Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
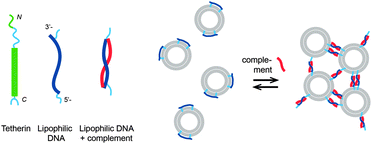
DNA-controlled aggregation of virus like particles – mimicking a tetherin-like mechanism†
Daniela
Serien
ab,
Christiane
Grimm
a,
Jürgen
Liebscher
cd,
Andreas
Herrmann
a and
Anna
Arbuzova
*a
aHumboldt-Universität zu Berlin, Institut für Biologie, Invalidenstr. 42, 10115 Berlin, Germany. E-mail: arbuzova@cms.hu-berlin.de
bThe University of Tokyo, Institute of Industrial Science, 4-6-1 Komaba, Meguro-ku, Tokyo 153-8505, Japan
cHumboldt-Universität zu Berlin, Institut für Chemie, Brook-Taylor-Str. 2, 12489 Berlin, Germany
dNational Institute of Research and Development for Isotopic and Molecular Technologies, Donath 65-103, Cluj-Napoca, Romania
First published on 24th June 2014
Abstract
Mimicking cellular processes by functional comparable structures helps to understand their molecular mechanism. We report on an oligonucleotide modified with two α-tocopherol anchors mimicking tetherin, a cellular protein reducing spreading of viruses. The lipophilic DNA was incorporated into vesicles and virus like particles; their aggregation was induced by the complement addition.
Nucleic acid is a versatile tool for bionanotechnology:1–3 DNA-controlled assembly of structures is used to fabricate new materials and for development of drug delivery systems.4–7 Recently, Chou et al. demonstrated that superstructures (built through DNA-controlled assembly of colloidal particles) showed improved accumulation in tumor.5 The conjugation of a nucleic acid to a hydrophobic anchor allows attaching a molecular recognition unit to a lipid membrane.8–10 Controlled assembly of vesicles, nanodiscs, and lipid layers, as well as hybridization-induced aggregation and fusion of lipid vesicles carrying lipophilic modifications were reported.11–20
Lipophilic nucleic acids have been used to mimic protein functions, e.g. channels21 and SNARE proteins mediating fusion.13,14,22 Here we suggest using a lipophilic oligonucleotide with two anchors to mimic tetherin/BST-2/CD317 function. Tetherin is an unusual cell protein with two membrane anchors – a transmembrane domain close to the N- and a glycophosphatidylinositol (GPI) anchor at the C-terminus; it was found on the cell surface and in endosomes. It is constitutively expressed in many human organs and cell lines, e.g. in primary T lymphocytes and macrophages.23,24 Tetherin was found to prevent the release of enveloped viruses from infected cells.25 Viruses budding from the infected cells expressing tetherin stay first connected to the cell surface via tetherin, then are taken up and digested.25,26 Tetherin-based chains of the viruses attached to the cell surface were observed by immunoelectron microscopy.27 It was suggested that one of the membrane anchors of the tetherin stays in the cellular and the other is incorporated into the viral membrane.25,26 This mechanism is reminiscent of the mechanism suggested for the aggregation of vesicles induced by lipophilic DNA (Scheme 1).11,12 A lipophilic DNA with a hydrophobic anchor at each end binds first to the membrane of one liposome, as tetherin is incorporated into the plasma membrane via both N- and C-termini. Addition of the complementary DNA strand leads to the formation of a rigid DNA double helix (between the anchors). As the helix becomes oriented perpendicular to the lipid membrane driven by entropy, one of the anchors is pulled out of the membrane of the initial vesicle and might insert into another lipid vesicle.11,12 Indeed, vesicles aggregated upon addition of lipophilic oligonucleotides of 17–27 nucleic bases (about 2–3 helical turns) modified with cholesteryl or palmitoyl anchors at both ends and the unmodified complementary strands.11,12
Using α-tocopherol-modified thymine phosphoamidides, we have shown that the lipophilic anchor can be incorporated at any position of an oligonucleotide.28,29 We observed that α-tocopherol-modified oligonucleotides incorporated spontaneously into preformed lipid vesicles within seconds, preferentially into the liquid-disordered phase, without significant perturbation of the lipid bilayer recommending such lipophilic oligonucleotides for functionalization of biological membranes.30 Oligonucleotides modified with α-tocopherol at the terminal phosphate moiety were recently shown to incorporate spontaneously into cell membranes.31 Here, we report investigations of a lipophilic oligonucleotide with two α-tocopherol anchors (L) separated by 18 bases and having additional 5 bases after the lipid anchor closer to the 3′end, TL(dT)18L(dT)5, termed LiNA. Sequences of the oligonucleotides are provided in Table 1.
| Name | Sequence |
|---|---|
| TL(dT)18L(dT)5 | 5′-TLT TTT TTT TTT TTT TTT TTL TTT TT-3′ |
| (dA)20 | 5′-AAA AAA AAA AAA AAA AAA AA-3′ |
| (dT)20 | 5′-TTT TTT TTT TTT TTT TTT TT-3′ |
| 3′Rh(dA)20 | 5′-AAA AAA AAA AAA AAA AAA AA_TMR-3′ |
| 3′Rh(dT)20 | 5′-TTT TTT TTT TTT TTT TTT TT_TMR-3′ |
The tocopherol anchors were attached to the nucleobase29,32,33 rather than to the terminal phosphate moiety.31,34 The structure of the anchor is shown in the ESI.† We characterized the LiNAs and their potential to function in a tetherin-like manner with model lipid vesicles and then verified it on HIV virus like particles (VLPs). Lacking regulatory proteins and genetic material, VLPs resemble morphologically native viruses but are noninfectious.35
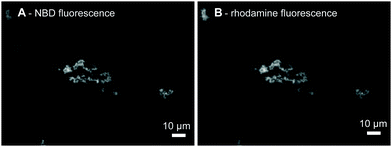
We first show using fluorescence microscopy that the LiNA induced aggregation of model vesicles triggered by hybridization with a fluorescently labeled complementary strand, 3′Rh(dA)20. Upon mixing of the LiNA, its complement (200 nM each), and POPC liposomes labeled with the fluorescent lipid analogue N-NBD-DPPE (400 μM total lipid), micrometer size aggregates were formed (Fig. 1A). For such high lipid concentrations, the resulting aggregates became visible even by the naked eye after a few minutes and precipitated if left undisturbed for several hours. Apparent co-localization of N-NBD and rhodamine signals revealed that in the presence of the LiNA 3′Rh(dA)20 was accumulated within the aggregates (Fig. 1A and B). If either of the oligonucleotides was absent, no aggregation was observed: in the absence of LiNA rhodamine fluorescence was homogeneously distributed as shown in Fig. S1A, ESI.† Addition of a non-complementary strand neither induced any aggregation of the vesicles. The labeled non-complementary strands, 3′Rh(dT)20, and the N-NBD-DPPE labeled vesicles were also distributed homogeneously in the presence and in the absence of the LiNA (Fig. S1B, ESI,† and not shown, respectively). Thus, hybridization of the LiNA and the complementary strand, 3′Rh(dA)20, induced aggregation of lipid vesicles. We presume that in agreement with the mechanism suggested before11,12,36 addition of 3′Rh(dA)20 led to the formation of a rigid DNA helix between the anchors; as a consequence one of the α-tocopherol anchors was pulled out of the vesicle, where the LiNA was incorporated before, and could insert into another lipid vesicle.
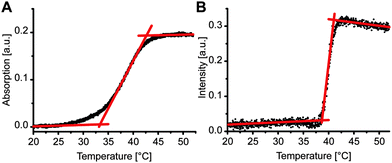
We compared transition temperature of the LiNA with unlabeled complementary strand, (dA)20, in the absence and the presence of POPC liposomes. In the absence of liposomes, dehybridization of LiNA/(dA)20 was measured by following the increase in absorbance at 270 nm upon heating (Fig. 2A). The melting temperature, at which half of the hybrids de-hybridized, was about 38 °C. This melting temperature value indicates that according to the empirically based prediction algorithm 17 to 18 bases of TL(dT)18L(dT)5 in between the two lipid anchors are involved in DNA hybridization (e.g. OligoAnalyzer; Integrated DNA Technologies). For samples with vesicles, LiNA, and (dA)20, a well-defined sharp transition, corresponding to the disassembly of the vesicle aggregates, was observed at 39.9 ± 0.2 °C by light scattering (Fig. 2B).
The width of the transition of 2.3 °C was much smaller than that of 10 °C observed in the absence of vesicles. Similarly, a smaller width for the disaggregation of DNA-linked 13 nm gold particles in comparison to that measured for unmodified DNA was reported previously and explained by the higher DNA density on the particles.37 The sharp transition observed here can be explained by the effect of multiple connections between the vesicles (combinatorial entropic factors38,39) and a reduction of local salt concentration upon partial dehybridization.37,40 In more detail, only at the contact sites of vesicles the distance between vesicle membranes is small enough to enable the insertion of the two α-tocopherol anchors of a LiNA/(dA)20 hybrid into the hydrophobic environment of the two opposing membranes. Outside the contact sites only one hydrophobic anchor of the stiff hybrid could insert into a membrane while the other would be exposed to the aqueous phase being energetically unfavorable. Hence, contact sites ensemble a trap enriching the hybrids locally. As a consequence, when raising temperature apparently cooperative dehybridization will occur caused by a change of DNA length and a reduction of local salt concentration upon partial dehybridization. The steep and sharp transition of vesicle disaggregation is in agreement with previous reports.11,40,41
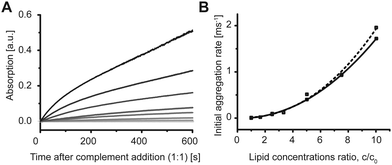
Aggregation of the vesicles upon hybridization with an unlabeled complement was then measured by 90° light scattering (Fig. S2 and S3, ESI†) and UV absorption (Fig. 3A). We studied the dependence of the aggregation kinetics on the lipid concentration and observed that the rate of the aggregation increased with the increasing concentration (Fig. 3A). Therefore, not only the amount of DNA per vesicle,17 but also the lipid concentration is crucial for the rate of the aggregation of the model vesicles. The initial rates were proportional to the ratio of the concentrations and lowest tested concentration c0 by the power of b = 2.2 ± 0.2 as shown in Fig. 3B. The second-order kinetics indicates that collision of two vesicles might be the rate limiting step for the initial aggregation (see ESI† for details).
Finally, based on the properties of the LiNA reported here, we suggest that a LiNA may act as a synthetic structure mimicking the function of tetherin which has membrane anchors at both N and C-termini and was shown to tether budding HIV viruses to the surface of the infected cells.25–27 To this end, we investigated whether the LiNA hybridization can induce aggregation of HIV virus-like particles (VLPs).35,42 VLPs are of 100–120 nm diameter and resemble native-like virions. Gag protein is required for the VLP formation; VLPs used here contained a fusion protein of Gag with YFP or mCherry.
We observed that addition of (dA)20 to VLPs pre-incubated with the LiNA led to an increase in light scattering (Fig. 4A). The scattering experiments required 1 mL sample volume for stirring, therefore, only a very low concentration of VLPs could be used. The increase in scattering is rather low, nevertheless, it is comparable with the increase in scattering for the similarly low concentration of lipid vesicles (see Fig. 3A, 55 μM). Using higher VLP concentrations (in a smaller volume), aggregates were also detected using a microscope 10 min after addition of the complement to VLPs pre-incubated with LiNA for 30 min (Fig. 4B and C). The results show that tocopherol anchors could easily insert into the lipid envelope of the VLPs and cause aggregation due to DNA-hybridization and accompanied exposure of hydrophobic anchors as described above for model vesicles. Indeed this VLP aggregation is reminiscent of the chain-like aggregation of HIV viruses attached on the cell surface observed by electron microscopy.27
While the density of spike proteins in the HIV envelope is rather low, other enveloped viruses have a very high density of spike proteins, e.g. hemagglutinin, the major surface protein of the Influenza viruses, covers more than 80% of the surface and is at least 13 nm high.43,44 To trigger aggregation of such viruses it may require longer DNA-strands as used here. This conclusion is supported by a recent report of Selden et al.45 who showed that a long linker of at least 60 nucleic bases was necessary for cell–cell attachment. Recently, cholesterol-modified PEG presented on a glass surface via biotin/avidin was used to immobilize lipid vesicles, viruses, bacteria, and yeast cells.46 Hence, also for a controlled and more efficient virus aggregation, a longer lipophilic nucleic acid with a longer sequence or a linker might be required.
Lipophilic nucleic acids modified with two palmitoyl chains or cholesterol partitioning into raft domains specifically were recently reported by us and others.10,47–49 Application of different lipophilic anchors allows sequestration of the LiNAs in domains and a remote control of the functional moieties by temperature48 or enzymatic cleavage.49 As tetherin was found to localize in rafts,50 a similarly built LiNA with a longer sequence and two anchors partitioning into raft-like domains, e.g. cholesterol or palmitoyl anchors, might even better mimic tetherin.
Taken together, we have shown here that aggregation of HIV virus-like particles, likewise to the aggregation of lipid vesicles, can be achieved using the lipophilic nucleic acid with two lipophilic anchors each close to one end of the sequence.
Experimental
The sequence of the addition of the oligonucleotides was not essential for the aggregation process (Fig. S2, ESI†); however, the initial aggregation rates were slightly higher when vesicles were pre-incubated with the LiNA. HIV virus like particles labeled with mCherry or YFP were isolated as described elsewhere42 and were a kind gift of Andrea Gramatica. For details of VLPs see ESI.† Total protein concentration of the VLP stocks was about 0.8 mM.This work was supported by the grants of the Deutsche Forschungsgemeinschaft (AR 783/1-1 (AA); SFB 765 (AH)), the Federal Ministry of Education and Research (BMBF INUNA 0312027 (JL, AH)), and the Humboldt University Gender Equal Opportunities Fund (52516/0105 (AA)). We are grateful to Andrea Gramatica for providing the virus-like particles.
Notes and references
- P. X. Guo, Nat. Nanotechnol., 2010, 5, 833–842 CrossRef CAS PubMed.
- Y. Krishnan and F. C. Simmel, Angew. Chem., Int. Ed., 2011, 50, 3124–3156 CrossRef CAS PubMed.
- N. C. Seeman, Nature, 2003, 421, 427–431 CrossRef PubMed.
- Y. Wu, K. Sefah, H. Liu, R. Wang and W. Tan, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 5–10 CrossRef CAS PubMed.
- L. Y. Chou, K. Zagorovsky and W. C. Chan, Nat. Nanotechnol., 2014, 9, 148–155 CrossRef CAS PubMed.
- O. Pokholenko, A. Gissot, B. Vialet, K. Bathany, A. Thiéry and P. Barthélémy, J. Mater. Chem. B, 2013, 1, 5329–5334 RSC.
- A. Aimé, N. Beztsinna, A. Patwa, A. Pokolenko, I. Bestel and P. Barthélémy, Bioconjugate Chem., 2013, 24, 1345–1355 CrossRef PubMed.
- H. Rosemeyer, Chem. Biodiversity, 2005, 2, 977–1063 CAS.
- D. Berti, C. Montis and P. Baglioni, Soft Matter, 2011, 7, 7150–7158 RSC.
- M. Schade, D. Berti, D. Huster, A. Herrmann and A. Arbuzova, Adv. Colloid Interface Sci., 2014, 208, 235–251 CrossRef CAS PubMed.
- U. Jakobsen, A. C. Simonsen and S. Vogel, J. Am. Chem. Soc., 2008, 130, 10462–10463 CrossRef CAS PubMed.
- G. Zhang, F. Farooqui, O. Kinstler and R. L. Letsinger, Tetrahedron Lett., 1996, 37, 6243–6246 CrossRef CAS.
- G. Stengel, R. Zahn and F. Höök, J. Am. Chem. Soc., 2007, 129, 9584–9585 CrossRef CAS PubMed.
- G. Stengel, L. Simonsson, R. A. Campbell and F. Höök, J. Phys. Chem. B, 2008, 112, 8264–8274 CrossRef CAS PubMed.
- T. Maruyama, H. Yamamura, M. Hiraki, Y. Kemori, H. Takata and M. Goto, Colloids Surf., B, 2008, 66, 119–124 CrossRef CAS PubMed.
- A. Bunge, M. Loew, P. Pescador, A. Arbuzova, N. Brodersen, J. Kang, L. Dähne, J. Liebscher, A. Herrmann, G. Stengel and D. Huster, J. Phys. Chem. B, 2009, 113, 16425–16434 CrossRef CAS PubMed.
- P. A. Beales and T. K. Vanderlick, J. Phys. Chem. A, 2007, 111, 12372–12380 CrossRef CAS PubMed.
- P. A. Beales, C. L. Bergstrom, N. Geerts, J. T. Groves and T. K. Vanderlick, Langmuir, 2011, 27, 6107–6115 CrossRef CAS PubMed.
- P. A. Beales and T. K. Vanderlick, Adv. Colloid Interface Sci., 2014, 207, 290–305 CrossRef CAS PubMed.
- B. van Lengerich, R. J. Rawle, P. M. Bendix and S. G. Boxer, Biophys. J., 2013, 105, 409–419 CrossRef CAS PubMed.
- J. R. Burns, K. Göpfrich, J. W. Wood, V. V. Thacker, E. Stulz, U. F. Keyser and S. Howorka, Angew. Chem., Int. Ed., 2013, 1–5 Search PubMed.
- Y.-H. M. Chan, B. van Lengerich and S. G. Boxer, Biointerphases, 2008, 3, FA17 CrossRef CAS PubMed.
- E. Erikson, T. Adam, S. Schmidt, J. Lehmann-Koch, B. Over, C. Goffinet, C. Harter, I. Bekeredjian-Ding, S. Sertel, F. Lasitschka and O. T. Keppler, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 13688–13693 CrossRef CAS PubMed.
- S. J. D. Neil, T. Zang and P. D. Bieniasz, Nature, 2008, 451, 425–430 CrossRef CAS PubMed.
- J. Hammonds and P. Spearman, Cell, 2009, 139, 456–457 CrossRef CAS PubMed.
- S. Venkatesh and P. D. Bieniasz, PLoS Pathog., 2013, 9, e1003483 CAS.
- J. Hammonds, J.-J. Wang, H. Yi and P. Spearman, PLoS Pathog., 2010, 6, e1000749 Search PubMed.
- A. Kurz, A. Bunge, A.-K. Windeck, M. Rost, W. Flasche, A. Arbuzova, D. Strohbach, S. Müller, J. Liebscher, D. Huster and A. Herrmann, Angew. Chem., Int. Ed., 2006, 45, 4440–4444 CrossRef CAS PubMed.
- W. Flasche, C. Cismas, A. Herrmann and J. Liebscher, Synthesis, 2004, 2335–2341 CAS.
- A. Bunge, A. Kurz, A.-K. Windeck, T. Korte, W. Flasche, J. Liebscher, A. Herrmann and D. Huster, Langmuir, 2007, 23, 4455–4464 CrossRef CAS PubMed.
- T. Tokunaga, S. Namiki, K. Yamada, T. Imaishi, H. Nonaka, K. Hirose and S. Sando, J. Am. Chem. Soc., 2012, 134, 9561–9564 CrossRef CAS PubMed.
- N. Brodersen, J. Li, O. Kaczmarek, A. Bunge, L. Löser, D. Huster, A. Herrmann and J. Liebscher, Eur. J. Org. Chem., 2007, 6060–6069 CrossRef CAS.
- O. Kaczmarek, N. Brodersen, A. Bunge, L. Löser, D. Huster, A. Herrmann, A. Arbuzova and J. Liebscher, Eur. J. Org. Chem., 2008, 1917–1928 CrossRef CAS.
- K. Nishina, T. Unno, Y. Uno, T. Kubodera, T. Kanouchi, H. Mizusawa and T. Yokota, Mol. Ther., 2008, 16, 734–740 CrossRef CAS PubMed.
- L. Deml, C. Speth, M. P. Dierich, H. Wolf and R. Wagner, Mol. Immunol., 2005, 42, 259–277 CrossRef CAS PubMed.
- U. Jakobsen and S. Vogel, Bioconjugate Chem., 2013, 24, 1485–1495 CrossRef CAS PubMed.
- R. Jin, G. Wu, Z. Li, C. A. Mirkin and G. C. Schatz, J. Am. Chem. Soc., 2003, 125, 1643–1654 CrossRef CAS PubMed.
- S. Angioletti-Uberti, P. Varilly, B. M. Mognetti, A. V. Tkachenko and D. Frenkel, J. Chem. Phys., 2013, 138, 021102 CrossRef PubMed.
- P. Varilly, S. Angioletti-Uberti, B. M. Mognetti and D. Frenkel, J. Chem. Phys., 2012, 137, 094108 CrossRef PubMed.
- P. A. Beales and T. K. Vanderlick, Biophys. J., 2009, 96, 1554–1565 CrossRef CAS PubMed.
- N. Dave and J. Liu, ACS Nano, 2011, 5, 1304–1312 CrossRef CAS PubMed.
- M. Perlman and M. D. Resh, Traffic, 2006, 7, 731–745 CrossRef CAS PubMed.
- C. Böttcher, K. Ludwig, A. Herrmann, M. van Heel and H. Stark, FEBS Lett., 1999, 463, 255–259 CrossRef.
- A. K. Harris, J. R. Meyerson, Y. Matsuoka, O. Kuybeda, A. Moran, D. Bliss, S. R. Das, J. W. Yewdell, G. Sapiro, K. Subbarao and S. Subramaniam, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 4592–4597 CrossRef CAS PubMed.
- N. S. Selden, M. E. Todhunter, N. Y. Jee, J. S. Liu, K. E. Broaders and Z. J. Gartner, J. Am. Chem. Soc., 2012, 134, 765–768 CrossRef CAS PubMed.
- P. Kuhn, K. Eyer, T. Robinson, F. I. Schmidt, J. Mercer and P. S. Dittrich, Integr. Biol., 2012, 4, 1550–1555 RSC.
- P. A. Beales, J. Nam and T. K. Vanderlick, Soft Matter, 2011, 7, 1747–1755 RSC.
- M. Loew, R. Springer, S. Scolari, F. Altenbrunn, O. Seitz, J. Liebscher, D. Huster, A. Herrmann and A. Arbuzova, J. Am. Chem. Soc., 2010, 132, 16066–16072 CrossRef CAS PubMed.
- M. Schade, A. Knoll, A. Vogel, O. Seitz, J. Liebscher, D. Huster, A. Herrmann and A. Arbuzova, J. Am. Chem. Soc., 2012, 134, 20490–20497 CrossRef CAS PubMed.
- P. G. Billcliff, R. Rollason, I. Prior, D. M. Owen, K. Gaus and G. Banting, J. Cell Sci., 2013, 126, 1553–1564 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental details, structure of the α-tocopherol anchor, and results of the control measurements and details of the rate of the initial aggregation and virus like particles. See DOI: 10.1039/c4nj00724g |
| This journal is © The Royal Society of Chemistry and the Centre National de la Recherche Scientifique 2014 |