid solution at 4 °C for 24 hours. Micro-shrinkage temperature measurements were carried out on both CICF and pure collagen. It was found that without curcumin, the shrinkage temperature of collagen was 55 °C, which was increased to 78 °C when curcumin was incorporated in the collagen. It was concluded from this finding that curcumin increases the thermal stability of collagen. This group previously reported that quercetin also increased the thermal stability of collagen, allowing them to draw the conclusion that polyphenols are generally capable of enhancing the temperature stability of collagen and matrices of different types.
Previous studies have demonstrated that natural phenolic compounds suffer from biodegradation, change of structure, and loss of biological activity when heated. Based on this, Liu et al. set out to investigate the thermal stability of curcumin.12 Specifically, the relationship between the pre-incubation temperature of curcumin and its inhibitory effect on amyloid fibrillation of hen egg-white lysozyme (HEWL) was studied. Unmodified HEWL was used as a control agent, along with HEWL modified with curcumin pre-incubated at 10, 25, 40, 55, and 70 °C for 329 hours. The result of the thioflavin T (ThT) fluorescence assay is shown in Fig. 3, where a decrease in fluorescence corresponds to a decrease in amyloids.
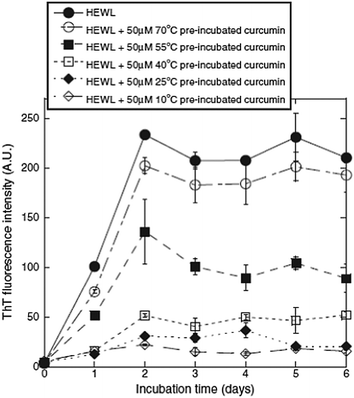 | ||
| Fig. 3 ThT assay results, showing that the ability of curcumin to inhibit the amyloid fibrillation of HEWL, decreased with increasing incubation to and temperature (inhibition is detected as a decrease in fluorescence intensity). Reprinted from ref. 12, with permission from Elsevier. | ||
As shown in Fig. 3, the unmodified HEWL exhibited the highest amyloid content, and the HEWL sample modified with curcumin pre-incubated at 70 °C provided similar results. This can be attributed to the lack of biologically active curcumin following incubation at this temperature. However, at a pre-incubation temperature of 55 °C, it was evident that not all of the curcumin degraded, as the fluorescence decreased by approximately 50%. Therefore, a clear dependence of curcumin stability on temperature was observed. This trend was further verified using transmission electron microscopy (TEM), where the images clearly illustrated the presence of high amounts of amyloid fibrils in the unmodified HEWL, and that modified with curcumin pre-incubated at 70 °C. However, the samples containing curcumin pre-incubated at lower temperatures exhibited fewer amyloid fibrils, with the amount decreasing with decreasing pre-incubation temperature. Similar results were obtained in the cell viability study, leading the authors to conclude that curcumin is most stable in the temperature range of 10–55 °C and degraded almost completely at 70 °C within 24 hours.
Thus, it can be generally concluded that curcumin is most stable at low temperatures; however, its stability is still maintained at room temperature and physiological temperature, allowing it to be used in medical applications. At very high temperatures however, curcumin degrades into different products, which will be discussed briefly below.
Degradation products of curcumin
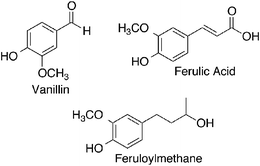
Under certain conditions curcumin becomes unstable, as previously discussed, and degrades, yielding other compounds. The three degradation products often found are vanillin, ferulic acid, and feruloylmethane.13 In the previously described study by Wang et al. it was noted that when curcumin degraded in different media, the resulting degradation products were characterized by HPLC-mass spectrometry to be vanillin, ferulic acid, and feruloylmethane (Fig. 4).10 However, in this study there was also an unknown product present in larger amounts. It was hypothesized that this product was trans-6-(4′-hydroxy-3′-methoxyphenyl)-2, 4-dioxo-5-hexenal.A study conducted by Schieffer investigated the degradation products of curcumin obtained after exposure to light.14 Curcumin is extremely sensitive to light and undergoes degradation in its presence. The study concluded that curcumin underwent photolytic degradation to produce several products with the most abundant being p-hydroxybenzaldehyde, vanillic acid, vanillin, and ferulic acid. The difference in some of the products proposed in this study versus those obtained by Wang et al. suggests that the thermal and photochemical degradation pathways of curcumin may be different, though it is generally agreed that the degradation of curcumin is much more rapid in the presence of light.
Singh et al. explored the photodegradation of curcumin in the presence of a TiO2 catalyst.15 It was reported that curcumin formed a complex with TiO2, which was evidenced by the presence of a broad, red-shifted absorption band. Photodegradation was carried out in two solvent media, water–methanol and water–acetonitrile, and the degradation products were evaluated using gas chromatography. The results were similar for both media, with the predominant degradation product being vanillin, along with minimal amounts of CO2, 4-hydroxybenzaldehyde, and methoxyvanillin.
It is widely proposed that vanillin and ferulic acid are the major degradation products and in fact contribute to the therapeutic effects of curcumin. In fact, there is a variety of work concerning the therapeutic effects of vanillin.13,16–18 Despite the potential therapeutic effects of vanillin, the recent work of Wang et al. outlined above,10 casts doubt on the proposition that vanillin is in fact a major degradation product of curcumin. In the case of this study, vanillin and ferulic acid were only minor degradation products, and there existed a major degradation product with currently unconfirmed identity.
Gordon and Schneider, and Griesser et al. also performed similar degradation reactions in order to challenge the belief that vanillin, ferulic acid, and feruloylmethane are the major degradation products of curcumin.19,20 The results obtained using HPLC suggested the presence of one major product, while vanillin, ferulic acid, and feruloylmethane were close to undetectable. Further analysis revealed a molecular weight of 400 g mol−1, indicating insertion of oxygen to curcumin. Based on 1D and 2D homo- and heteronuclear NMR experiments, the unknown compound was identified as a bicyclopentadione form of curcumin.19 Fig. 5 illustrates the proposed autoxidation mechanism leading to this product. It was proposed that this degradation mechanism accounts for curcumin's antioxidant properties, and not the alternative degradation products.
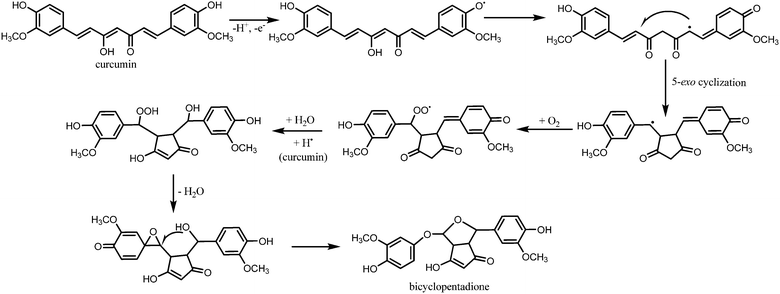 | ||
| Fig. 5 Proposed mechanism for the autoxidation of curcumin.19 | ||
Ketron et al. also investigated the oxidative properties of the bicyclopentadione form of curcumin, exploring its effects on human type II topoisomerases.21 A series of experiments were performed to evaluate the effect of the bicyclopentadione form of curcumin, stable curcumin, vanillin, ferulic acid, and feruloylmethane on the human enzymes. The results revealed that the bicyclopentadione form of curcumin poisons human topoisomerases, in contrast to curcumin and its three fragmentation products, which did not reveal a significant effect. Thus, while vanillin, ferulic acid, and feruloylmethane may exhibit therapeutic effects, it still remains to be confirmed which therapeutic effects of curcumin arise from the parent molecule, and which arise from the various possible degradation products.
Effects of curcumin on cancer cells
Although curcumin has been used as a traditional medicine to treat several conditions including inflammation, respiratory infection, and blood clotting, there is rapidly growing interest in its effects on cancer. The motivation for the use of curcumin arises not only from its potential therapeutic effects, but also from the knowledge that it may be better tolerated by patients, in comparison with most chemotherapeutic drugs that suffer from numerous harmful side effects such as nausea and vomiting, diarrhea, hair loss,22 and more serious long term conditions such as liver failure.23 Several studies comparing the incidences of cancer and cancer related deaths in India and the West revealed lower risk of cancer in India (Table 1).24 It is proposed that one of the major reasons for the drastic difference in cancer occurrences in these regions is the increased intake of dietary agents such as curcumin, and other plant derivatives in Asia.| Cancer | USA | India | ||
|---|---|---|---|---|
| Cases | Deaths | Cases | Deaths | |
| Lung | 660 | 580 | 38 | 37 |
| Breast | 660 | 160 | 79 | 41 |
| Prostate | 690 | 130 | 20 | 9 |
| Colon/rectum | 530 | 220 | 30 | 18 |
| Bladdar | 202 | 43 | 15 | 11 |
| Thyroid | 55 | 5 | 12 | 3 |
| Leukemia | 100 | 70 | 19 | 17 |
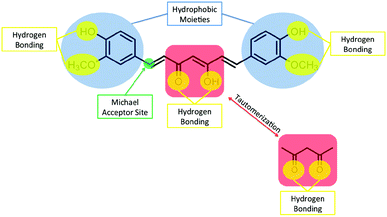
Why does curcumin possess anti-cancer properties? Generally, cancer with its diverse origins has several molecular markers involved in its onset and progression. Curcumin is capable of interfering with several biochemical pathways involved in the proliferation and survival of cancer cells by directly and indirectly binding to different targets. Curcumin has been shown to interact with various targets including transcription factors, growth factors, DNA, RNA, and several proteins that are involved in cell signal transduction pathways.25,26 There are several features of curcumin's chemical structure that make it a favourable and versatile binding partner for a wide variety of molecular targets (Fig. 6). For example, curcumin possesses two hydrophobic phenyl groups connected by a relatively flexible linker. This allows the molecule to assume different conformations that can maximize π–π and van der Waals interactions with aromatic and other hydrophobic amino acid residues of proteins. The phenolic hydroxyl and methoxy groups, as well as the ketone and enol groups present on the ends and in the middle of the molecule respectively can participate in strong and directed hydrogen-bonding interactions. The possibility for keto-enol tautomerism introduces additional functionality, with the possibility to arrange donor and acceptor groups for hydrogen bonding in multiple ways. Furthermore, the α,β-unsaturated ketone moiety can serve as a Michael acceptor for nucleophilic attack by the thiol moieties of cysteine residues or the Se− moieties of selenocysteine. The proposed pathways by which curcumin exhibits anti-cancer properties have been described in detail in several recent reviews25–29 and some of these mechanisms are described below.
Curcumin's mechanism of action can occur both intrinsically (mitochondrial) and extrinsically (mediated via cell surface transmembrane death receptors).30 The intrinsic pathway is generally initiated by activation of tumour suppressor p53 (p53), which is a cell cycle regulator, as well as through members of the B-cell lymphoma 2 (Bcl-2) family. Up-regulation of p53 activates Bcl-2 homologous antagonist killer (Bak) and Bcl-2 associated x protein (Bax), which are pro-apoptotic members of the Bcl-2 family. Bak and Bax promote apoptosis by forming pores in the mitochondrial membrane releasing cytochrome c into the cytoplasm, thereby activating a caspase cascade.30 Curcumin is capable of inhibiting Bcl-2 and B-cell lymphoma extra large (Bcl-xL) which is another pro-survival protein of the Bcl-2 family. Luthra et al. showed using molecular docking studies that curcumin may inhibit Bcl-2 by directly binding to cavity 2 of the protein through interactions with multiple amino acids.31 Guo et al. showed that curcumin significantly inhibited growth of human colorectal carcinoma LoVo cells.32 Nuclear and annexin V/PI staining confirmed that curcumin induced apoptosis in LoVo cells, in addition to decreasing the mitochondrial membrane potential and activating caspase-3 and -9 at concentrations of 0–30 μM.
Nuclear factor (erythroid-derived 2)-like 2 (Nrf2) is a transcription factor involved in the primary defence pathway against the effects of oxidative stress.33–35 Under normal conditions, Nrf2 is located in the cytoplasm with its activity suppressed by the chaperon protein Kelch like-ECH-associated protein 1 (Keap1). Under conditions of oxidative stress, which alter the thiol groups on Keap 1, the Nrf2-Keap1 interaction is disturbed, causing Nrf2 to be transported to the nucleus where it binds to the anti-oxidative response element (ARE) and activates the transcription of genes encoding for detoxifying enzymes such as hemeoxygenase-1 (HO-1), which protect against various forms of stress.36 It has been proposed that Michael addition of curcumin to thiols on Keap1 may result in a conformational change that disrupts the Nrf2-Keap1 complex, promoting Nrf2 binding to AREs, but this still requires structural verification.28
Another transcription factor commonly studied as a target for cancer treatment is nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB). This transcription factor is involved in cell response to free radicals, cytokines, ultraviolet radiation, and viral or bacterial antigens. Because NF-κB is present in the cytoplasm in an inactive form until stimulated, it is considered a first responder transcription factor as it does not require synthesis for activation. Upon stimulation, NF-κB is translocated to the nucleus triggering the expression of certain genes that suppress apoptosis and promote proliferation and metastasis.
In the work of Youns et al. the effects of curcumin were studied on human pancreatic cell lines expressing different levels of cyclooxygenase-2 (COX-2), an enzyme that plays an important role in response to inflammation.37 Curcumin was capable of inhibiting proliferation and inducing apoptosis through the extrinsic pathway mediated through tumour necrosis factor (TNF) receptor activation. The TNF pathway triggers caspase-8 and -3 activation, which are pro-apoptotic, but also induces NF-κB, which regulates the expression of the COX-2 gene. Curcumin also resulted in down-regulation of COX-2, suggesting the suppression of NF-κB activation. Using molecular docking studies, curcumin has been shown to bind to TNF-α through various van der Waals and hydrogen bonding interactions, which may inhibit the binding of TNF-α to its receptor, thus preventing the activation of NF-κB.38
In another study, curcumin's ability to chemosensitize breast cancer cells to 5-fluorouracil was investigated.39 5-Fluorouracil has been noted to up-regulate the NF-κB pathway inducing the expression of its regulated genes, leading to chemoresistance. IκB kinase (IKK) is a signal-induced enzyme, which phosphorylates IκBα, an NF-κB inhibitory protein. Upon phosphorylation, IκBα is detached from NF-κB, leaving NF-κB free to translocate to the nucleus and induce the expression of different genes. The authors showed that curcumin inhibited the phosphorylation of IκBα by IKK, preventing the dissociation of IκBα from NF-κB, keeping NF-κB dormant. As a result, chemoresistance towards 5-fluorouracil treatment was significantly decreased.
Activated protein-1 (AP-1), composed of Jun, FOS, and ATF protein dimers,40 is another transcription factor commonly used as a target for cancer treatment as it is responsible for differentiation, proliferation, apoptosis, and oncogenic transformations.41 Activation of AP-1 by stimuli such as growth factors, bacterial/viral infections, cytokines, stress signals, or oncogenic factors promotes the binding of AP-1 to the TPA responsive element (TRE) causing the expression of several genes such as cyclin-D1, MMP, VEGF, and uPA, some of which mirror genes activated by NF-κB and are involved in the angiogenesis and growth of cancer cells.24 Curcumin has been proposed to inhibit this pathway via direct interaction with the AP-1 DNA-binding site.42 Spectroscopic evidence has shown that curcumin can in general bind to DNA through O2 of thymine in the minor groove, N7 of guanine and adenine in the major groove and to the backbone phosphate moieties with a binding constant of 4.3 × 104 M−1.43
Curcumin was shown to suppress binding of AP-1 to DNA in human leukemia cells,44 and transformed keratinocytes.45 Furthermore, curcumin possesses cytotoxic activities towards human papillomavirus (HPV)-16 and HPV-18 infected cells associated with the development of cervical cancer, in addition to promotion of cervical cancer cell apoptosis, and inhibition of AP-1-DNA binding, thereby preventing HPV gene expression.46 The down regulation of AP-1 by curcumin has also been reported for prostate cancer.47,48 It was reported that exogenous hydrogen peroxide induces prostate cancer LNCaP cell proliferation via activation of AP-1 and consequent stimulation of heparin affin regulatory peptide (HARP) gene.49 This cascade is halted by curcumin inhibition of the hydrogen peroxide induced HARP expression.
The Janus kinase/signal transducer and activator of transcription (JAK/STAT) signalling pathway is another cascade noted to be involved in the progression of cancer.50 JAK belongs to the non-protein receptor tyrosine kinase family, and STATs are latent cytoplasmic transcription factors. The JAK/STAT pathway plays a role in immune response, mediating interactions of cytokines, hormones, and growth factors and their respective receptors. Upon binding of a ligand to its receptor, JAK is activated and the increased kinase activity induces phosphorylation of tyrosine residues on the receptor. Tyrosine phosphorylation creates phosphotyrosine sites on the receptor, which STATs are capable of binding to. The STATs recruited to these binding sites are themselves phosphorylated, creating phosphotyrosine sites for other STATs, thereby creating dimers. STAT dimers accumulate in the nucleus and activate their target genes. Kroon et al. studied the effect of this pathway on prostate cancer cells, specifically with respect to STAT3 which of the seven known STAT proteins has been reported to be closely associated to a variety of human cancers.51
Yang et al. investigated the effect of curcumin on the JAK/STAT signalling pathway in small lung cancer cells.52 The authors reported that curcumin was capable of suppressing cell proliferation and migration through inhibition of STAT3 phosphorylation, thus suppressing downstream target genes. To further ensure the role of this cascade on the cancer cells, the authors showed that activation of STAT3 induced by interleukin-6 (IL-6), a pro-inflammatory cytokine, lead to increased proliferation and cell survival. These results supported the conclusion that this pathway can be a potential target for cancer treatment with curcumin. Comparable results were demonstrated earlier on glioblastomas by Weissenberger et al. in 2010.53
In addition to the above, other molecular signalling pathways reported to be associated with cancer cell proliferation have also been studied in the context of curcumin. The effect of curcumin on both the phosphoinositide 3-kinase (PI3K)/protein kinase B (AKT)/mammalian target of rapamycin (mTOR) pathway and the mitogen-activated protein kinases (MAPK) pathway has been investigated by Ono et al.54 PI3K, a family of enzymes involved in cell growth, survival, and proliferation, are activated via ligand/receptor interactions where they are recruited to the inner cell membrane. PI3K activates AKT, which in turn activates mTOR through direct phosphorylation, promoting cell survival.55 MAPK are activated by a series of phosphorylation cycles in response to a variety of stimuli. The authors were able to demonstrate that curcumin inhibited the phosphorylation of mTOR by AKT, suppressing its downstream targets and inducing apoptosis. Interestingly, curcumin enhanced MAPK activity. Specifically, ERK1/2, a subgroup of MAPK, activation by curcumin promoted apoptosis in colorectal carcinoma cells. As determined by Western blotting, both pathways were associated with G2/M phase arrest, and poly (ADP-ribose) polymerase (PARP) cleavage, which is characteristic of cells undergoing apoptosis.
The exportin 1 (CRM1)-dependent nuclear export pathway is a less commonly investigated cascade. CRM-1 facilitates the transport of large molecules across the nuclear membrane to the cytoplasm.56 Furthermore, the structure of CRM1 allows the binding of Ran protein bound to GTP promoting the binding of different cargo proteins to CRM1 in response to a nuclear export signal (NES). Nui et al. showed that curcumin targets CRM1 through the Michael addition reaction of the α, β-unsaturated carbonyl of curcumin and the nucleophilic Cys528of CRM1.57 Mass spectrometry revealed that curcumin bound directly to a CRM1-derived peptide containing the Cys528 moiety, making it capable of arresting this cascade. Furthermore, the authors investigated the effect of curcumin on cargo proteins, as many cancer-promoting proteins are trafficked by CRM1. It was found that curcumin treatment caused the up-regulation of tumour suppressor cargo proteins p73 and p27, and a down-regulation of the pro-survival COX-2 and Cyclin D1.
In other work, the TNF-related apoptosis-inducing ligand (TRAIL) pathway and its enhancement by curcumin has been investigated.58 TRAIL is a protein that acts as a ligand, binding to death receptor (DR)-4 and −5, to activate caspase-8. Caspase-8 cascade activates procaspase-3, -6, and -7 downstream to induce apoptosis. It was reported that breast cancer cells pre-incubated with curcumin became sensitized to TRAIL, promoting cell death.58 However, the enhancement of TRAIL sensitization was cell-type dependent, with the extent of sensitization differing from one breast cancer cell line to another.
Steroid receptors such as estrogen receptors (ER), progesterone receptors (PR), and androgen receptors (AR) function as transcription factors, regulating their respective target genes. Abnormal expression of steroid receptors' transcription coregulators causes the deviation away from their normal functions, thereby participating in the onset and progression of cancers.59 For this reason, anti-steroid receptor compounds are used clinically to treat cancer due to their apoptosis-inducing properties.
Curcumin analogues were shown to behave as androgen receptor antagonists in human prostate cancer as studied by Ohtsu et al.60 Curcumin was identified as a binding ligand for the nuclear vitamin D receptor (VDR) correlated with human colon cancer, playing a role in cancer chemoprevention.61 In particular, curcumin was capable of competing with 1,25-dihydroxyvitamin D3 (1,25D) for the direct binding to VDR. Occupation of the VDR binding site by curcumin caused conformational changes to the receptor, transforming it to its transcriptionally active form, thereby inducing the expression of tumour suppressor genes. Moreover, the use of curcumin in breast cancer treatment showed that it can interact with breast cell lines expressing varying cellular receptor phenotypes involved in breast cancer, thus inducing anti-proliferative effects.62,63
In addition to anti-proliferative and apoptotic effects, curcumin possess chemosensitizing,64,65 radiosensitizing,64,66 and radioprotective effects.66,67 Furthermore, the effect of curcumin on newly identified pathways associated with microRNA, stem cells, and autophagy have been studied.26 MicroRNA are small, noncoding RNAs that negatively regulate the expression of genes by binding to the 3′ region of the untranslated target mRNA, thereby inhibiting translation or inducing mRNA degradation. Because microRNA possess both oncogenic and tumour suppressing properties, anti-cancer effects were achieved upon treatment with curcumin, through the up- and down-regulation of certain microRNA.68–70 Cancer stem cells—undifferentiated cancer cells that can generate tumours by self-renewal—have been shown to drive and sustain human cancers, and treatment with curcumin has been demonstrated to interfere with the self-renewal of cancer stem cells.71–73 Moreover, autophagy is the catabolic or “self-eating” mechanism involving the sequestration of unnecessary, dysfunctional components or organelles into autophagosomes and their degradation in lysosomes.74 This process is altered in cancer cells, promoting cell survival. Curcumin was shown to trigger autophagy in pleural mesothelioma,75 oral cell carcinoma,76 and glioma cells.77,78 With the many molecular targets of curcumin, indeed it can serve as a potential therapeutic for the destruction of cancer cells.
Clinical studies
Thus far, more than 20 clinical trials with curcumin for various cancers including colorectal, pancreatic, breast, prostate, multiple myeloma, lung, head and neck, and cancer lesions have been completed. The results of these studies have been summarized in recent review articles,25,26,79 so we describe here only a few recent noteworthy examples.At a dose of 360 mg, three times per day, curcumin was found to increase body weight, increase the number of apoptotic cells, and enhance the expression of p53 in colorectal cancer patients after diagnosis and before surgery.80 However, the health improvement related to increase p53 expression still needs to be studied. Curcumin was also shown to reduce the formation of aberrant crypt foci (ACF), the precursor of colorectal polyps, in smokers at a dose of 4 g per day, suggesting a potential cancer chemopreventive role for curcumin.81 A recent study demonstrated that following doses of 2.35 g per day for 14 days prior to biopsy or colonic resection, curcuminoids were detectable in colonic tissue, mucosa and urine of most patients, with minimal side effects observed.82 The efficacy of curcumin in combination with gemcitabine against advanced pancreatic cancer was also recently evaluated in two different trials.83,84 Kanai et al. found that curcumin at a 8 g per day in combination with gemcitabine was safe and well-tolerated.84 However, Epelbaum and coworkers found that some patients experienced abdominal pain at this dose, and did not obtain a highly promising therapeutic response.83 Curcumin in combination with docetaxel in patients with advanced and metastatic breast cancer was also recently studied in a Phase 1 trial.85 It was found that the maximum tolerable dose of curcumin was 8 g per day and in combination with the standard dose of docetaxel, the recommended dose was 6 g per day for seven consecutive days every three weeks. The safety, tolerability, and efficacy of curcumin in patients with asymptomatic, relapsed, or plateau phase multiple myeloma has also evaluated by Vadhan-Raj et al.86 At doses of 4 to 8 g per day for a year, curcumin was found to down-regulate the activation of NF-κB and STAT3, and suppress COX-2 expression, but further studies are required to demonstrate efficacy against multiple myeloma. In patients with familial ademonmatous polyposis, curcumin was reported to provide improvements in patients with precancerous lesions at a dose of 12 g per day (0.5% curcumin).87
Based on a search of https://www.clinicaltrials.gov, there are currently more than 15 clinical trials ongoing for the treatment of various cancers with curcumin. Curcumin is approved by the United States Food and Drug Administration as being generally regarded as safe (GRAS). It is marketed in various forms as a dietary supplement in many countries. However, there is not yet an approved drug license for curcumin. This can likely be attributed in part to the multi-targeting property of curcumin.88 It also results from the extremely poor solubility in aqueous media and very low bioavailability of curcumin.89,90 The next section of the review discusses potential means of increasing the solubility and bioavailability of curcumin in order for the drug to make the transition from the lab into clinical use.
Enhancement of curcumin solubility and bioavailability
Curcumin has been reported to be safe at doses up to 12 g per day.91–93 However, because curcumin undergoes rapid metabolism and clearance when administered, it has very poor bioavailability.94 A study demonstrated that when curcumin was administered orally to rats at a dose of 500 mg kg−1, a peak concentration of 1.8 ng mL−1 was detected in the plasma, while curcumin given intravenously showed no trace of the drug in plasma within 1 hour of administration.95 In another study, an intraperitoneal injection of 0.1 g kg−1 in mice showed a plasma concentration of 2.25 μg mL−1 after 15 minutes of administration, with concentrations of 177.04, 26.06, 26.9, and 7.51 μg mL−1 in the intestines, spleen, liver, and kidneys, respectively.96 A human study revealed that a total oral dose of 3.6 g per day of curcumin resulted in nanomolar amounts of curcumin in plasma samples on day one at the 1 hour mark, with similar amounts on day 2, 8, and 29.97 How can the bioavailability of curcumin be enhanced knowing that concentrations of at least 10−5 to 10−4 M are required for the drug to have any therapeutic impact?95 One approach can involve the co-administration of adjuvants that can block the metabolic processing of curcumin. For example, the coadministration of curcumin and piperine, a known inhibitor of intestinal and hepatic glucuronidase, involved in curcumin glucuronidation, was shown to increase the bioavailability of curcumin by 2000%.98 There has also been ongoing research on drug delivery systems that may be used to transport curcumin to the desired site, or simply enhance its solubility.Preparation of curcumin co-crystals
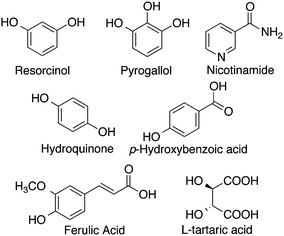
A promising approach to solubility enhancement is the development of co-crystals. Co-crystals are a class of solid drugs formed using an active pharmaceutical ingredient (API), and a solubilizing agent.99 The resulting co-crystals generally possess enhanced physicochemical properties such as solubility and stability, due to increased hydrogen bonds in the system.93Sanphui and coworkers studied co-crystals synthesized using curcumin as the API with two different solubilizing agents, resorcinol and pyrogallol (Fig. 7), by a liquid-assisted grinding method.93 The melting point of the co-crystals was reported to be between that of pure curcumin and the solubilizing agent, with curcumin-pyrogallol having a lower melting point than curcumin-resorcinol. Curcumin-resorcinol co-crystals exhibited a dissolution rate 5-fold that of curcumin alone, while curcumin-pyrogallol co-crystals had a dissolution rate 12-fold higher than that of curcumin.
In another study, curcumin co-crystals were synthesized with five different coformers including nicotinamide, ferulic acid, hydroquinone, p-hydroxybenzoic acid, and L-tartaric acid (Fig. 7).90 All five co-crystal solids were noted to be eutectic, having lower melting points than both constituents alone, and dissolved faster than pure curcumin, with curcumin–nicotinamide having the fastest dissolution rate. Undoubtedly, curcumin co-crystals can serve as a solution to increase the bioavailability of curcumin when administered orally. With the wide variety of coformers such as therapeutic drugs, salts, or even natural spices, that can be used, curcumin co-crystals can potentially be synthesized to treat several conditions.
Delivery systems for curcumin
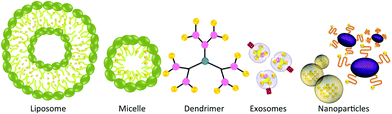
Alternative to co-crystals, which simply enhance the dissolution rate of the drug, methods have been developed to transport curcumin throughout the body to the desired sites for therapeutic applications. A variety of nano-vehicles including liposomes, exosomes, micelles, nanoparticles, and dendrimers have been used to encapsulate and deliver curcumin, resulting in enhanced water solubility, stability and bioactivity (Fig. 8).Liposomes can be described as phospholipid bilayers surrounding an aqueous core, and have been investigated for the delivery of a wide variety of different pharmaceutical agents.100 In fact, lipid-based drug delivery systems are available in the clinic; one such product is Doxil, which is a PEGylated liposomal doxorubicin.101 Li et al. have studied the in vitro and in vivo effects of liposome-encapsulated curcumin on human pancreatic carcinoma cells.102 The liposomes were prepared from a 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 weight ratio of lipid (1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) and/or 1,2-dimyristoyl-sn-glycero-3-phosphor-rac-(1-glycerol) sodium salt (DMPG)) to curcumin. It was demonstrated that liposomal curcumin was capable of down-regulating NF-κB machinery, resulting in suppressed growth and increased apoptosis of various human pancreatic cancer cell lines in vitro. In addition, tumour suppressive and anti-angiogenic effects were observed for both BxPC-3 and MiaPaCa2 cell lines in murine models. In a recent study, Saengkrit et al. investigated the cellular uptake of curcumin-loaded didecyldimethylammonium bromide (DDAB)-modified liposomes compared to non-modified liposomes on cervical cancer cells.103 Liposomes were prepared using various ratios of soybean lecithin, non-ionic surfactant, cholesterol and DDAB by means of the conventional thin film hydration method. The authors reported that cell uptake was enhanced with DDAB-containing liposomes, however the cytotoxicity of these cationic liposomes was high and more work is required to optimize this. Furthermore, in vitro release studies showed that curcumin was released faster from DDAB-containing liposomes, which the authors hypothesized to be a consequence of reduced interaction forces between lipid chains due to the cationic charges of DDAB.
1 weight ratio of lipid (1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) and/or 1,2-dimyristoyl-sn-glycero-3-phosphor-rac-(1-glycerol) sodium salt (DMPG)) to curcumin. It was demonstrated that liposomal curcumin was capable of down-regulating NF-κB machinery, resulting in suppressed growth and increased apoptosis of various human pancreatic cancer cell lines in vitro. In addition, tumour suppressive and anti-angiogenic effects were observed for both BxPC-3 and MiaPaCa2 cell lines in murine models. In a recent study, Saengkrit et al. investigated the cellular uptake of curcumin-loaded didecyldimethylammonium bromide (DDAB)-modified liposomes compared to non-modified liposomes on cervical cancer cells.103 Liposomes were prepared using various ratios of soybean lecithin, non-ionic surfactant, cholesterol and DDAB by means of the conventional thin film hydration method. The authors reported that cell uptake was enhanced with DDAB-containing liposomes, however the cytotoxicity of these cationic liposomes was high and more work is required to optimize this. Furthermore, in vitro release studies showed that curcumin was released faster from DDAB-containing liposomes, which the authors hypothesized to be a consequence of reduced interaction forces between lipid chains due to the cationic charges of DDAB.
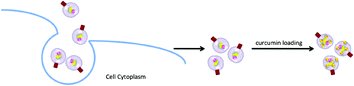
An alternative to synthetic phospholipid vesicles is the naturally occurring analogue, referred to as exosomes. Exosomes are small, endocytic membrane vesicles that are excreted by many cells. They are generally formed by budding from the membrane of multivesicular endosomes found in different cell types, and thus contain different protein families (Fig. 9). Exosomes are usually 30–100 nm in diameter and are capable of floating on sucrose gradients making them easy to separate from other contaminants.104 Due to their small size and biocompatibility, these vesicles can potentially be used for the delivery of pharmaceuticals.
Sun et al. studied the anti-inflammatory effects of curcumin when encapsulated in exosomes.105 Curcumin was mixed at 22 °C with exosomes derived from EL-4 cells, and the mixture was purified by sucrose gradient separation. A loading capacity of 2.9 g curcumin per 1 g exosomes was reported and the solubility of curcumin was five-fold higher with exosome-entrapment in comparison to free curcumin. Furthermore, a stability study was conducted and it was found that in phosphate buffered saline at 37 °C, in the absence of exosomes, 75% of curcumin was degraded over 2.5 hours, while only 20% degradation was observed for exosome-encapsulated curcumin. An in vivo study was carried out on mice with a dose of 100 mg kg−1 of curcumin administered orally or intraperitoneally. Exosomal curcumin was present in peripheral blood at concentrations five to ten-fold higher than free curcumin, with no detectable amount of free curcumin circulating in the blood. The in vivo anti-inflammatory effects of curcumin were also evaluated, using a lipopolysaccharide-induced septic shock model. A significant survival advantage and lower cytokine levels were demonstrated for mice treated with exosomal curcumin in comparison with free curcumin or curcumin-free exosome and saline controls.
Mohanty et al. proposed a polymeric micelle as a drug delivery system for curcumin encapsulation to be used for cancer therapy.106 Curcumin-containing micelles were synthesized using a methoxy poly(ethylene glycol) (mPEG)/poly-ε-caprolactone (PCL) formulation. The micelles were loaded with curcumin via a dialysis method at room temperature. The micellar solution was then freeze dried to obtain a dry solid form of curcumin-containing micelles. The authors reported a curcumin encapsulation efficiency of 60%, with a micelle size of 110 nm. The loaded micelles showed sustained release of curcumin lasting for one week. Furthermore, curcumin uptake was investigated on PANC-1 pancreatic cancer cell line using both curcumin-loaded micelles, and unmodified curcumin for comparison. Curcumin concentrations of 10, 20, and 30 μM were studied. The authors observed that cell uptake of curcumin-loaded micelles at a concentration of 10 μM was ∼3-fold higher than that of unmodified curcumin. However, at the highest curcumin concentration (30 μM), the micellar uptake was only ∼2-fold higher than unmodified curcumin. This led the authors to conclude that cell uptake is more efficient at lower concentrations, rationalizing that at higher concentrations of curcumin-loaded micelles, saturation may occur, causing decreased entry of micelles into the cells.
In another study, Podaralla et al. investigated a micellar formulation of curcumin prepared from mPEG conjugated to zein, a hydrophobic plant protein.107 Curcumin was encapsulated in mPEG-zein micelles by dissolving mPEG-zein and curcumin (100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 wt/wt) in 90% ethanol, followed by dialysis to remove the remaining ethanol and free curcumin. The authors reported a micelle size range of 95–125 nm, and release of curcumin over a period of ∼24 hours in vitro. The micellar system resulted in 1000–2000-fold enhancement in curcumin water solubility and a ∼6-fold increase in stability, as evaluated by UV-visible spectroscopy. The uptake of curcumin-loaded micelles by ovarian cancer cells was 2–3-fold higher than free curcumin, leading the authors to conclude that this delivery system is highly promising for the delivery of anti-cancer drugs. They also suggested that the core or shell could potentially be modified by cross-linking in order to further sustain the release.
2 wt/wt) in 90% ethanol, followed by dialysis to remove the remaining ethanol and free curcumin. The authors reported a micelle size range of 95–125 nm, and release of curcumin over a period of ∼24 hours in vitro. The micellar system resulted in 1000–2000-fold enhancement in curcumin water solubility and a ∼6-fold increase in stability, as evaluated by UV-visible spectroscopy. The uptake of curcumin-loaded micelles by ovarian cancer cells was 2–3-fold higher than free curcumin, leading the authors to conclude that this delivery system is highly promising for the delivery of anti-cancer drugs. They also suggested that the core or shell could potentially be modified by cross-linking in order to further sustain the release.
Gao et al. studied biodegradable mPEG-poly(lactide) copolymer (mPEG-PLA) micelles for curcumin delivery in colon cancer therapy.108 The micelles were prepared by a self-assembly method, and were reported to have a narrow size distribution with an average diameter of 30 nm. In vitro release studies carried out in PBS containing 0.5% w/w Tween-80 at physiological temperature showed that free curcumin was rapidly released reaching maximum release (83%) within 12 hours. However, curcumin-loaded micelles showed more sustained release, reaching approximately 60% curcumin release in the same time period. The authors also reported enhanced uptake and apoptosis of colon cancer cells demonstrated by curcumin-loaded micelles compared to free curcumin alone.
Nanoparticle vehicles for the encapsulation and transportation of curcumin have also been developed. For example, in the formulation THERACURMIN®, curcumin powder and glycerin was added to a solution of polysaccharides from ghatti trees, then the mixture was processed by wet grinding and high-pressure homogenization to produce a stable colloidal dispersion of nanoparticles with diameters of 190 nm.109 In clinical trials, the area under the blood concentration–time curve was found to be 27-fold higher for this formulation than for curcumin powder.109 In addition, in pancreatic or biliary tract cancer patients receiving gemcitabine no increase in adverse effects was observed for THERACURMIN® at a curcumin dose of 200 or 400 mg per day.110
O'Toole et al. have used chitosan-based particles to encapsulate curcumin.111 The authors used a spray drying method to encapsulate curcumin inside chitosan/Tween 20 particles where the ratio of chitosan/Tween 20 was varied. The particle size was noted to be 285 ± 30 nm, with a curcumin encapsulation efficiency of nearly 100%. In release experiments, a burst release profile was observed with all of the curcumin released over a period of 2 hours. While suitable for applications in which a rapid release of curcumin is desired, additional work may be required to prolong the curcumin release for some applications.
Misra and Sahoo have co-encapsulated curcumin with doxorubicin in poly(D,L-lactide-co-glycolide) nanoparticles.112 Doxorubicin is used to treat a variety of cancers, including leukemia; however, a number of cancer cells, including the chronic myeloid leukemia blasts such as K562 cells are resistant to doxorubicin due to its sequestration into cytoplasmic vesicles and the induction of multi-drug resistance (MDR). Along with its other anti-cancer properties, as curcumin has been demonstrated to down-regulate MDR transporters, it was of particular interest to investigate the potential beneficial effects of incorporating both drugs into a single nanoparticle. The particles were prepared by a single emulsion, solvent evaporation technique, which resulted in particles with diameters of ∼250 nm. Incorporation of the drugs into the nanoparticles resulted in ∼8-fold higher uptake than for the free drugs in solution. The dual drug nanoparticle formulation also resulted in increased nuclear retention of doxorubicin. This was found to correspond to lower levels of expression of resistance genes MDR1 and BCl-2 in K562 cells, attributed to curcumin inhibition. Combined, these properties resulted in increased in vitro cytotoxicity for the dual drug nanoparticles in comparison to doxorubicin nanoparticles or the dual drugs in solution.
Curcumin was also encapsulated in another nanoparticle system developed by Mohanty and Sahoo.113 Curcumin was loaded in glycerol monooleate (GMO)/Pluronic F-127 particles using an emulsification technique upon the addition of 0.5% w/v polyvinyl alcohol. The nanoparticles displayed an average diameter of 192 ± 7 nm and had a spherical morphology. HPLC studies revealed an encapsulation efficiency of 90 ± 3%, and in vitro release experiments demonstrated an initial burst of 46% of drug released in 24 hours, after which the remaining drug was released over a period of 10 days. Similar to the cell uptake of curcumin-loaded micelles investigated by Mohanty et al., the authors found that cell uptake was concentration dependent, with lower concentrations of curcumin-loaded nanoparticles exhibiting better cell uptake than unmodified curcumin, in addition to more effective anti-proliferative activity.
Sindhu et al. synthesized spherical gold nanoparticles using curcumin alone as a reducing agent.114 The particles were spherical, with an average size of 58 nm and a zeta potential of −23 mV. The authors reported that the particles were stable at room temperature for up to 6 months, and that they were non-toxic in vitro. Mesoporous silica nanoparticles (MSN) (type MCM-41) have also been used to encapsulate curcumin within the pores of the nanoparticles in order to enhance its solubility.115 The authors demonstrated that the solubility of curcumin encapsulated in the silica nanoparticles (0.53 μg mL−1) was increased by 71% compared to that of curcumin alone (0.31 μg mL−1) and curcumin-MSN physical mixture (0.36 μg mL−1). The authors noted that in vitro curcumin release was much more rapid when encapsulated in the MSN, reaching 29% over 72 hours due to the formation of curcumin nano-aggregates in the pores, compared to 8.9% and 9.0% as demonstrated by curcumin and curcumin-MSN physical mixture, respectively. This resulted in enhanced cytotoxic effects for curcumin-MSN on human breast cancer cells compared to free curcumin and curcumin-MSN physical mixture.
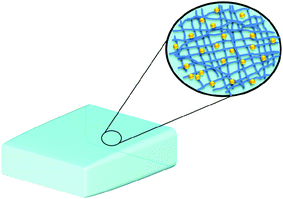
Though not formally nanomaterials in their simplest form, hydrogels, composed of networks of polymer chains, are high water-content materials that can possess strength and flexibility similar to human tissue (Fig. 10).116 Because there exists a large array of water-soluble polymers that can be used for the preparation of hydrogels having a wide variety of physical morphologies (e.g. particles, slabs, and films) and properties, hydrogels can potentially be used for many different biomedical applications including drug delivery. Curcumin-encapsulating hydrogels containing 0.5, 1, or 2 wt% of a 20 amino acid peptide referred to as MAX8, have been synthesized via a self-assembly method upon the addition of salt solution buffered to pH 7.4 or cell culture medium at the same pH, where curcumin-encapsulation and hydrogel formation occurs concurrently.117 Hydrogels containing curcumin displayed solid-like properties even after shear thinning, where they were capable of re-healing quickly according to the oscillatory rheology and shear stiffness study conducted. This suggested their potential as injectable materials for localized curcumin delivery. The release of curcumin occurred over a period of 14 days and could be modulated to some extent as a function of the MAX8 peptide concentration. In addition, through in vitro experiments, it was demonstrated that the presence of curcumin in the hydrogels inhibited the growth of human medulloblastoma cells on the hydrogel. The bioactivity of the released curcumin was also confirmed by its ability to inhibit growth of the same cell line.
An emerging field of hydrogels, called “smart” hydrogels have gained immense interest. Smart hydrogels are capable of dramatically changing their properties in response to stimuli such as temperature, pH, and chemicals.118 Chen et al. reported a curcumin-loaded thermosensitive hydrogel for brain targeting applications through intranasal administration.119 Hydrogels were synthesized using Pluronic F127 and Poloxamer 188. The curcumin-loaded hydrogels underwent sol–gel transition in the temperature range 32–35 °C, therefore undergoing gelation at physiological temperatures. In vitro release studies revealed that 80% of curcumin was released within 6 hours, and in vivo studies showed that curcumin-loaded hydrogels took approximately one hour to pass from the nasal cavity of rats to their oropharynx. In addition, brain tissue distribution analyses showed the presence of curcumin in the cerebellum, cerebrum, hippocampus, olfactory bulb, and plasma after the administration of 250 μg kg−1 of the nasal hydrogel. This suggests the application of these curcumin hydrogels for targeting of brain tumours.
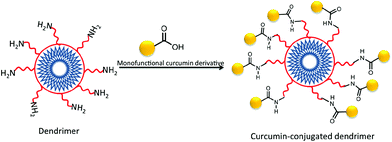
In addition to assemblies of phospholipids and polymers into which curcumin can be physically encapsulated, dendrimers have also been widely studied for drug delivery and have often been used for the preparation of covalent drug conjugates.120 Dendrimers are highly branched polymers with precise architectures that result from their step-wise synthesis.121 Because these structures are prepared on a generation-by-generation basis, they can be tailored for use in many applications. Shi et al. used dendrimers for the enhancement of curcumin's bioavailability and its effects in the dissolution of amyloid fibrils.122 The group produced monofunctional derivatives of curcumin where one of the phenolic groups of curcumin was modified with azide, alkyne, or carboxylic acid. These monofunctional derivatives of curcumin were then used to produce other forms of curcumin, among them a polyamidoamine (PAMAM) dendrimer–curcumin conjugate (Fig. 11). The PAMAM–curcumin conjugate was synthesized using a fourth generation PAMAM dendrimer with a cystamine core and amine surface groups. Curcumin monocarboxylic acid was coupled to the amine termini using 1,3-dicyclohexyl-carbodiimide, N-hydroxysuccinimide, and triethanolamine. The biological activity of curcumin was not disrupted by its chemical modification as reported; in fact the chemical properties of curcumin were enhanced. The water-soluble PAMAM–curcumin conjugate was able to stain and dissolve amyloid fibrils in vitro. The enhanced water solubility of curcumin by attachment to dendrimers suggests promising therapeutic applications.
In another study, with the aim of understanding curcumin's wound healing properties despite its rapid degradation in aqueous solution, the stabilization of curcumin by four different plasma proteins, human serum albumin (HSA), fibrinogen, immunoglobulin G (IgG), and transferrin was investigated.123 It was reported that curcumin degradation was suppressed with a yield of 94.2 ± 13.9% when stabilized with HSA, and 93.6 ± 14.0% with fibrinogen. Transferrin did not suppress curcumin degradation as efficiently (47.8 ± 14.1%), and IgG had no effect on degradation. The impressive degradation suppression of HSA and fibrinogen were attributed to the hydrophobic interactions between the proteins and curcumin, where binding of curcumin takes place in the hydrophobic pocket in the protein. This eliminates interactions with surrounding water molecules inhibiting curcumin degradation by hydrolysis.
With the potential application of the wide range of above vehicles as drug delivery systems come several challenges. Vesicles often require complex preparation procedures, and are often too large for drug delivery applications. The large size (>200 nm) results in clearance by the reticuloendothelial system (RES) thereby eliminating the drug from the circulatory system.124 Nanoparticles are also often too large to exhibit long circulation times in vivo, frequently having diameters >200 nm. In addition, they have some tendency to exhibit an undesirable “burst” release of the drug. Micelles on the other hand are smaller, and can sometimes exhibit long circulation times in vivo. However, as they are prepared by self-assembly approaches, their behaviour in vivo in the presence of membranes and hydrophobic serum proteins, will not always be the same as their in vitro behaviour in simpler model systems. In addition, the metabolic fate of these carriers is often difficult to determine, and some may not be biodegradable. Nevertheless, the work this far has demonstrated the great potential of drug delivery systems to overcome some of curcumin's problematic properties such as its extremely low aqueous solubility, stability, and bioavailability.
Conclusions and future prospects
Although there is still much to be investigated with respect to curcumin and its therapeutic properties, it has great potential in cancer therapy and prevention. Curcumin is proposed to exert its anti-cancer effects through a variety of mechanisms including the down-regulation of NF-κB, the suppression of AP-1 activity, various effects on STAT proteins, as well as other mechanisms. The controversy regarding curcumin's chemical structure in aqueous solution at neutral pH appears to be solved, with the dominance of the keto-enol form well supported by NMR spectroscopy as well as theoretical studies. However, there is still debate as to whether it is curcumin itself or its degradation products that provide the observed biological activities. This issue highlights one of the main challenges in working with curcumin – its susceptibility to rapid degradation in a wide range of environments and its sensitivity to pH and light. Other major challenges include curcumin's extremely low aqueous solubility and bioavailability. While curcumin has been safely administered at high doses in many clinical trials, has achieved the status of GRAS, and is marketed as a dietary supplement in many countries, it has yet to be licensed as a drug, likely due to the above-mentioned limitations as well as its multi-target property.To address the solubility and bioavailability limitations, a variety of approaches have been explored to improve its solubility and to deliver it to cancer cells. The formation of co-crystals with a variety of molecules has resulted in significant enhancements in the curcumin dissolution rate. Liposomes, micelles, nanoparticles, and other delivery systems have generally led to enhanced curcumin solubility, stability, cell uptake, and improved biological activities, and are starting to be evaluated in clinical trials.
Despite the immense progress made on the study and application of curcumin over the last couple of decades, many questions and challenges still exist. For example, it will be critical to determine whether the biological properties of curcumin indeed result from the parent compound or from its degradation products. This is important not only for the fundamental understanding of this drug but also because the drug delivery systems tend to stabilize curcumin, an advantage that only persists if this is the bioactive molecule. It will also be important to continue the development of viable drug delivery systems for curcumin. Here, the critical aspects will involve the enhancement of curcumin solubility and bioavailability, as well as approaches for selectively targeting these delivery systems to the disease sites. With this promising progression, the use of curcumin for the treatment of cancer and other diseases can be envisioned in the near future.
References
- H. Hatcher, R. Planalp, J. Cho, F. M. Torti and S. V. Torti, Cell. Mol. Life Sci., 2008, 65, 1631 CrossRef CAS PubMed
.
- B. K. Tiwari and G. P. Agrawal, World J. Pharm. Pharm. Sci., 2012, 1, 161 CAS
.
- B. B. Aggarwal, C. Sundaram, N. Malani and H. Ichikawa, Adv. Exp. Med. Biol., 2007, 595, 1 CrossRef
.
- B. B. Aggarwal and S. Shishodia, Ann. N. Y. Acad. Sci., 2004, 1030, 434 CrossRef CAS PubMed
.
- F. Payton, P. Sandusky and W. L. Alworth, J. Nat. Prod., 2007, 70, 143 CrossRef CAS PubMed
.
- H. H. Tonnesen, J. Karlsen and A. Mostad, Acta Chem. Scand., Ser. B, 1982, 36, 475 CrossRef PubMed
.
- S. V. Jovanovic, S. Steenken, C. W. Boone and M. G. Simic, J. Am. Chem. Soc., 1999, 121, 9677 CrossRef CAS
.
- L. Shen and H. F. Ji, Spectrochim. Acta, Part A, 2007, 67, 619 CrossRef PubMed
.
- H. H. Tonnesen and J. Karlsen, Z. Lebensm.-Unters. Forsch., 1985, 180, 402 CrossRef CAS
.
- Y. J. Wang, M. H. Pan, A. L. Cheng, L. I. Lin, Y. S. Ho, C. Y. Hsieh and J. K. Lin, J. Pharm. Biomed. Anal., 1997, 15, 1867 CrossRef CAS
.
- D. Gopinath, M. R. Ahmed, K. Gomathi, K. Chitra, P. K. Sehgal and R. Jayakumar, Biomaterials, 2004, 25, 1911 CrossRef CAS
.
- K. N. Liu, C. M. Lai, Y. T. Lee, S. N. Wang, R. P. Chen, J. S. Jan, H. S. Liu and S. S. Wang, Biochim. Biophys. Acta, Gen. Subj., 2012, 1820, 1774 CrossRef CAS PubMed
.
- L. Shen and H. F. Ji, Trends Mol. Med., 2012, 18, 138 CrossRef CAS PubMed
.
- G. W. Schieffer, J. Liq. Chromatogr. Relat. Technol., 2002, 25, 3033 CrossRef PubMed
.
- U. Singh, S. Verma, H. N. Ghosh, M. C. Rath, K. I. Priyadarsini, A. Sharma, K. K. Pushpa, S. K. Sarkar and T. Mukherjee, J. Mol. Catal. A: Chem., 2010, 318, 106 CrossRef CAS PubMed
.
- K. Lirdprapamongkol, H. Sakurai, S. Suzuki, K. Koizumi, O. Prangsaengtong, A. Viriyaroj, S. Ruchirawat, J. Svasti and I. Saiki, In Vivo, 2010, 24, 501 CAS
.
- S. L. Wu, J. C. Chen, C. C. Li, H. Y. Lo, T. Y. Ho and C. Y. Hsiang, J. Pharmacol. Exp. Ther., 2009, 330, 370 CrossRef CAS PubMed
.
- C. Zhang, X. Li, L. Lian, Q. Chen, O. Abdulmalik, V. Vassilev, C. S. Lai and T. Asakura, Br. J. Haematol., 2004, 125, 788 CrossRef CAS PubMed
.
- O. N. Gordon and C. Schneider, Trends Mol. Med., 2012, 18, 361–363 CrossRef CAS PubMed
.
- M. Griesser, V. Pistis, T. Suzuki, N. Tejera, D. A. Pratt and C. Schneider, J. Biol. Chem., 2011, 286, 1114 CrossRef CAS PubMed
.
- A. C. Ketron, O. N. Gordon, C. Schneider and N. Osheroff, Biochemistry, 2013, 52, 221 CrossRef CAS PubMed
.
- T. G. Burish and R. A. Jenkins, Health Psychol., 1992, 11, 17 CrossRef CAS
.
- W. M. Lee, N. Engl. J. Med., 1993, 329, 1862 CrossRef CAS PubMed
.
- T. Dorai and B. B. Aggarwal, Cancer Lett., 2004, 215, 129 CrossRef CAS PubMed
.
- A. Shehzad, J. Lee and Y. S. Lee, BioFactors, 2013, 39, 56 CrossRef CAS PubMed
.
- S. C. Gupta, G. Kismali and B. B. Aggarwal, BioFactors, 2013, 39, 2 CrossRef CAS PubMed
.
- J. Epstein, I. R. Sanderson and T. T. Macdonald, Br. J. Nutr., 2010, 103, 1545 CrossRef CAS PubMed
.
- S. C. Gupta, S. Prasad, J. H. Kim, S. Patchva, L. J. Webb, I. K. Priyadarsini and B. B. Aggarwal, Nat. Prod. Rep., 2011, 28, 1937 RSC
.
- A. B. Kunnumakkara, P. Anand and B. B. Aggarwal, Cancer Lett., 2008, 269, 199 CrossRef CAS PubMed
.
- A. Shehzad and Y. S. Lee, BioFactors, 2013, 39, 27 CrossRef CAS PubMed
.
- P. M. Luthra, R. Kumar and A. Prakash, Biochem. Biophys. Res. Commun., 2009, 384, 420 CrossRef CAS PubMed
.
- L. D. Guo, X. J. Chen, Y. H. Hu, Z. J. Yu, D. Wang and J. Z. Liu, Phytother. Res., 2013, 27, 422 CrossRef CAS PubMed
.
- J. W. Kaspar, S. K. Niture and A. K. Jaiswal, Free Radical Biol. Med., 2009, 47, 1304 CrossRef CAS PubMed
.
- S. K. Niture, J. W. Kaspar, J. Shen and A. K. Jaiswal, Toxicol. Appl. Pharmacol., 2010, 244, 37 CrossRef CAS PubMed
.
- A. M. Pickering, R. A. Linder, H. Zhang, H. J. Forman and K. J. Davies, J. Biol. Chem., 2012, 287, 10021 CrossRef CAS PubMed
.
- S. Shishodia, BioFactors, 2013, 39, 37 CrossRef CAS PubMed
.
- M. Youns and G. M. Fathy, J. Cell. Biochem., 2013, 114, 2654 CrossRef CAS PubMed
.
- S. T. Wua, J.-C. Sun, K.-J. Lee and Y.-M. Sun, Int. J. Eng. Sci. Technol., 2010, 2, 4263 Search PubMed
.
- B. S. Vinod, J. Antony, H. H. Nair, V. T. Puliyappadamba, M. Saikia, S. S. Narayanan, A. Bevin and R. J. Anto, Cell Death Dis., 2013, 4, e505 CrossRef CAS PubMed
.
- J. Hess, P. Angel and M. Schorpp-Kistner, J. Cell Sci., 2004, 117, 5965 CrossRef CAS PubMed
.
- E. F. Wagner, Ann. Rheum. Dis., 2002, 61(suppl 2), ii40-2 Search PubMed
.
- A. Bierhaus, Y. Zhang, P. Quehenberger, T. Luther, M. Haase, M. Muller, N. Mackman, R. Ziegler and P. P. Nawroth, Thromb. Haemostasis, 1997, 77, 772 CAS
.
- S. Nafisi, M. Adelzadeh, Z. Norouzi and M. N. Sarbolouki, DNA Cell Biol., 2009, 28, 201 CrossRef CAS PubMed
.
- S. S. Han, Y. S. Keum, H. J. Seo and Y. J. Surh, J. Biochem. Mol. Biol., 2002, 35, 337 CrossRef CAS
.
- S. Balasubramanian and R. L. Eckert, J. Biol. Chem., 2007, 282, 6707 CrossRef CAS PubMed
.
- C. S. Divya and M. R. Pillai, Mol. Carcinog., 2006, 45, 320 CrossRef CAS PubMed
.
- K. Nakamura, Y. Yasunaga, T. Segawa, D. Ko, J. W. Moul, S. Srivastava and J. S. Rhim, Int. J. Oncol., 2002, 21, 825 CAS
.
- M. H. Teiten, F. Gaascht, S. Eifes, M. Dicato and M. Diederich, Genes Nutr., 2010, 5, 61 CrossRef CAS PubMed
.
- C. Polytarchou, M. Hatziapostolou and E. Papadimitriou, J. Biol. Chem., 2005, 280, 40428 CrossRef CAS PubMed
.
- T. Decker, Jak-Stat Signaling:From Basics to Disease. Springer, Vienna, 2012 Search PubMed
.
- P. Kroon, P. A. Berry, M. J. Stower, G. Rodrigues, V. M. Mann, M. Simms, D. Bhasin, S. Chettiar, C. Li, P. K. Li, N. J. Maitland and A. T. Collins, Cancer Res., 2013, 73, 5288 CrossRef CAS PubMed
.
- C. L. Yang, Y. Y. Liu, Y. G. Ma, Y. X. Xue, D. G. Liu, Y. Ren, X. B. Liu, Y. Li and Z. Li, PLoS One, 2012, 7, e37960 CAS
.
- J. Weissenberger, M. Priester, C. Bernreuther, S. Rakel, M. Glatzel, V. Seifert and D. Kogel, Clin. Cancer Res., 2010, 16, 5781 CrossRef CAS PubMed
.
- M. Ono, T. Higuchi, M. Takeshima, C. Chen and S. Nakano, Anticancer Res., 2013, 33, 1861 CAS
.
- J. Rodon, R. Dienstmann, V. Serra and J. Tabernero, Nat. Rev. Clin. Oncol., 2013, 10, 143 CrossRef CAS PubMed
.
- K. T. Nguyen, M. P. Holloway and R. A. Altura, Int. J. Biochem. Mol. Biol., 2012, 3, 137 CAS
.
- M. Niu, S. Wu, L. Mao and Y. Yang, Traffic, 2013, 14, 1042 CrossRef CAS PubMed
.
- S. Park, D. H. Cho, L. Andera, N. Suh and I. Kim, Mol. Cell. Biochem., 2013, 383, 39 CrossRef CAS PubMed
.
- X. Gao, B. W. Loggie and Z. Nawaz, Mol. Cancer, 2002, 1, 7 CrossRef PubMed
.
- H. Ohtsu, Z. Xiao, J. Ishida, M. Nagai, H. K. Wang, H. Itokawa, C. Y. Su, C. Shih, T. Chiang, E. Chang, Y. Lee, M. Y. Tsai, C. Chang and K. H. Lee, J. Med. Chem., 2002, 45, 5037 CrossRef CAS PubMed
.
- L. Bartik, G. K. Whitfield, M. Kaczmarska, C. L. Lowmiller, E. W. Moffet, J. K. Furmick, Z. Hernandez, C. A. Haussler, M. R. Haussler and P. W. Jurutka, J. Nutr. Biochem., 2010, 21, 1153 CrossRef CAS PubMed
.
- J. D. Altenburg, A. A. Bieberich, C. Terry, K. A. Harvey, J. F. Vanhorn, Z. Xu, V. Jo Davisson and R. A. Siddiqui, BMC Cancer, 2011, 11, 149 CrossRef CAS PubMed
.
- M. A. Tessel, N. L. Krett and S. T. Rosen, Curr. Opin. Oncol., 2010, 22, 592 CrossRef CAS PubMed
.
- A. K. Garg, T. A. Buchholz and B. B. Aggarwal, Antioxid. Redox Signaling, 2005, 7, 1630 CrossRef CAS PubMed
.
- P. Limtrakul, Adv. Exp. Med. Biol., 2007, 595, 269 CrossRef
.
- G. C. Jagetia, Adv. Exp. Med. Biol., 2007, 595, 301 CrossRef
.
- N. Sebastià, A. Montoro, A. Montoro, M. Almonacid, J. I. Villaescusa, J. Cervera, E. Such, M. A. Silla and J. M. Soriano, Radiat. Meas., 2011, 46, 962 CrossRef PubMed
.
- M. Sun, Z. Estrov, Y. Ji, K. R. Coombes, D. H. Harris and R. Kurzrock, Mol. Cancer Ther., 2008, 7, 464 CrossRef CAS PubMed
.
- S. Ali, A. Ahmad, A. Aboukameel, B. Bao, S. Padhye, P. A. Philip and F. H. Sarkar, Cancer Lett., 2012, 319, 173 CrossRef CAS PubMed
.
- S. Ali, A. Ahmad, S. Banerjee, S. Padhye, K. Dominiak, J. M. Schaffert, Z. Wang, P. A. Philip and F. H. Sarkar, Cancer Res., 2010, 70, 3606 CrossRef CAS PubMed
.
- M. Kakarala, D. E. Brenner, H. Korkaya, C. Cheng, K. Tazi, C. Ginestier, S. Liu, G. Dontu and M. S. Wicha, Breast Cancer Res. Treat., 2010, 122, 777 CrossRef CAS PubMed
.
- K. J. Lim, S. Bisht, E. E. Bar, A. Maitra and C. G. Eberhart, Cancer Biol. Ther., 2011, 11, 464 CrossRef CAS
.
- L. Lin, Y. Liu, H. Li, P. K. Li, J. Fuchs, H. Shibata, Y. Iwabuchi and J. Lin, Br. J. Cancer, 2011, 105, 212 CrossRef CAS PubMed
.
- Y. Ohsumi, Nat. Rev. Mol. Cell Biol., 2001, 2, 211 CrossRef CAS PubMed
.
- Y. Yamauchi, Y. Izumi, K. Asakura, Y. Hayashi and H. Nomori, Phytother. Res., 2012, 26, 1779 CrossRef CAS PubMed
.
- J. Y. Kim, T. J. Cho, B. H. Woo, K. U. Choi, C. H. Lee, M. H. Ryu and H. R. Park, Arch. Oral Biol., 2012, 57, 1018 CrossRef CAS PubMed
.
- H. Aoki, Y. Takada, S. Kondo, R. Sawaya, B. B. Aggarwal and Y. Kondo, Mol. Pharmacol., 2007, 72, 29 CrossRef CAS PubMed
.
- W. Zhuang, L. Long, B. Zheng, W. Ji, N. Yang, Q. Zhang and Z. Liang, Cancer Sci., 2012, 103, 684 CrossRef CAS PubMed
.
- S. C. Gupta, S. Patchva and B. B. Aggarwal, AAPS J., 2013, 15, 195 CrossRef CAS PubMed
.
- Z.-Y. He, C.-B. Shi, H. Wen, F.-L. Li, B.-L. Wang and J. Wang, Cancer Invest., 2011, 29, 208 CrossRef CAS PubMed
.
- R. E. Carroll, R. V. Benya, D. K. Turgeon, S. Vareed, M. Neuman, L. Rodriguez, M. Kakarala, P. M. Carpenter, C. McLaren, F. L. Meyskens Jr and D. E. Brenner, Cancer Prev. Res., 2011, 4, 354 CrossRef CAS PubMed
.
- G. R. Irving, L. M. Howells, S. Sale, I. Kralj-Hans, W. S. Atkin, S. K. Clark, R. G. Britton, D. J. Jones, E. N. Scott, D. P. Berry, D. Hemingway, A. S. Miller, K. Brown, A. J. Gescher and W. P. Steward, Cancer Prev. Res., 2013, 6, 119 CrossRef CAS PubMed
.
- R. Epelbaum, M. Schaffer, B. Vizel, V. Badmaev and G. Bar-Sela, Nutr. Cancer, 2010, 62, 1137 CrossRef CAS PubMed
.
- M. Kanai, K. Yoshimura, M. Asada, A. Imaizumi, C. Suzuki, S. Matsumoto, T. Nishimura, Y. Mori, T. Masui, Y. Kawaguchi, K. Yanagihara, S. Yazumi, T. Chiba, S. Guha and B. Aggarwal, Cancer Chemother. Pharmacol., 2011, 68, 157 CrossRef CAS PubMed
.
- M. Bayet-Robert, F. Kwiatkowski, M. Leheurteur, F. Gachon, E. Planchat, C. Abrial, M. A. Mouret-Reynier, X. Durando, C. Barthomeuf and P. Chollet, Cancer Biol. Ther., 2010, 9, 8 CrossRef CAS
.
- S. Vadhan-Raj, D. Weber, M. Wang, S. Giralt, R. Alexanian, S. Thomas, X. Zhou, P. Patel, C. Bueso-Ramos, R. Newman and B. Aggarwal, Blood, 2007, 110, 357a Search PubMed
.
- A. Shehzad, F. Wahid and Y. S. Lee, Arch. Pharm., 2010, 343, 489 CrossRef CAS PubMed
.
- G. Liang, Curr. Pharm. Des., 2013, 19, 1965 CAS
.
- Y. Gao, Z. Li, M. Sun, C. Guo, A. Yu, Y. Xi, J. Cui, H. Lou and G. Zhai, Drug Delivery, 2011, 18, 131 CrossRef CAS PubMed
.
- N. R. Goud, K. Suresh, P. Sanphui and A. Nangia, Int. J. Pharm., 2012, 439, 63 CrossRef CAS PubMed
.
- P. Anand, A. B. Kunnumakkara, R. A. Newman and B. B. Aggarwal, Mol. Pharmaceutics, 2007, 4, 807 CrossRef CAS PubMed
.
- P. Basnet and N. Skalko-Basnet, Molecules, 2011, 16, 4567 CrossRef CAS PubMed
.
- P. Sanphui, N. R. Goud, U. B. R. Khandavilli and A. Nangia, Cryst. Growth Des., 2011, 11, 4135 CAS
.
- S. K. Vareed, M. Kakarala, M. T. Ruffin, J. A. Crowell, D. P. Normolle, Z. Djuric and D. E. Brenner, Cancer Epidemiol., Biomarkers Prev., 2008, 17, 1411 CrossRef CAS PubMed
.
- C. Ireson, S. Orr, D. J. Jones, R. Verschoyle, C. K. Lim, J. L. Luo, L. Howells, S. Plummer, R. Jukes, M. Williams, W. P. Steward and A. Gescher, Cancer Res., 2001, 61, 1058 CAS
.
- M. H. Pan, T. M. Huang and J. K. Lin, Drug Metab. Dispos., 1999, 27, 486 CAS
.
- R. A. Sharma, S. A. Euden, S. L. Platton, D. N. Cooke, A. Shafayat, H. R. Hewitt, T. H. Marczylo, B. Morgan, D. Hemingway, S. M. Plummer, M. Pirmohamed, A. J. Gescher and W. P. Steward, Clin. Cancer Res., 2004, 10, 6847 CrossRef CAS PubMed
.
- G. Shoba, D. Joy, T. Joseph, M. Majeed, R. Rajendran and P. S. S. R. Srinivas, Planta Med., 1998, 64, 353 CrossRef CAS PubMed
.
- J. Lu and S. Rohani, J. Pharm. Sci., 2010, 99, 4042 CAS
.
- D. D. Lasic and D. Needham, Chem. Rev., 1995, 95, 2601 CrossRef CAS
.
- T. M. Allen and P. R. Cullis, Science, 2004, 303, 1818 CrossRef CAS PubMed
.
- L. Li, F. S. Braiteh and R. Kurzrock, Cancer, 2005, 104, 1322 CrossRef CAS PubMed
.
- N. Saengkrit, S. Saesoo, W. Srinuanchai, S. Phunpee and U. R. Ruktanonchai, Colloids Surf., B, 2014, 114, 349 CrossRef CAS PubMed
.
- C. Théry, L. Zitvogel and S. Amigorena, Nat. Rev. Immunol., 2002, 2, 569 Search PubMed
.
- D. Sun, X. Zhuang, X. Xiang, Y. Liu, S. Zhang, C. Liu, S. Barnes, W. Grizzle, D. Miller and H. G. Zhang, Mol. Ther., 2010, 18, 1606 CrossRef CAS PubMed
.
- C. Mohanty, S. Acharya, A. K. Mohanty, F. Dilnawaz and S. K. Sahoo, Nanomedicine, 2010, 5, 433 CrossRef CAS PubMed
.
- S. Podaralla, R. Averineni, M. Alqahtani and O. Perumal, Mol. Pharmaceutics, 2012, 9, 2778 CrossRef CAS PubMed
.
- X. Gao, F. Zheng, G. Guo, X. Liu, R. Fan, Z.-y. Qian, N. Huang and Y.-q. Wei, J. Mater. Chem. B, 2013, 1, 5778 RSC
.
- H. Sasaki, Y. Sunagawa, K. Takahashi, A. Imaizumi, H. Fukuda, T. Hashimoto, H. Wada, Y. Katanasaka, H. Kakeya, M. Fujita, K. Hasegawa and T. Morimoto, Biol. Pharm. Bull., 2011, 34, 660 CAS
.
- M. Kanai, Y. Otsuka, K. Otsuka, M. Sato, T. Nishimura, Y. Mori, M. Kawaguchi, E. Hatano, Y. Kodama, S. Matsumoto, Y. Murakami, A. Imaizumi, T. Chiba, J. Nishihira and H. Shibata, Cancer Chemother. Pharmacol., 2013, 71, 1521 CrossRef CAS PubMed
.
- M. G. O'Toole, R. M. Henderson, P. A. Soucy, B. H. Fasciotto, P. J. Hoblitzell, R. S. Keynton, W. D. Ehringer and A. S. Gobin, Biomacromolecules, 2012, 13, 2309 CrossRef CAS PubMed
.
- R. Misra and S. K. Sahoo, Mol. Pharmaceutics, 2011, 8, 852 CrossRef CAS PubMed
.
- C. Mohanty and S. K. Sahoo, Biomaterials, 2010, 31, 6597 CrossRef CAS PubMed
.
- K. Sindhu, A. Rajaram, K. J. Sreeram and R. Rajaram, RSC Adv., 2014, 4, 1 RSC
.
- S. Jambhrunkar, S. Karmakar, A. Popat, M. Yu and C. Yu, RSC Adv., 2014, 4, 709 RSC
.
- T. R. Hoare and D. S. Kohane, Polymer, 2008, 49, 1993 CrossRef CAS PubMed
.
- A. Altunbas, S. J. Lee, S. A. Rajasekaran, J. P. Schneider and D. J. Pochan, Biomaterials, 2011, 32, 5906 CrossRef CAS PubMed
.
- L.-W. Xia, R. Xie, X.-J. Ju, W. Wang, Q. Chen and L.-Y. Chu, Nat. Commun., 2013, 4, 3226/1 CAS
.
- X. Chen, F. Zhi, X. Jia, X. Zhang, R. Ambardekar, Z. Meng, A. R. Paradkar, Y. Hu and Y. Yang, J. Pharm. Pharmacol., 2013, 65, 807 CrossRef CAS PubMed
.
- S. Svenson, Eur. J. Pharm. Biopharm., 2009, 71, 445 CrossRef CAS PubMed
.
- J. M. J. Fréchet and D. A. Tomalia, Dendrimers and Other Dendritic Polymers, Wiley and Sons, Chichester and New York, 2001 Search PubMed
.
- W. Shi, S. Dolai, S. Rizk, A. Hussain, H. Tariq, S. Averick, W. L'Amoreaux, A. El Idrissi, P. Banerjee and K. Raja, Org. Lett., 2007, 9, 5461 CrossRef CAS PubMed
.
- M. H. M. Leung and T. W. Kee, Langmuir, 2009, 25, 5773 CrossRef CAS PubMed
.
- B. Wang, X. He, Z. Zhang, Y. Zhao and W. Feng, Acc. Chem. Res., 2012, 46, 761 CrossRef PubMed
.
| This journal is © The Royal Society of Chemistry 2014 |